Abstract
Key message
TaHsfA6b-4D relocalizes intracellularly upon heat stress and play a significant role in linking the heat stress response to unfolded-protein response so as to maintain cellular homeostasis.
Abstract
Heat stress transcription factors (Hsfs) play a crucial role in protecting the plants against heat stress (HS). In case of wheat, TaHsfA6b-4D (earlier known as TaHsfA2d) has been identified as a seed preferential transcription factor and its role has been shown in various abiotic stresses such as heat, salt and drought stress. In the present study, a homeologue of TaHsfA6b gene (TaHsfA6b-4A) was identified and was found to be transcriptionally inactive but it localized to the nucleus. Interestingly, TaHsfA6b-4D localized to the endoplasmic reticulum-Golgi complex and peroxisomes under non-stress conditions, but was observed to accumulate in the nucleus upon HS. The expression of TaHsfA6b-4D was upregulated by dithiothreitol (DTT), which is a known ER stress inducer. Consistent with this, Arabidopsis transgenic plants overexpressing TaHsfA6b-4D performed better on DTT containing media, which further corroborated with the increased expression of ER stress marker genes in these transgenic plants in comparison to the wild type plants. Thus, these studies together suggest that TaHsfA6b-4D may relocalize intracellularly upon heat stress and may play a significant role in linking the unfolded-protein response with heat stress response so as to maintain protein homeostasis inside the cell under heat stress.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Wheat is one of the most important cereal crops that is grown worldwide. Being a cool season crop, heat stress becomes one of the major abiotic stresses that affect its yield. It is predicted that with the increase of every 1 °C in temperature leads to the drop of 4% of wheat productivity. To cope with high temperature stress, plants have well conserved heat stress response (HSR) pathway wherein heat responsive genes, which majorly includes heat shock proteins, accumulate to prevent heat-induced damage (Wahid 2007). The expression of these heat shock proteins is in turn controlled by heat stress transcription factors (Hsfs) by binding to the Heat Stress Element (HSE) present in their promoter regions (Iba 2002). Thus, Hsfs function as the terminal components of the signaling pathway controlling the expression of genes responsive to heat stress (Nover et al. 2001).
Like other transcription factors, Hsf family also has a conserved domains with significant diversification in their structure. In eukaryotes Hsf proteins are composed of N-terminal DNA binding domain (DBD) followed by oligomerization domain (HR-A/B or OD), nuclear localization signal (NLS), nuclear export signal (NES), repressor domain (RD) and C terminal activator domain (CTAD) (Chauhan et al. 2013; Lohani et al. 2019). DBD is characterized by the presence of central helix-turn-helix motif, which is known to interact with HSE within the promoter region of target genes (Scharf et al. 2012). The OD domain (which is composed of heptad repeats of hydrophobic amino acids) is connected to DBD domain by a flexible linker of 15–80 amino acid residues and helps in formation of homo and hetro-dimers (Baniwal et al. 2004; Scharf et al. 2012). Interestingly, Hsfs are able to maintain a balance between nucleus and cytoplasm with the help of NLS and NES, which are present at the C-terminus. Moreover, the class A Hsfs have been reported to possess the AHA motifs which are the short peptides (formed by aromatic, hydrophobic and acidic amino acid residues), responsible for imparting the transcriptional activity of these Hsfs (Scharf et al. 2012). On the other hand, class B Hsfs are well known to have the repressor domain that is formed by tetrapeptide LFGV amino acid residues at their C-terminus (Scharf et al. 2012).
Organisms like Saccharomyces cerevisiae, nematodes and Drosophila possess only one Hsf gene which is required for HSR and also for the growth and development, whereas vertebrates are reported to have four Hsfs (Greg et al. 1988; Sorger and Pelham 1988; Miller and Mittler 2006). In plants, three classes of Hsfs have been reported (class A, B and C), depending upon the distinctive flexible linker and OD (HR-A/B) domain architecture. Class A and C Hsfs have an extended HR-A/B due to insertion of 21 and 7 residues whereas class B Hsfs have HR-A/B regions that are more similar to non-plant Hsfs (Nover et al. 2001). Moreover, there are reports that suggest that class A and C Hsfs exists as trimer and HsfB1 forms dimer in case of tomato (Nover et al. 2001). Thus, variation in structure and domain architecture of Hsf contributes to their different biological functions. Class A Hsfs are known to impart thermotolerance in plants whereas class B act as a repressor of certain HS induced genes (Röth et al. 2017). Apart from this, genome wide studies have been done wherein 21 Hsfs in Arabidopsis (Scharf et al. 2012), 64 in brassica (Zhu et al. 2017), 24 in tomato (Scharf et al. 2012), 25 in rice (Chauhan et al. 2011), and 25 in maize (Yong-Xiang et al. 2011) have been reported. Recently, in wheat 82 Hsf members have been identified (Duan et al. 2019).
The protein unfolding response (UPR) is one of the conserved pathways in eukaryotes and is activated by the accumulation of misfolded proteins in the ER. Environmental stresses like heat stress which disrupt protein folding in ER leads to the activation of UPR (Bao and Howell 2017). In case of Arabidopsis, membrane associated transcription factors i.e. AtbZIP28, AtbZIP17 and AtbZIP60 play an important role in UPR by activating the downstream target genes (Deng and Howell 2013). In contrast to ER-UPR, protein unfolding in cytosol trigger HSR and HsfA2 has been shown as a specific Hsf involved in the HSR response. A recent study by Kataoka et al. has highlighted how the coordination between AtbZIP28 and AtHsfA2 help in regulation of heat stress signals (Kataoka et al. 2017). Interestingly HsfA2 has been reported to be regulated by alternative splicing and nonsense-mediated decay (Sugio et al. 2009). It was further demonstrated that a heat inducible splice variant of HsfA2 (HsfA2-III) is involved in self-regulation of HsfA2 transcription in Arabidopsis (Liu et al. 2013). In case of rice, it has been shown that alternative splicing of OsHsfA2a helps in regulating stress-specific cellular adaptation responses (Wang et al. 2013). A subsequent report by Cheng et al. highlighted that the alternatively spliced variant of OsHsfA2d (i.e., OsHsfA2dI) functions in HS induced UPR by regulating the expression of OsBiP1(Cheng et al. 2015).
In case of wheat, it has already been reported that TaHsfA2d and TaHsfA2-1 provides thermotolerance in Arabidopsis thaliana (Chauhan et al. 2013; Liu et al. 2020). However, TaHsfA2d later has been renamed as TaHsfA6b by Scharf et al. (2012) and Poonia et al. (2020). Its overexpression in barley attributed thermotolerance to the transgenic plants by reducing the oxidative load generated during the HS (Poonia et al. 2020). In the present study, an attempt has been made to further explore the role of TaHsfA6b (earlier known as TaHsfA2d) in HSR. With the help of updated wheat reference genome, it was found to be located at chromosme 4D. Interestingly, similar to the splice variant of TaHsfA2d identified in Arabidopsis and rice, a homeologue of TaHsfA6b-4D was identified, which was found to be coded by chromosome 4A. The transcriptional inactivity of TaHsfA6b-4A and its nuclear localization suggested its possible role in regulating the function of TaHsfA6b-4D. Apart from this, subcellular localization studies have showed the presence of TaHsfA6b-4D in organelles such as ER-Golgi complex and peroxisomes under control conditions. However, it was found to accumulate inside the nucleus under HS which suggested its possible role in ER-UPR. This was further supported by the fact that the expression of TaHsfA6b-4D peaked in wheat seedlings after treatment of DTT. The TaHsfA6b-4D overexpression lines of Arabidopsis thaliana were found to survive better in comparison to wild type (WT) plants in the presence of DTT, which is a known protein unfolding inducer. Thus, our results suggest that TaHsfA6b-4D might play an important role in linking UPR with HSR and might be regulated by its homeologue i.e. TaHsfA6b-4A.
Materials and methods
Expression analysis of TaHsfA6b-4D and TaHsfA6b-4A
For semi quantitative PCR, forward primer was made from the end of first exon and reverse primer was made from the start of the last exon. This primer combination was used for checking the expression of TaHsfA6b-4D and TaHsfA6b-4A under different stresses by semi quantitative PCR. Amplification was performed using High Fidelity Phusion enzyme (NEB, England). PCR amplicons were separated on 2% agarose and bands were extracted from gel using gel extraction kit (Qiagen, Germany). Each band was given for sequencing. For expression analysis TaHsfA6b-4D and TaHsfA6b-4A, plants were subjected to heat stress at 42 °C for 2 h and salt (200 mM NaCl), drought (200 mM mannitol) and cold (4 °C) for 24 h. Further, expression was checked at different time period for temperature stress.
Phylogenetic analysis and protein structure prediction
The protein sequences of all the plant species were downloaded from Ensembl and TAIR databases and were used for the construction of the phylogenetic tree. Multiple sequence alignment was carried out by the CLUSTALW program, and the phylogenetic tree was constructed using the MEGA7 program by Neighbor-Joining method. Phyre2 web portal was used for prediction of three-dimensional structure of TaHsfA6b proteins of wheat and TaHsfA2d proteins of Arabidopsis and Oryza sativa.
Subcellular localization
To determine the subcellular localization of TaHsfA6b-4D, and TaHsfA6b-4A, complete ORF of two were amplified using HF Phusion enzyme (NEB) and cloned in pENTR D-topo vector followed by their mobilization into the destination vector i.e. pSITE-3CA, under CaMV35S promoter. Complete ORF of these genes were fused in frame with C terminal of YFP. PDS-1000 bombardment system (Bio-Rad, Canada) was used for bombardment of onion epidermal cells, at a pressure of 1100 psi with gold particles coated with plasmid construct (Lee et al. 2008). Transformed onion peels were kept at 27 °C for 16 h in dark and fluorescence was observed in confocal microscope (Leica, Germany).
Transactivation assay of TaHsfA6b-4D and TaHsfA6b-4A
To study the transactivation activity of TaHsfA6b-4D, the full length coding sequence and its deletion constructs (DBD, DBD + OD, AHA, NLS) were amplified using specific primers and cloned into the vector pGBKT7 (Clontech, USA), using the NdeI and SmaI restriction sites. Similarly full length sequence of TaHsfA6b-4A was cloned into pGBKT7 vector. The recombinant plasmids were transformed into the yeast strain AH109 harboring the HIS3 reporter gene. Positive clones were selected on histidine lacking media. Dropout assay was performed on different synthetic media with different concentration of 3-AT (Sigma Aldrich, USA) and incubated for 5 days. Full-length protein transactivation assay of TaHsfA6b-4D was carried out at different stringent media such as -HW with increasing concentration of 3-AT.
TaHsfA6b-4D cloning and characterization in Arabidopsis thaliana
For generation of Arabidopsis overexpression plants, full CDS of 1026 bp was amplified from Triticum aestivum (PBW343 wheat cultivar). The amplified product was then cloned in an entry vector (pENTRTM/D-TOPO) and then in destination vector pMDC32 under CaMV35S promoter following Gateway™ cloning strategy (Directional TOPO cloning kit and LR clonase Enzyme mix II kit, Invitrogen Inc. USA). The GV3101 strain of Agrobacterium tumefaciens harboring pMDC32- TaHsfA6b-4D was used for transformation in Arabidopsis thaliana through floral dip method (Clough and Bent 1998). The T1 seeds were selected on MS-agar plates supplemented with 50 µg/µl of hygromycin and the resistant plants were transferred to pots. Further, the overexpressing transgenics were confirmed by PCR using hygromycin and gene specific primers. Selected plants were further grown up to T3 homozygous stage. The plants were confirmed by PCR (Fig. S1). For characterization in Arabidopsis one-week old transgenic plants were transferred to MS media 2.5 mM DTT (Hossain et al. 2016). Phenotype of plants was observed after four days.
Estimation of chlorophyll content
Ten-day-old WT and transgenic plants were transferred to media containing DTT (2.5 mM). After four days 50 mg of leaf tissue was taken in a tube containing 5 ml of DMSO. Tubes were incubated overnight for chlorophyll bleaching. Absorbance was taken at 645 and 663 nm in a UV-Vis spectrophotometer (Hitachi U-2810, Tokyo, Japan) and chlorophyll content were estimated accordingly to Arnon ( 1949).
RNA isolation and expression analysis
For expression analysis in wheat, 10-day old seedlings of PBW343 were subjected to DTT (7.5 mM for 3 and 6 h). Control and treated samples were frozen in liquid nitrogen for RNA isolation. Total RNA was isolated using the RNeasy plant mini kit (Qiagen, Germany) according to the manufacturer’s instructions, including on-column DNase I treatment to remove genomic DNA contamination. 2 µg of the total RNA was used as template to synthesize cDNA employing the High Capacity cDNA Archive kit (Applied Biosystems, USA) and mixed with 200 nm of each primer and SYBR Green PCR Master Mix (Applied Biosystems) for real-time PCR analysis, using the ABI Prism 7000 sequence detection system and software (PE Applied Biosystems) according to the manufacturer’s protocol. Housekeeping gene TaGAPDH were used as internal control in wheat. Primers used for this experiment are listed in Table S1.
Results
Identification of TaHsfA6b-4D and TaHsfA6b-4A gene
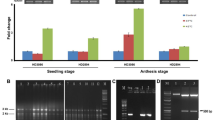
It has been reported in rice and Arabidopsis that HsfA2d undergoes alternate splicing and the splice variants might play a role in regulating its function (Liu et al. 2013; Cheng et al. 2015). To see if a similar situation exists in case of wheat, we checked for the splicing of TaHsfA6b-4D. For this expression of TaHsfA6b-4D was analyzed by semi quantitative PCR by using the forward primer from the end of first exon and the reverse primer from the start of the last exon. Similar strategy has been used by Liu et al. (2013) and Cheng et al. (2015) for identification of HsfA2 splice variants. Initially as expected, two variants of TaHsfA6b-4D were observed (Fig. 1A). Interestingly, under heat stress conditions predominantly lower band corresponding to TaHsfA6b-4D was observed whereas in cold stress an upper band was observed, which was speculated as its Isoform at that time (Fig. 1B). In salt and drought stress both the variants were seen (Fig. 1B). When expression was checked in a time course manner to see if Isoform (upper band) appeared later during heat stress (Fig. 1D), it was found that only TaHsfA6b-4D existed in heat stress. This speculated TaHsfA6b-4D Isoform was then amplified and cloned for sequencing purpose. Upon sequence analysis, it was found that the TaHsfA6b-Isoform was coded by the homoelogue chromosome i.e. 4A and the protein had a premature STOP codon which leads to the formation of a truncated protein (Fig. 2A). At the protein level, it was found to have only the DNA binding domain (Fig. 2B) and the other characteristic domains were missing. Thus, this Isoform was actually found to be TaHsfA6b homeologue, here after referred to as TaHsfA6b-4A. Moreover, the expression pattern of these TaHsfA6b homeologues was analyzed in different developmental stages of the wheat plant (Fig S2). Both the homeologs showed differential expression pattern in various tissues such as stigma, ovary, anther and leaf. Thus, it appears that both the homeologues show differential expression pattern not only under stress but also in various developmental tissues.
Gene structure and protein domain analysis. A Gene structure of TaHsfA6b-4D and TaHsfA6b-4A gene. Exons are depicted in pink color, intron are depicted by solid line. Orange color depicts the UTR region. B Protein domain analysis ofTaHsfA6b-4D and TaHsfA6b-4A. Heatster software was used to predict the protein domains
Structural and phylogenetic analysis of TaHsfA6b-4D and TaHsfA6b-4A
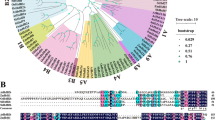
To analyze the evolution of the TaHsfA6b-4D and TaHsfA6b-4A, we constructed a Neighbor-Joining (NJ) tree based on a total of 21 sequences (containing both splice variants and HsfA2d gene) obtained from different plants. TaHsfA6b and its homeologue were found to be more similar to the predicted HsfA2d in Aegilops tauschii (Fig. 3A). This was found to be in accordance to the fact that TaHsfA6b-4D was located on D genome, which is derived from Aegilops tauschii. Domain analysis showed that the TaHsfA6b-4A had only DNA binding domain (Fig. 3B), similar to the case seen in Arabidopsis and rice. Apart from this, the three-dimensional structures of Arabidopsis and rice HsfA2d and their splice variants were predicted and compared with that of TaHsfA6b and its homeologue (Fig. S3). The structure of TaHsfA6b-4A was found to be similar to the splice variants of Arabidopsis and rice which suggests that even in case of wheat a truncated protein is formed which might have a role in regulation of TaHsfA6b-4D.
Analyzing the transcriptional activation and localization of TaHsfA6b-4A
Sequencing of the TaHsfA6b-4A revealed that the CDS of TaHsfA6b-4A was found to retain an intron which was found to be present in TaHsfA6b-4D. As protein sequence analysis showed that it lacked NLS and AHA domains, therefore, it did not show trancriptional activity in yeast cells as well (Fig. 4C). Apart from this, localization studies in onion peel cells demonstrated its localized primarily in the nucleus (Fig. 5A).
Transcriptional activation assay of TaHsfA6b-4A and TaHsfA6b-4D in yeast. A and B Growth of yeast cells harbouring histidine reporter gene were transformed with fusion constructs pDEST- GBKT7::TaHsfA6b-4D and its deletions were analyzed on SD/-Trp (-T) medium and on SD/-Trp/-His (-HW) medium. Yeast cells transformed with the empty vector pGBKT7 (EV) alone were used as a control. C Transcriptional activation assay of TaHsfA6b-4A in yeast. Growth of yeast cells harbouring histidine reporter gene were transformed with fusion constructs pDEST- GBKT7::TaHsfA6b-4A were analysed on SD/-Trp (-T) medium and on SD/-Trp/-His (-HW) medium. Yeast cells transformed with the empty vector pGBKT7 (EV) alone were used as a control
Subcellular localisation of TaHsfA6b-4D and TaHsfA6b-4A in onion epidermal cell. A CDS of TaHsfA6b-4D and TaHsfA6b-4A was cloned in frame with YFP protein and observed under confocal microscope to locate the genes. TaHsfA6b-4D protein can be seen in the form of speckles inside the cell under control conditions. B Subcellular localisation of TaHsfA6b-4D under heat stress. TaHsfA6b-4D total protein accumulated in the nucleus under HS condition. YFP Yellow fluorescent protein
Transcriptional activation assay of TaHsfA6b-4D in yeast
Earlier, full length protein of TaHsfA6b-4D was reported to exhibit transactivation activity in yeast (Chauhan et al. 2013) and we also observed the same result. It was found that TaHsfA6b-4D full length protein possessed very strong transactivation activity. As shown in Fig. 4A, even at 100 mM 3-AT concentration, TaHsfA6b-4D was able to activate the HIS gene transcription. To gain further insight, as to which domain of the protein is responsible for its strong transactivation, different domains of the protein (such as DNA binding domain, oligomerization domain, NLS and AHA motif) were analyzed for the transactivation activity in yeast. Amongst the deletion constructs, DBD and OD domains of the protein did not show any transactivation on SD-HW media. Only the protein having NLS and AHA domains were found to possess transactivation activity (Fig. 4B). Thus, it appears that TaHsfA6b-4D could act as trans-activator intracellularly in the cell.
Subcellular localization of TaHsfA6b-4D
To characterize the function, subcellular localization of TaHsfA6b-4D was performed. For this purpose, the pSITE-3CA::TaHsfA6b-4D constructs were used to bombard epidermal peels of onion. As TaHsfA6b-4D protein contained a nuclear localization signal we hypothesized that it should be present inside the nucleus. However, it was found to be localized in the cytoplasm in the form of speckles (Fig. 5A). The localization of TaHsfA6b-4D under HS conditions was checked by keeping the onion cells at 42 °C for 2 h. It was observed that under HS condition, most of the TaHsfA6b-4D protein accumulated inside the nucleus of the cells (Fig. 5B). Thus, it appears that during HS TaHsfA6b-4D translocated to the nucleus from the cytoplasm where it activated its target genes. To find the exact localization of TaHsfA6b-4D, its co-localization was performed with certain organelle specific markers. Interestingly, it was observed that TaHsfA6b-4D protein localized in the ER-Golgi complex and peroxisomes as depicted in the Fig. 6. To corroborate these results, we also checked for the presence of ER and peroxisomes localization signals in the protein sequence of TaHsfA6b-4D (Fig. S4). Only ER localization signals were found to be present in the protein sequence of TaHsfA6b-4D.
Subcellular localisation of TaHsfA6b-4D along with different organelle markers. CDS of TaHsfA6b-4D was cloned in frame with YFP protein and observed in onion epidermal cells under confocal microscope. Different organelle markers were used to locate the gene in onion epidermis cells. ER endoplasmic reticulum marker, YFP yellow fluorescent protein
Developmental role of TaHsfA6b-4D
Recently, different Hsfs have been shown to constitutively express in different developmental stages of wheat (Guo et al. 2020). To gain further understanding regarding the functional role of TaHsfA6b-4D, its overexpressing transgenic plants, were grown on MS media under control conditions (Fig. 7). They showed not only enlongated roots but also had larger number of lateral roots as compared to WT plants. Therefore, length of roots and total number of lateral roots were calculated in control conditions and were found to be more in comparison to WT plants (Fig. 7A, B).
Phenotype of transgenic plants overexpressing TaHsfA6b-4D under control conditions. A Plants grown under control conditions. Phenotype was observed after ten-day. B Root length and number of lateral roots were calculated in WT and transgenic lines under control conditions. Asterisks on top of the error bars represents the significance levels (Students t-test; p value ≤ 0.05)
Role of TaHsfA6b-4D in unfolded protein response
Subcellular localization study indicated the presence of TaHsfA6b-4D in the ER-Golgi complex, which led to the speculation that it might have a role in the UPR. Therefore, the expression of TaHsfA6b-4D was checked in wheat after DTT treatment. DTT is a strong reducing agent for protein, therefore it is used as an ER stress inducer in plants (Yu et al. 2019). When the wheat seedlings were subjected to DTT treatment, expression of TaHsfA6b-4D was significantly increased within 3 h of treatment (Fig. 8A). To further confirm this result, TaHsfA6b-4D overexpression lines of Arabidopsis thaliana were subjected to DTT containing media and the phenotype was observed after four days, as shown in Fig. 8B. Transgenic plants grown on DTT containing media performed better as compared to WT plants. Also, the chlorophyll content of transgenic plants were high as compared to WT plants (Fig. 8C). This analysis clearly indicated the involvement of TaHsfA6b-4D in UPR.
Expression and phenotypic analysis of TaHsfA6b-4D. A 10-day-old seedlings of Triticum aestivum cv. PBW343 variety were subjected to DTT (7.5 mM) for 3 and 6 h. Expression was checked using qRT PCR. Three biological and three technical replicates were used in the experiment. TaGAPDH was used as an internal housekeeping gene. B Phenotype of transgenic Arabidopsis plants overexpressing TaHsfA6b-4D. 1-week-old plants were subjected to DTT treatment. Phenotype was observed after four-day. C Chlorophyll content of WT and transgenic plants after DTT treatment. Asterisks on top of the error bars represent the significance levels (Students t-test; p value ≤ 0.05)
To further assess the ER stress in Arabidopsis transgenic plants, the overexpression lines were checked with various ER stress marker genes under control and DTT conditions. It was observed that the relative expression of AtBip3, AtPDI, calnexin and Calreticulin genes were found to be higher in overexpression lines than the WT under both control as well in DTT treated seedlings (Fig. 9). These data together indicated that TaHsfA6b-4D may play a major role in regulating the expression of stress response genes in UPR which is a major constituent of stress response in HS.
Expression analysis of ER stress marker genes in 10-day-old WT and transgenic plants of Arabidopsis. Plants were subjected to 2.5 mM DTT for 4 h. Transcript levels were normalized to WT and AtActin was used as housekeeping gene. Error bars indicate values ± SD. Asterisks on top of the error bars represent the significance levels (Students t-test; p value ≤ 0.05)
Discussion
HS is known to be one of the major causes which leads to the accumulation of misfolded proteins inside the cell. Accumulation of misfolded proteins in the ER leads to ER stress which elicits the UPR whereas the protein accumulation in the cytosol leads to the activation of HSR (Li et al. 2020). Both of these responses help to maintain cellular homeostasis. While both HSR and UPR occur independently in two different compartments of the cell, there have been reports that highlight the connection between the two (Kataoka et al. 2017; Li et al. 2020). In the present study, an attempt has been made to identify the potential role of TaHsfA6b in linking UPR and HSR. Our experimental analysis under cold and heat stress, helped to identify TaHsfA6b homeologues (i.e. TaHsfA6b-4D and TaHsfA6b-4A) which showed differential expression pattern under both the stresses (Fig. 1A). TaHsfA6b-4D was found to heat responsive whereas TaHsfA6b-4A was found to be cold responsive. Also, they were found to differentially express in various developmental tissues of the wheat (Fig. S2). Although, from sequence analysis TaHsfA6b-4A resembled like the splice variant of TaHsfA6b-4D, but they both were found to be coded from homeologous chromosomes. This scenario was found to be different from that reported in case of rice and Arabidopsis wherein splice variant of HsfA2d are generated from the same locus (Liu et al. 2013; Wang et al. 2013). The localization and transactivation studies showed its presence in the nucleus although it lacked the transactivation potential (Figs. 4 and 5). This could be because of the premature stop codon in the protein sequence leading to the generation of protein only having the DNA binding domain (Fig. 2). Interestingly, in case of Arabidopsis, the heat inducible splice variant of HsfA2 (i.e. HsfA2II) also formed a truncated protein having only DNA bindng protein and inturn self-regulated the transcription of HsfA2 by binding to its promoter (Liu et al. 2013). Thus, it is probable that TaHsfA6b-4A might also regulate the expression of TaHsfA6b-4D, although future experiments needs to validate this. Moreover, the phylogenetic analysis depicted the presence of Isoform like protein in other polyploid species as well, such as barley (Fig. 3A). This hinted towards the evolutionary conserved nature of TaHsfA6b isofroms/homeologues in other crop species.
It is known from earlier studies that change in environmental conditions leads to the changes in protein localization inside the cell (Fernandez-Bautista et al. 2017; Meena et al. 2020). Subcellular localization of HsfA2 has been extensively studied in case of Arabidopsis, rice and tomato and it showed varied pattern of localization and regulation in these plants. Tomato, HsfA2 has been known to localize in the cytoplasm under control conditions but it showed accumulation in the nucleus under HS conditions (Scharf et al. 1998). Whereas in case of Arabidopsis, HsfA2 localized in both the nucleus and cytoplasm (Meiri and Breiman 2009). In case of rice, HsfA2d was found to localize to the nucleus only (Cheng et al. 2015). Surprisingly, the localization of TaHsfA6b-4D revealed its occurrence in the ER-Golgi complex and in the peroxisomes (Fig. 6). This corroborated with the fact that it harbored an integral ER localization signal (Fig. S4). However, the peroxisomal localization signal was found to be absent in the peptide sequence. Therefore, it is speculated that either there exists a poor peroxisomal localization signal in the TaHsfA6b-4D peptide sequence or it interacts with some other protein which takes it inside the peroxisomes.
Hsfs are known to regulate various heat responsive genes by binding to HSE in their promoter region. Using yeast transactivation studies, TaHsfA6b-4D was found to possess strong transactivation potential which was majorly contributed by the AHA and the NLS motifs (Fig. 4). Moreover, under HS condition, TaHsfA6b-4D translocated to nucleus which could probably occur due to its temperature dependent conformational transition (Fig. 5B). In case of tomato, interaction of HsfA2 with HsfA1 leads to its translocation into the nucleus under HS conditions (Heerklotz et al. 2001). Thus, the results indicate the presence of a heat regulated intracellular distribution of TaHsfA6b-4D inside the cell.
ER is known to be involved in the synthesis and folding of proteins inside the cell, which is achieved by the combinatorial effect of certain HSPs, foldases and lectins (Gidalevitz et al. 2013). During stressful conditions the demand of protein folding increases and exceeds the capacity of the system. This results in the accumulation of unfolded protein in the ER and leads to the UPR (Fernandez-Bautista et al. 2017). Thus, ER stress forms a major constituent of various stresses such as heat, drought and salt (Park and Park 2019). The presence of TaHsfA6b-4D in the ER-Golgi complex led to the speculation of it being involved in the UPR. This was further investigated by checking the expression of TaHsfA6b-4D after DTT treatment which causes protein unfolding inside the cell. TaHsfA6b-4D was found to be upregulated within 3 h of the DTT treatment (Fig. 8A). Further, when the performance of Arabidopsis transgenics overexpressing TaHsfA6b-4D gene was analyzed on DTT containing media, the transgenics plants performed better as compared to WT (Fig. 8B). Even the expression of ER stress marker genes such as AtBip3, AtPDI, Calnexin and Calreticulin was higher in the transgenics (Fig. 9). Thus, this data clearly indicated the involvement of TaHsfA6b-4D in the UPR. In support of this idea, recent report by Li et al. (2020) has demonstrated that the expression of HSFTF-13 (HsfA6b family member) was found to be downregulated in bzip60-2 Arabidopsis mutant. Also, HSFTF-13 was observed to be regulated by bZIP60. Further the authors have suggested that bZIP60 plays an important role in linking UPR with the HSR (Li et al. 2020). Therefore, it is probable that in case of wheat as well, TaHsfA6b-4D might be involved in linking HSR with UPR.
Apart from stress, role of Hsfs have been well documented in plant development as well. In case of poplar, the transcripts of PtHsfs of the B4 subfamily showed higher expression during leaf expansion stage (Liu et al. 2019). In Arabidopsis also, overexpression of HsfB4 caused shorter root length (Begum et al. 2013). Similar to this case, overexpression of TaHsfA6b-4D in Arabidopsis caused elongated roots and more lateral root development in the transgenics as compared to WT (Fig. 7). Thus, apart from stress responses, TaHsfA6b-4D might play a role in root developmental processes as well.
In conclusion, hexaploid crops like wheat, possess homeologues which could function similar to splice variants reported in Arabidopsis and Oryza sativa. TaHsfA6b-4D was found to be one of the heat responsive Hsf member which apart from being involved in HS also played an important role in plant root development. Moreover, it also appeared to be involved in the UPR and therefore could play an important role in maintaining the cellular homeostasis. However, in future it will be of interest to explore how TaHsfA6b-4D regulate the expression of ER stress marker genes and how it connects HSR with UPR. Also, investigating its downstream target genes involved in plant development processes becomes another area of subsequent research.
Data availability
Enquiries about data availability should be directed to the authors.
Abbreviations
- AHA:
-
Aromatic, hydrophobic and acidic amino acid residues
- DBD:
-
DNA binding domain
- DTT:
-
Dithiothreitol
- ER:
-
Endoplasmic reticulum
- HS:
-
Heat stress
- Hsf:
-
Heat stress transcription factor
- HSR:
-
Heat stress response
- NLS:
-
Nuclear localization signal
- OD:
-
Oligomerization domain
- UPR:
-
Unfolded protein response
- WT:
-
Wildtype
References
Arnon DI (1949) Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol 24:1–15
Baniwal SK, Bharti K, Chan KY, Fauth M, Ganguli A, Kotak S, Mishra SK, Nover L, Port M, Scharf KD, Tripp J, Weber C, Zielinski D, von Koskull-Döring P (2004) Heat stress response in plants: a complex game with chaperones and more than twenty heat stress transcription factors. J Biosci 29:471–487
Bao Y, Howell S, H (2017) The unfolded protein response supports plant development and defense as well as responses to abiotic stress. Front Plant Sci 8:344
Begum T, Reuter R, Schöffl F (2013) Overexpression of AtHsfB4 induces specific effects on root development of Arabidopsis. Mech Dev 130:54–60
Chauhan H, Khurana N, Agarwal P, Khurana P (2011) Heat shock factors in rice (Oryza sativa L.): genome-wide expression analysis during reproductive development and abiotic stress. Mol Genet Genomics 286:171–187
Chauhan H, Khurana N, Agarwal P, Khurana JP, Khurana P (2013) A seed preferential heat shock transcription factor from wheat provides abiotic stress tolerance and yield enhancement in transgenic Arabidopsis under heat stress environment. PLoS ONE 8:e79577
Cheng Q, Zhou Y, Liu Z, Zhang L, Song G, Guo Z, Wang W, Qu X, Zhu Y, Yang D (2015) An alternatively spliced heat shock transcription factor, OsHSFA2dI, functions in the heat stress-induced unfolded protein response in rice. Plant Biol 17:419–429
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Deng Y, Howell H, Stephen, (2013) Protein kinase and ribonuclease domains of IRE1 confer stress tolerance, vegetative growth, and reproductive development in Arabidopsis. Proc Natl Acad Sci USA 173:2221–2230
Duan S, Liu B, Zhang Y, Li G, Guo X (2019) Genome-wide identification and abiotic stress-responsive pattern of heat shock transcription factor family in Triticum aestivum L. BMC Genomics 20:257
Fernandez-Bautista N, Fernandez-Calvino L, Munoz A, Castellano MM (2017) HOP3, a member of the HOP family in Arabidopsis, interacts with BiP and plays a major role in the ER stress response. Plant Cell Environ 40:1341–1355
Gidalevitz T, Stevens F, Argon Y (2013) Orchestration of secretory protein folding by ER chaperones. Biochim Biophys Acta 1833:2410–2424
Greg W, Donald S, Carl S, Parker (1988) Isolation of the gene encoding the S. cerevisiae heat shock transcription factor. Cell 54:841–853
Guo XL, Yuan SN, Zhang HN, Zhang YY, Zhang YJ, Wang GY, Li YQ, Li GL (2020) Heat-response patterns of the heat shock transcription factor family in advanced development stages of wheat (Triticum aestivum L.) and thermotolerance-regulation by TaHsfA2-10. BMC Plant Biol 20:364
Heerklotz D, Döring P, Bonzelius F, Winkelhaus S, Nover L (2001) The balance of nuclear import and export determines the intracellular distribution and function of tomato heat stress transcription factor HsfA2. Mol Cell Biol 21:1759–1768
Hossain M, Henríquez-Valencia C, Gómez-Páez M, Medina J, Orellana A, Vicente-Carbajosa J, Zouhar J (2016) Identification of novel components of the unfolded protein response in Arabidopsis. Front Plant Sci 7:650
Iba K (2002) Acclimative response to temperature stress in higher plants: approaches of gene engineering for temperature tolerance. Annu Rev Plant Biol 53:225–245
Kataoka R, Takahashi M, Suzuki N (2017) Coordination between bZIP28 and HSFA2 in the regulation of heat response signals in Arabidopsis. Plant Signal Behav 12:e1376159
Lee LY, Fang MJ, Kuang LY, Gelvin SB (2008) Vectors for multi-color bimolecular fluorescence complementation to investigate protein-protein interactions in living plant cells. Plant Methods 4:24
Li Z, Tang J, Srivastava R, Bassham DC, Howell SH (2020) The Transcription factor bZIP60 links the unfolded protein response to the heat stress response in maize. Plant Cell 32:3559–3575
Liu J, Sun N, Liu M, Liu J, Du B, Wang X, Qi X (2013) An autoregulatory loop controlling Arabidopsis HsfA2 expression: role of heat shock-induced alternative splicing. Plant Physiol 162:512–521
Liu B, Hu J, Zhang J (2019) Evolutionary divergence of duplicated hsf genes in Populus. Cells 8:438
Liu Z, Li G, Zhang H, Zhang Y, Zhang Y, Duan S, Sheteiwy MSA, Zhang H, Shao H, Guo X (2020) TaHsfA2-1, a new gene for thermotolerance in wheat seedlings: characterization and functional roles. J Plant Physiol 246–247:153135
Lohani N, Golicz AA, Singh MB, Bhalla PL (2019) Genome-wide analysis of the Hsf gene family in Brassica oleracea and a comparative analysis of the Hsf gene family in B. oleracea, B. rapa and B. napus. Funct Integr Genomics 19:515–531
McDonald KG, Sutton GB, Ellison WF (1983) The effect of time of sowing on the grain yield of irrigated wheat in the Namoi Valley, New South Wales. Aust J Agric Res 34:229–240
Meena S, Deb S, Samtani H, Khurana P (2020) Dissecting the molecular function of Triticum aestivum STI family members under heat stress. Front Genet 11:873
Meiri D, Breiman A (2009) Arabidopsis ROF1 (FKBP62) modulates thermotolerance by interacting with HSP90.1 and affecting the accumulation of HsfA2-regulated sHSPs. Plant J 59:387–399
Miller G, Mittler R (2006) Could heat shock transcription factors function as hydrogen peroxide sensors in plants? Ann Bot 98:279–288
Nover L, Bharti K, Döring P, Mishra KS, Ganguli A, Scharf DK (2001) Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need? Cell Stress & Chaperones 6:177–189
Park CJ, Park JM (2019) Endoplasmic reticulum plays a critical role in integrating signals generated by both biotic and abiotic stress in plants. Front Plant Sci 10:399
Poonia AK, Mishra SK, Sirohi P, Chaudhary R, Kanwar M, Germain H, Chauhan H (2020) Overexpression of wheat transcription factor (TaHsfA6b) provides thermotolerance in barley. Planta 252:1–14
Röth S, Mirus O, Bublak D, Scharf K, Schleiff E (2017) DNA-binding and repressor function are prerequisites for the turnover of the tomato heat stress transcription factor HsfB1. Plant J 89:31–44
Scharf K, Berberich D, Ebersberger T, Nover I L (2012) The plant heat stress transcription factor (Hsf) family: structure, function and evolution. Biochim Biophys Acta 1819:104–119
Scharf KD, Heider H, Höhfeld I, Lyck R, Schmidt E, Nover L (1998) The tomato Hsf system: HsfA2 needs interaction with HsfA1 for efficient nuclear import and may be localized in cytoplasmic heat stress granules. Mol Cell Biol 18:2240–2251
Sorger PK, Pelham HR (1988) Yeast heat shock factor is an essential DNA-binding protein that exhibits temperature-dependent phosphorylation. Cell 54:855–864
Sugio A, Dreos R, Aparicio F, Maule AJ (2009) The cytosolic protein response as a subcomponent of the wider heat shock response in Arabidopsis. Plant Cell 21:642–654
Wahid A (2007) Physiological implications of metabolite biosynthesis for net assimilation and heat-stress tolerance of sugarcane (Saccharum officinarum) sprouts. J Plant Res 120:219–228
Wang H, Bian M, Yang Z, Lin C, Shi W (2013) Preliminary functional analysis of the isoforms of OsHsfA2a (Oryza sativa L.) generated by alternative splicing. Plant Mol Biol Rep 31:38–46
Yong-Xiang L, Hai-Yang J, Zhang-Xin C, Xiu-Li T, Zhu SW, Bei-Jiu C (2011) Genome-wide identification, classification and analysis of heat shock transcription factor family in maize. BMC Genomics 12:1–14
Yu X, Wang T, Zhu M, Zhang L, Zhang F, Jing E, Ren Y, Wang Z, Xin Z, Lin T (2019) Transcriptome and physiological analyses for revealing genes involved in wheat response to endoplasmic reticulum stress. BMC Plant Biol 19:1–22
Zhu X, Huang C, Zhang L, Liu H, Yu J, Hu Z, Hua W (2017) Systematic analysis of Hsf family genes in the Brassica napus genome reveals novel responses to heat, drought and high CO2 stresses. Front Plant Sci 8:1174
Funding
SM and HS are thankful to the Department of Biotechnology (DBT) and University Grant Commission (UGC) for fellowships. This work has been supported by grants from DBT and JC Bose fellowship award, Science and Engineering Research Board, Government of India, for research support.
Author information
Authors and Affiliations
Contributions
SM and HS conducted the experiments. PK conceived the idea and provided logistic support.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
11103_2022_1252_MOESM1_ESM.tif
Supplementary material 1 (TIF 868.2 kb)—Confirmation of Arabidopsis transgenics: (A) PCR with TaHsfA6b-4D specific primers, (B) hygromycin specific primers, and (C) overexpression checked with qRT-PCR using TaHsfA2d primers
11103_2022_1252_MOESM2_ESM.tif
Supplementary material 2 (TIF 472.5 kb)—Expression analysis of TaHsfA6b homeologues in different developmental stages of wheat. Heatmap showing the relative expression profile of TaHsfA6b-4D and TaHsfA6b-4A in different tissues of wheat
11103_2022_1252_MOESM3_ESM.tif
Supplementary material 3 (TIF 1668.2 kb)—Comparison of three-dimensional structure of protein of TaHsfA6b-4D and TaHsfA6b-4A from Triticum aestivum, HsfA2.1 and HsfA2.3 from Arabidopsis thaliana and HsfA2dI and HsfA2dII from Oryza sativa. The structures were predicted using Pyre2 web portal
11103_2022_1252_MOESM4_ESM.tif
Supplementary material 4 (TIF 256.0 kb)—Depiction of NLS and ER signal motifs. The presence of ER retention signals, bilysine motifs KXKXX and RXR motif, in the TaHsfA6b-4D protein sequences are depicted by the highlighted residues in the red color
Rights and permissions
About this article
Cite this article
Meena, S., Samtani, H. & Khurana, P. Elucidating the functional role of heat stress transcription factor A6b (TaHsfA6b) in linking heat stress response and the unfolded protein response in wheat. Plant Mol Biol 108, 621–634 (2022). https://doi.org/10.1007/s11103-022-01252-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-022-01252-1