Abstract
Key message
The understanding of roles of bZIP factors in biological processes during plant development and under abiotic stresses requires the detailed mechanistic knowledge of behaviour of TFs.
Abstract
Basic leucine zipper (bZIP) transcription factors (TFs) play key roles in the regulation of grain development and plant responses to abiotic stresses. We investigated the role and molecular mechanisms of function of the TabZIP2 gene isolated from drought-stressed wheat plants. Molecular characterisation of TabZIP2 and derived protein included analyses of gene expression and its target promoter, and the influence of interacting partners on the target promoter activation. Two interacting partners of TabZIP2, the 14-3-3 protein, TaWIN1 and the bZIP transcription factor TaABI5L, were identified in a Y2H screen. We established that under elevated ABA levels the activity of TabZIP2 was negatively regulated by the TaWIN1 protein and positively regulated by the SnRK3/CIPK protein kinase WPK4, reported previously to be responsive to nutrient starvation. The physical interaction between the TaWIN1 and the WPK4 was detected. We also compared the influence of homo- and hetero-dimerisation of TabZIP2 and TaABI5L on DNA binding. TabZIP2 gene functional analyses were performed using drought-inducible overexpression of TabZIP2 in transgenic wheat. Transgenic plants grown under moderate drought during flowering, were smaller than control plants, and had fewer spikes and seeds per plant. However, a single seed weight was increased compared to single seed weights of control plants in three of four evaluated transgenic lines. The observed phenotypes of transgenic plants and the regulation of TabZIP2 activity by nutrient starvation-responsive WPK4, suggest that the TabZIP2 could be the part of a signalling pathway, which controls the rearrangement of carbohydrate and nutrient flows in plant organs in response to drought.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Drought is a major cause of crop losses throughout the world. Plants have developed various molecular, cellular and physiological mechanisms to withstand drought and other environmental stresses (Hrmova and Lopato 2014). Abscisic acid (ABA) is a main abiotic stress hormone, which significantly affects plant growth and regulation under osmotic stress, which is a key component of strong drought. Under stress, ABA activates cascades of stress-responsive regulatory genes, which change the plant transcriptome by activation or modulation of the activity of a variety of stress-related transcription factors (TFs). The main receptor of elevated ABA concentrations is the pyrabactin resistance/pyrabactin resistance-like/regulatory component of the ABA receptor (PYR/PYL/RCAR) (Danquah et al. 2014; Gonzalez-Guzman et al. 2012). For instance, Arabidopsis mutants with overexpressed PYL5, PYL8, or PYL9 manifest an improved drought tolerance, which is accompanied by enhanced responses to ABA (Saavedra et al. 2010; Ma et al. 2009).
The binding of ABA to PYR/PYL/RCAR proteins induces changes in their conformation, which enable them to bind and deactivate protein phosphatases 2C (PP2C), converting them to negative regulators of ABA-mediated signalling (Schweighofer et al. 2004). In the absence of ABA, PP2Cs keep sucrose nonfermenting1-related protein kinase2 (SnRK2) in an inactive non-phosphorylated state, thereby preventing the initiation of downstream phosphorylation cascades. SnRK2s belong to the family of plant-specific Ser/Thr kinases. In the presence of ABA, SnRK2 remains in an active phosphorylated state, which at least partially leads to auto-phosphorylation. In its phosphorylated state, SnRK2 plays the role of a positive regulator of ABA-mediated signalling (Danquah et al. 2014; Umezawa et al. 2010). Phosphorylated SnRK2s either initiate a mitogen-activated protein kinase (MAPK) pathway (MAP3K → MAP2K → MAPK), or they activate a calcium dependent protein kinase (CDPK) or casein kinase II (CKII), which in turn phosphorylate target basic leucine zipper (bZIP) TFs (Colcombet and Hirt 2008). Examples of direct physical interactions of SnRKs and bZIP TFs, and direct activation of bZIP TFs by SnRK2s have also been reported (Chae et al. 2007; Kobayashi et al. 2005; Tang et al. 2012; Wang et al. 2013).
The bZIP family is one of the largest and most diverse TF families (Jakoby et al. 2002). Numerous data demonstrate the involvement of bZIP TFs in responses to ABA signalling, and abiotic and biotic stresses such as drought, salt and cold stress, including pathogen defense. In addition, bZIP TFs participate in many physiological and developmental processes, such as organ formation and development (Walsh et al. 1998; Chuang et al. 1999; Abe et al. 2005; Silveira et al. 2007; Shen et al. 2007), regulation of seed development and seed maturation (Yamamoto et al. 2006; Alonso et al. 2009), and photomorphogenesis, light, hormone and sugar signalling (Gangappa et al. 2013; Kang et al. 2010; Li et al. 2011; Thalor et al. 2012; Prasad et al. 2012). bZIP TFs contain a highly conserved bZIP domain, which is composed of a region enriched in basic amino acid residues and a leucine zipper (Hurst 1994; Jakoby et al. 2002; Sornaraj et al. 2016). The basic region consists of approximately 16 residues with an invariant N-x7-R/K motif, which contains a nuclear localisation signal and is responsible for DNA binding. The leucine zipper forms an amphipathic α-helix consisting of seven repeats of leucine or other hydrophobic residues (Ile, Val, Phe, or Met) (Landschulz et al. 1988; Ellenberger et al. 1992; Sornaraj et al. 2016). Previous studies have suggested that plant bZIP proteins, which are induced by elevated concentrations of ABA and abiotic stresses, bind to DNA sequences known as abscisic acid responsive elements (ABREs), containing an ACGT core cis-element, such as G-box (CACGTG), C-box (GACGTC) and A-box (TACGTA) (Izawa et al. 1993; Foster et al. 1994). Notably, hetero-dimers, formed through their leucine zippers, often bind specific DNA elements tighter, and are more efficient in the activation of target genes (Sornaraj et al. 2016).
Post-translational modifications such as phosphorylation are the main mechanism of activity regulation of bZIP TFs, although components of this regulation are still poorly understood. For example, it was shown that ABA-activated SnRK2 can phosphorylate Ser/Thr residues at R-X-X-S/T sites in the conserved regions of the Arabidopsis bZIP TF, AREB1 (Fujita et al. 2009; Furihata et al. 2006). However, a substitution of Ser/Thr residues for Asp decreased but did not abolish the activation properties of AREB1, suggesting the presence of other functional phosphorylation sites.
The AREB/ABF/ABI5 subfamily of the Group A bZIP TFs in Arabidopsis has been well studied because of its prominent role in the ABA signalling pathway and abiotic stress responses. ABI5 was initially identified as an ABA signalling component with an essential role in seed germination and an ABA-dependent arrest of seedling development after germination (Lopez-Molina and Chua 2000; Lopez-Molina et al. 2002). In contrast, ABFs and AREBs function mostly during later vegetative stages of plant development (Choi et al. 2000; Uno et al. 2000; Kang et al. 2002; Kim et al. 2004; Fujita et al. 2011). AREB/ABF/ABI5 members are major targets of SnRK2 protein kinases during ABA signalling (Fujii and Zhu 2009; Fujita et al. 2009). Yoshida et al. (2015) demonstrated that four Arabidopsis bZIP TFs (ABF1, AREB1/ABF2, ABF3 and AREB2/ABF4) were downstream genes of the ABA-dependent drought signalling pathway, and were possible substrates of SnRK2s. Based on the analysis of triple mutants in AREB1, AREB2 and ABF3, SnRK2s were suggested to be the master regulators of drought responses in Arabidopsis (Yoshida et al. 2010). Furthermore, SnRK2-AREB/ABF have been confirmed to function as major regulatory components of ABA-dependent gene expression in response to abiotic stresses in Arabidopsis (Kim et al. 2004; Fujita et al. 2009; Furihata et al. 2006; Fujii et al. 2009; Yoshida et al. 2010).
The roles and mechanisms of post-translational regulation of AREB/ABF/ABI5-like TFs have also been studied in grasses (Kobayashi et al. 2008a, b; Ohnishi et al. 2008; Tang et al. 2012; Xu et al. 2014; Wang et al. 2016; Zou et al. 2008). For instance, drought, heat, and elevated concentrations of ABA induced the expression of ABI5-like OsbZIP46 from rice (Oryza sativa L.). Overexpression of the native OsbZIP46 gene increased ABA sensitivity but had no positive effect on drought tolerance. Using a series of truncated proteins, it was identified that the region of the OsbZIP46 activation domain, designated as domain D, affected transactivation negatively. The deletion of domain D led to a constitutively active form of OsbZIP46 (OsbZIP46CA1). Overexpression of OsbZIP46CA1 in rice significantly increased tolerance to drought and osmotic stress (Tang et al. 2012). SnRK2 from rice (OSRK1) phosphorylated the rice bZIP factor OREB1 in vitro at multiple sites of a variety of functional domains. OSRK1 showed a strong substrate preference for bZIP TF, and an uncommon cofactor requirement for Mn2+ over Mg2+. MALDI-TOF analysis identified one of the phosphorylation sites at Ser44 of OREB1 and mutation of this residue significantly decreased the substrate specificity of OSRK1. The authors concluded that the interaction between SnRK2 family kinases and ABF TFs may constitute an important part of a cross-talk mechanism in stress signalling networks in rice (Chae et al. 2007). Several wheat bZIP TFs from the group A were characterised and their involvement in stress response was demonstrated in transgenic plants (Kobayashi et al. 2008b; Xu et al. 2014; Zhang et al. 2015; Gahlaut et al. 2016; Wang et al. 2016). However, other wheat components of ABA- and stress-activated pathways and their relations to reported bZIP factors were poorly elucidated. Further, in most studies, the expression of wheat bZIP TFs was performed in heterologous systems and no transgenic wheat plants were generated; therefore, no evaluations of influences of transgenes on the grain yield were performed.
In this work, we have characterised the gene encoding wheat bZIP TFs from Group A, designated TabZIP2. TabZIP2 was overexpressed in transgenic wheat lines under the strong drought-inducible ZmRab17 promoter. Comparative analyses of growth and yield components of transgenic and control plants, grown under slowly developing drought, suggested that transgenic plants under strong stress sacrificed the development of certain plant tissues for the acceleration of grain development. We revealed that TabZIP2 was partially activated by the SnRK3-type protein kinase, which together with TabZIP2 and 14-3-3 proteins may constitute a portion of a signalling pathway that under drought controls the rearrangements of nutrient flows in wheat plant organs.
Materials and methods
Cloning of TabZIP2 cDNA
Eighteen independent clones containing the full-length coding regions of TabZIP2 cDNA were isolated in a Yeast-one-Hybrid (Y1H) screen using a cDNA library prepared from spikes and leaves of a drought-tolerant wheat genotype (T. aestivum cv. RAC875), subjected to drought and heat stresses (Amalraj et al. 2016). The cDNA library was prepared and screened following procedures described by Pyvovarenko and Lopato (2011), using as a bait three consecutive repeats of the synthetic ABRE (ABRE, GGCCCACGTGGCCC; the recognition sequence in bold) cis-element, containing G-box as a core element (Choi et al. 2000).
Plasmid construction and plant transformation
A 759 bp fragment of the TabZIP2 coding region (including stop codon) was cloned into the pENTR-D-TOPO vector and re-cloned into the pRab17 vector (Morran et al. 2011), which was generated by modification of the pMDC32 vector (Curtis and Grossniklaus 2003). This modification involved replacing the 35S promoter with the maize Rab17 promoter using HindIII—KpnI restriction sites. The pRab17-TabZIP2 construct was transformed into the Australian wheat, cv. Gladius, using a biolistic bombardment method (Ismagul et al. 2014). Genomic DNA was extracted from young leaf tissue using a freeze-dry method (Shavrukov et al. 2010). Transgene integration was confirmed by PCR using a forward primer from the 3′-end of the transgene coding region and a reverse primer from the 5′-end of the nos terminator. Transgene copy number was estimated in T1 progeny of selected transgenic lines using quantitative real-time PCR (Q-PCR) (Fletcher 2014). Homozygous transgenic lines were selected as described by Yang et al. (2017). Expression of a transgene and an endogene in unstressed and dehydrated leaves of transgenic lines was assessed by Q-PCR (Fletcher 2014) and primers defined in Table S1. Representatives of T3 progeny of homozygous Lines 1, 2 and 5 were used in Q-PCR experiments.
Analysis of transgenic plants
Transgenic wheat plants (T1–T3) were grown in either a growth room (drought tolerance tests) or a glasshouse to characterise plant phenotypes. Growth room temperatures were maintained at 24 °C during the 12-h day and 18 °C during the night, and the average relative humidity was 50% during the day and 80% during the night. Wild type (WT) plants were used as controls. Seeds were germinated in moist paper-lined Petri dishes at room temperature for 3 days and transferred to containers filled with a 1:1:1 mix of coco-peat, river sand, and clay soil collected near Adelaide (South Australia). For phenotyping under well-watered conditions, the plants were grown either in small pots (8 × 8 × 10 cm), one plant per pot (T1 generation) or in large, plot-sized containers (T2 generation). The size of each container was 120 l × 80 w × 40 d cm, and the distance between plants was eight cm. T2 plants of four homozygous lines (16 plants per line plus 16 WT plants) were used for the phenotyping in each of two containers. Each container had ten subplots, flanked by a border row (WT plants) on each short side of the container. Border plants were not used in the data analysis. The experimental design was identical for each container. No significant differences were found in plant growth between subplots in several preliminary experiments, and therefore, all replicates for each line and WT plants were used, to calculate confidence intervals and means for individual measurements. Containers were equipped with an automatic watering system, and four soil water tensiometers (gypsum blocks) were installed at 0.1 and 0.3 m soil depths and connected to a data logger for continuous monitoring of soil water tension. Plant height, number of tillers and spikes, plant biomass, seed number and seed weight were recorded for each plant.
The remainder of Materials and Methods is included in Supplementary Information.
Results
Phylogenetic analysis of TabZIP proteins
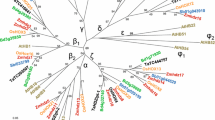
Full-length cDNA for the transcription factor TabZIP2 was isolated from a wheat cDNA library, as described in the Methods. TabZIP2 belongs to Group A of the bZIP family (Fig. 1, Table S2) and shares a high level of sequence identity at the protein level with homologues TabZIP3 from wheat (57%) and ZmbZIP1 from maize (58%), while the identity with the closest homologues from Arabidopsis, AtbZIP40 and AtbZIP13, is 33%. An interacting partner of TabZIP2, isolated in a Yeast-two-Hybrid (Y2H) screen of the same cDNA library and designated as TaABI5-like (TaABI5L), also clusters to group A of the bZIP TF family (Fig. 1); the sequence identity between TaABI5L and orthologous TabZIP60 is 96%, with 12 residue differences, which reside in unstructured N-termini. Detailed sequence comparisons of wheat and barley orthologues (TabZIP2, TaABI5L, TabZIP60 and HvABI5) revealed that these proteins are highly homologous (Figure S1). Notably, TaABI5L, TabZIP60 and HvABI5 have nearly identical pI values (9.25 and 9.7) and similar molecular masses of 38–39 kDa. This high relatedness is reflected by the identical positions of DNA-binding (37–39 residues), leucine zipper (38 residues) and conserved 14-3-3 binding motifs in C-termini (Figure S1); the PROMALS3D alignment analysis (Pei et al. 2008) returned sequence conservation scores of 9 in these regions (Figure S1). All four TFs showed the same distributions of secondary structural elements and domain boundaries for the DNA-binding and leucine zipper regions. We conclude that these cereal TFs are likely to retain nearly identical 3D structures in these regions, which would lead to common folding patterns and function. The main structural difference between TabZIP2 and TaABI5L proteins is in the length of unstructured N-terminal regions, of 155 (TabZIP2) to 260–270 residues (TaABI5L, TabZIP60 and HvABI5), which contain characteristic Pro, Gly, Ala and Ser repeats (Figure S1).
Evolutionary relationships of plant bZIP TFs. The species include Arabidopsis thaliana (At), Daucus carota (CAREB), Hordeum vulgare (Hv), Oryzae sativa (Os), Triticum aestivum (Ta), and Zea mays (Zm). The full list and details of the 81 protein sequences is included in Table S2. Wheat bZIP TFs studied in this paper (TabZIP2 and TaABI5L) are circled. Evolutionary analysis was conducted with MEGA, version 6.0 (Tamura et al. 2013) using the Neighbor-Joining method (Saitou and Nei 1987) following an alignment in MUSCLE (Edgar 2004). The optimal tree with the sum of branch length = 13.76526669 is shown. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances. The evolutionary distances were computed using the p-distance method (Saitou and Nei 1987) and are shown in units of the number of amino acid residue differences per site. All positions containing gaps and missing data were eliminated. Proteins are classified into nine groups based on sequence similarities of basic and other conserved motifs as described by Jakoby et al. (2002). Groups A, B, C, D, E, G, H, I and S are annotated with circles, triangles and squares
TabZIP2 expression in wheat tissues and transcriptional response to drought and cold
The TabZIP2 gene was strongly expressed in all tested tissues, with particularly high gene expression detected in leaves, floral tissues and endosperm of mature grain (Fig. 2a). Expression of TabZIP2 gene was not significantly affected by low temperature (4 °C) (Figure S2a). For this analysis, the cold-inducible TaCor410b gene (Eini et al. 2013) was used as a positive control. Only weak transcriptional activation of TabZIP2 was observed during slowly developing drought, while the control drought-inducible WZY2 gene (Zhu et al. 2014) was clearly activated under these conditions (Figure S2b). However, a several-fold increase of TabZIP2 transcripts was observed when drought conditions were severe, with rapid dehydration of young seedlings over two to three days. In the drought sensitive wheat cv. Bobwhite, the increase of transcript levels under severe drought (Fig. 2b, lower panel) was more pronounced than in the drought tolerant cv. Gladius (Figure S3). Additional comment to these results are included in Supporting Information.
Expression levels of the TabZIP2 gene in a variety of wheat tissues and in strongly dehydrated wheat leaves. Levels of expression were detected by Q-PCR and are shown as normalised transcript copy number per microgram of RNA. a TabZIP2 transcript levels in different tissue types under non-stressed conditions (cv. Chinese Spring); b expression of TabZIP2 in leaves of three-week-old seedlings subjected to rapid dehydration (cv. Bobwhite); leaves were collected at a wilting point from transgenic plants overexpressing TaDREB3, null-segregants, and control (WT) plants. Plants marked with letter ‘D’ were subjected to drought. Plants growing under control conditions (no drought) have no additional designations after names. Levels of transgene (TaDREB3) expression are shown in the upper panel. The error bars represent SD of three technical replicates
Drought-inducible expression of TabZIP2 in transgenic wheat alters grain number, size and yield under drought
Fifteen independent transgenic wheat lines were produced by stable transformation of the pRab17-TabZIP2 construct in the elite drought-tolerant Australian wheat cv. Gladius. Selected T0 transgenic lines with a single copy of the transgene showed elevated transgene transcription and slightly repressed expression of endogenous TabZIP2 (Figure S3). Expression levels of the TabZIP2 transgene under drought were six- to ten-fold higher than those of the endogene induced by drought. Under well-watered conditions, TabZIP2 transgene expression was undetectable in some lines, while other lines had basal levels of transgene expression that were three- to four-fold higher than those of the endogene. The expression data correlated well with plant phenotypes: no significant differences in plant growth and yield characteristics, compared to control (WT) plants, were observed under well-watered conditions (low or no transgene activation), except for grain size in several lines (larger or smaller than WT). However consistent significant changes in phenotypes compared to control (WT) plants occurred under drought (Figure S3). T2 generation plants of four independent transgenic lines together with untransformed control plants were grown to maturity in large containers under conditions designed to mimic to those in the field. Under drought, transgenic lines developed an average of five spikes fewer than control (WT) growing at well-watered conditions and one spike fewer than control plants growing under drought. They produced about 40–50% fewer seeds per plant than control plants grown under drought (Fig. 3). However, single grain weight was increased in three of the four analysed lines (in two of four lines the increase was significant) compared to WT plants grown under drought. Resulting seed yield per plant was significantly lower in drought-treated plants of two of the four transgenic lines compared to WT, while yields under well-watered conditions were not negatively affected (Fig. 3). No other abnormalities in plant growth and development, such as changes in leaf size and shape, flowering time, or shape of spikes or grains, were observed. In heterozygous lines (two of four lines used in this experiment), T2 null segregants were excluded from analyses of yield components.
Growth characteristics and yield components of control (WT) and homozygous T2 transgenic wheat (T. aestivum cv. Gladius), transformed with pRab17-TabZIP2. Plants were grown in large containers under well-watered conditions (left side of each panel) or constantly increasing drought (right side of each panel); 16 plants of each transgenic and control plants were used in each container. Values represent means ± SE; asterisks indicate lines that were significantly different to the WT control group within each treatment (‘*’ for P < 0.05, ‘**’ for P < 0.01 and ‘***’ for P < 0.005), as calculated by Student’s t tests. Null-segregants were removed from datasets before analysis
Identification and mapping of TdCor39, a target promoter of TabZIP2
A transient expression assay in wheat cell culture was used to find a potential target gene for TabZIP2. For this purpose, we used an in-house collection of stress-inducible promoters, cloned upstream of the reporter GUS gene. Co-bombardment of wheat cells with a 3:1 mix of the pUbi-TabZIP2 effector construct and each of the six reporter constructs revealed promoter of cold and drought responsive gene, TdCor39 (GenBank acesssion number AF058794; Kovalchuk et al. 2013), which was strongly activated by TabZIP2, and was chosen for further characterisation. No activation of the TdCor39 promoter was observed by other stress-related TFs (TaDREB3 and TaDREB2, Morran et al. 2011; TaERF4a; Eini et al. 2013) (Fig. 4a). Addition of 0.2 mM ABA to cell cultures one hour after bombardment with the pUbi-TabZIP2 construct led to several-fold further enhancement of the TdCor39 promoter activity (Fig. 4b), indicating that the promoter activation by TabZIP2 is regulated by ABA. Similar ABA enhancement of the TdCor39 promoter activation by TabZIP2 was not observed in cell cultures co-bombarded with the control pUbi-GFP construct, suggesting that post-translational activation of TabZIP2 rather than activation of endogenous bZIP TF(s) took place (Fig. 4b). The TdCor39 promoter was mapped using a combination of the TabZIP2 effector, 0.2 mM ABA and several reporter constructs with promoter deletions in the 5′ region cloned upstream of the GUS gene. Mapping revealed two functional ABRE elements and one functional C-repeat (CRT) element (Fig. 4c–e, Figure S4); functional cis-elements are indicated by short bulky arrows in Fig. 4c. The physical interaction of TabZIP2 homo-dimers with a number of documented ABREs, including the original ABRE used to clone TabZIP2 (ABRE in Fig. 5a) and the functional ABREs derived from the TdCor39 promoter (ABRE C in Fig. 5b), were confirmed by pull-down assays using biotinylated DNA cis-elements that bind catalytically tagged (by endo-1-4-β-d-glucanase, CelD) TFs. This method, known as catalytic tagging, allows estimation of the relative strength of each interaction. Using TdCor39 promoter segments we also established that the functional CRT element and its adjacent sequences were not able to interact directly with TabZIP2 (Fig. 5b).
Characterisation of the TdCor39 promoter activation by TabZIP2 using a transient expression assay in cultured wheat cells. Activation of the TdCor39 promoter by a stress-responsive TabZIP2, TaDREB3, TaERF4a and TaDREB2 TFs, and b TabZIP2 in the absence and presence of exogenous ABA. c Schematic representation of the TdCor39 promoter sub-cloned upstream of the GUS gene; the beginnings of 5′ promoter deletions (D2–D6) are shown with black right-pointing arrows. Bulky downwards-pointing arrows indicate functional ABRE and CRT cis-elements. d Mapping of functional cis-elements in the TdCor39 promoter using promoter deletions (D2–D6) as reporter constructs, and TabZIP2 activated with 0.2 mM ABA as an effector. pTdCor39-GUS constructs containing full-length promoter or 5′ promoter deletions were co-bombarded together with respective constructs for constitutive (pUbi) expression of TFs. Activation levels are shown as numbers of GUS foci. The promoter-GUS construct co-bombarded with pUbi-GFP was used as a negative control. e Precise mapping of the functional CRT element; full the full-length TdCor39 promoter. Further sequence information and positions of cis-elements are shown in Figure S4
DNA-binding specificities of TabZIP2 TF. The analysis is based on interactions of CelD-tagged TabZIP2 with biotinylated a ABRE cis-elements, and b fragments of the TdCor39 promoter containing functional cis-elements, as defined in Fig. 4c. Equimolar concentrations of proteins and DNA were used, and proteins without biotinylated cis-elements served as negative controls. Negative control values were subtracted from experimental values. DNA sequences of respective cis-elements are included on the right of each panel. Core sequences of tested ABREs are underlined and italicised, and CRT elements are underlined. Asterisks and twin asterisks represent significant differences compared to GCC-box control at P < 0.05 or P < 0.005 significance levels, respectively, calculated by t test. Paired Two Samples Means, using Microsoft Excel Professional Plus 2010, version 14.0.7177.5000
Identification of interaction partners of the TabZIP2 protein
Initially, the TabZIP2 protein was investigated in silico for the presence of known motifs and domains. Two domains, a DNA-binding domain and a dimerisation (leucine zipper) domain, were localised in the protein near to the C-terminus, and a short-conserved motif, known as the 14-3-3 binding sequence (Sirichandra et al. 2010) was found at the C-terminus of TabZIP2 (Figure S1). A potential activation domain (AD) was predicted to be positioned in N-terminal half of the protein (Fig. 6a). Two truncated forms of the TabZIP2 protein (D1 and D2) were generated and used in an ‘in-yeast activation assay’ to confirm the identity of the predicted TabZIP2 AD. It was shown that an AD was situated on the D1 fragment, which represents the N-terminal half of TabZIP2 (Fig. 6b). The D2 fragment demonstrated no self-activating properties. It was therefore used as a bait in a Y2H screen of a wheat cDNA library prepared from flag leaves and developing spikes of an Australian drought-tolerant bread wheat (T. aestivum genotype cv. RAC875,) subjected to high temperatures under both well-watered and drought stress conditions. After screening several million colonies and a second round of selection for positive clones, two groups of clones were isolated. The cloned inserts represented two different cDNA sequences.
The key residues underlying the function of TabZIP2. a Protein sequence of TabZIP2 with colour-coded DNA-binding (red), and leucine zipper (blue) regions with Leu and Val residues (involved in dimerisation) in green, and the C-terminal 14-3-3 binding motif (magenta). b Identification and a consequent removal of the protein segment containing the predicted activation domain that is responsible for self-activation of TabZIP2; the construct used as a bait in a Y2H screen of a wheat leaf cDNA library is marked with a green circle. c–e Identification (using a Y2H assay) of segments and residues of TabZIP2 that are important for TaWIN1 binding, and testing of TabZIP2 inability to bind TaWIN1 during dimerisation with TaABI5L. Interaction of TabZIP2 segments with TaWIN1 was dependent on the four of six residues of the 14-3-3 recognition motif (LRR--S). Red dots indicate the D2 segment (containing last nine C-terminal residues of the protein), and T247A and N248A mutations in the segment D2 did not disrupt interaction between D2 and TaWIN1. f Summary of involvement of LRR--S residues (bold magenta) that are required for TaWIN1 binding to TabZIP2 (D2), while T and N residues (regular green) are not required. g Identification of physical interactions between WPK4 and full-length TabZIP2 or TaWIN1. Yeast growth on selective media on the right-hand side of each panel indicates protein–protein interactions
The first group, comprising two independent clones, contained partial cDNA sequences of a bZIP TF. The cDNA fragments lacked 5′UTR and approximately 279 bp at the beginning of the coding region. The absent 5′ end was identified using NCBI EST databases and the full-length coding sequence (CDS) was isolated by nested PCR from the same wheat cDNA library. As with TabZIP2, the protein encoded by this cDNA belongs to the Group A bZIP TFs, but to a different sub-clade containing close homologues/orthologues of the Arabidopsis ABI5 protein, and therefore it was designated as TaABI5L (Fig. 1, Figure S1).
All inserts from the second group of clones, comprising eight independent clones, encoded the full-length CDS of the 14-3-3 protein TaWIN1, previously described by Ikeda et al. (2000). We also used as a bait full-length sequence of the SNF1-like WPK4 kinase, which was previously shown to physically interact with TaWIN1 (Ikeda et al. 2000). Beside several potential WPK4 substrates including TFs, we isolated clones containing TaWIN1 inserts, and these represented about 85% of all isolated positive clones. Interactions of two identified proteins with the D2 segment of TabZIP2, and the D2 segment lacking the 14-3-3 motif, designated D2-243, and mapping of residues in TabZIP2 critical for TaWIN1 or TaABI5L binding, are summarised in Fig. 6c–g. All baits, including mutant proteins, where one of the six residues of the 14-3-3 recognition motif was substituted by alanine (marked in magenta in Fig. 6), showed no self-activation after these baits were co-transformed with an empty vector (Fig. 6c). Interaction of the D2 segment of TabZIP2 with TaABI5L, which was expected to occur through their leucine zippers, was not affected by the absence of the 14-3-3 recognition motif (Fig. 6d). Interaction of TabZIP2 segments with TaWIN1 was dependent on four of the six residues LRR--S of the 14-3-3 recognition motif (Fig. 6e); this is summarised in Fig. 6f, where required interacting residues LRR--S of TabZIP2 are in magenta. No direct interaction of full-length TabZIP2 with WPK4 was detected, although interaction of WPK4 and TaWIN1, initially reported by Ikeda et al. (2000), was confirmed in our Y2H assay (Fig. 6g).
Dimerisation specificity of TabZIP2 and TaABI5L
It has been established that dimerisation via leucine zippers contributes to bZIP TF specificity and recognition in signalling networks (Sornaraj et al. 2016). To demonstrate dimerisation specificities of bZIP TFs from two sub-clades within group A (Fig. 1), a reporter-based subunit exchange pull-down assay was used, like that described in the section ‘Identification and mapping of TdCor39, a target promoter of TabZIP2’ (Xue 2005; Xue et al. 2006; Harris et al. 2016). This analysis is based on interactions of CelD-tagged TabZIP2 or TaABI5L with biotinylated forms, which are incubated together in equimolar ratios. These assays allow an exchange of subunits to be determined, where TabZIP2-CelD could interact with biotinylated subunits of TabZIP2 or TaABI5L after they are immobilised on a Streptavidin matrix, or when TaABI5L-CelD is used as a binding partner (Fig. 7a). Data, obtained with partially purified chimeric proteins (Figure S5), demonstrated that TabZIP2 dimerisation was stronger with TaABI5L than with itself (Fig. 7a, left panel). On the other hand, TaABI5L interacted with an equal probability with either itself or TabZIP2 (Fig. 7a, right panel).
Homo- and hetero-oligomerisation of TabZIP2 and TaABI5L proteins, and 3D structural models of their α-helical regions. a Homo- and hetero-dimerisation analysis of TabZIP2 with TaABI5L was performed using the catalytic tagging method. The analysis is based on the interaction of CelD-tagged TabZIP2 or TaABI5L with biotinylated TabZIP2 or TaABI5L, which were incubated together in equimolar ratios. Negative control values were subtracted, and absorbance values plotted as means of triplicate assays with standard deviations. An asterisk indicates significant differences between hetero- and homo-dimers for each biotinylated ZIP domain at P < 0.05 significance level, calculated by t-Tests, as specified in Fig. 5. b Molecular features of TabZIP2 and TaABI5L homo- and hetero-dimers with the ABRE cis-element. Ribbon representations show that both TFs consist of basic (grey) and leucine zipper (blue and salmon for TabZIP2 and TaABI5L, respectively) regions. Interactions between residues in both chains are indicated with dotted spheres that emulate Van der Waals radii
To examine the molecular mechanism behind these interactions, we generated 3D dimeric models of the α-helical regions of TabZIP2 and TaABI5L (Table S3), containing basic DNA binding and leucine zipper regions, in complex with the functional ABRE cis-element (GGCCCACGTGGCCC; recognition sequence in bold) (Fig. 7b, Figure S1). In these models, basic DNA binding regions are in grey, and the dimerisation leucine zipper regions of TabZIP2 and TaABI5L are in blue and salmon, respectively. Residues involved in interactions mediating dimerisation are shown in sticks and represent Van der Waals forces (between leucine and hydrophobic residues), hydrogen bonds (between polar residues) and electrostatic interactions (between charged residues).
Additional interactions were noted that stabilise intramolecular α-helical regions between Glu199 and Tyr194 (Fig. 7b, left panel), and between Lys314 and Glu311 (Fig. 7b, middle panel) in the homo-dimeric TabZIP2 and TaABI5L complexes, respectively. Close interactions of 2.8–3.2 Å were also formed between Glu215 and Gln216 in the TabZIP2 chain, and between Glu215 or Gln216 and Arg322 in the TabZIP2/TaABI5L hetero-dimeric chain (Fig. 7b, right panel). In the latter complex, we observed a deviation from a rigid α-helix of the TabZIP2 component, which didn’t seem to affect overall residue participation that formed the complex (Table S4). These observations indicated that the number of electrostatic interactions was higher in the hetero-dimeric TabZIP2/TaABI5L complex (Table S4), which may explain preferential hetero-dimerisation of TabZIP2 with TaABI5L, as it was observed in the pull-down assay (Fig. 7a).
As for binding interactions between the TF complexes and DNA (Fig. 7b), these were formed between positively charged Arg and Lys and phospho-diester backbones of ABRE, as described recently by Sornaraj et al. (2016).
3D modelling of the TaWIN1 homo-dimer reveals residues that mediate binding of bZIP TFs and protein kinases
To find out what residues might participate in protein–protein interactions between TaWIN1, bZIP TFs, and protein kinases, we generated a comparative model of TaWIN1 using a combination of two templates of the 14-3-3-like protein (Würtele et al. 2003) and the 14-3-3 epsilon isoform (Molzan et al. 2012). A ribbon representation of the model (Figure S6) shows that TaWIN1 forms a flattened horse-shoe-like structure, with interaction residues (Figure S6a) involved in binding bZIP TFs and protein kinases located within the cavity of dimeric TaWIN1. Residues involved in binding were analysed through multiple sequence alignments and structural analyses of 14-3-3 proteins, bZIP TFs and kinases (Zhang et al. 1997; Göransson et al. 2006; Taoka et al. 2011; Lozano-Durán and Robatzek 2015). Figure S6b shows putative TF-binding (red), kinase-binding (magenta), and both TF- and kinase-binding (blue) residues that are differentiated as Van der Waals spheres from the backbone of TaWIN1; the colour coding of spheres corresponds to that of the TaWIN1 protein sequence (Figure S6a). At this stage, it is impossible to predict the strength of binding of bZIP TF or a kinase, or if the presence of one protein would eliminate the simultaneous binding of the other protein.
The influence of TaWIN1 and WPK4 proteins on activation of the TdCor19 promoter by TabZIP2 in the presence or absence of ABA
The influence of TaWIN1 and WPK4 on TabZIP2 activity in the presence or absence of exogenously added ABA was studied in a transient expression assay using wheat cells. In the absence of ABA, both proteins either alone or together repressed activation of the TdCor39 promoter by TabZIP2 (Fig. 8a, left panel). As expected, addition of 0.2 mM ABA increased the activity of TabZIP2, by fivefold in this experiment. In the presence of ABA, TabZIP2 activity was suppressed by co-expression of the TaWIN1 protein, but increased by co-expression of the WPK4 kinase, by nearly twofold (Fig. 8a, right panel); this observation was confirmed in an additional independent experiment (Figure S7). Simultaneous co-expression of TaWIN1 and WPK4 led to the suppression of ABA-activated TabZIP2 activity (Fig. 8a).
Influence of TaWIN1 and WPK4 on activation of the TdCor39 promoter by TabZIP2 and its variants. Activation of the TdCor39 promoter a by TabZIP2 alone or with concurrent additions of TaWIN1 and WPK4, in the presence or absence of ABA, and b by TabZIP2 variants: TabZIP2-S249A and TabZIP2-del (deletion of last nine residues), in the absence or presence of ABA. The pTdCor39-GUS reporter construct was co-bombarded with effector construct(s). Promoter activation levels are shown as number of GUS foci. Co-bombardment with the reporter pUbi-GFP construct was used as a negative control with a basal level of the promoter activity. Asterisks and twin asterisks represent significant differences compared to pUbi-GFP control at P < 0.05 or P < 0.005 significance levels, respectively, and were calculated as specified in Fig. 5
We further detected that the deletion of the last nine residues in TabZIP2 (LRRTNSMEW; marked as pUbi-TabZIP2-del in Fig. 8b), decreased but did not entirely abolished TabZIP2 activation in the absence or presence of exogenous ABA (Fig. 8b). This deletion included the 14-3-3 binding motif (LRRTNS) with three additional C-terminal residues MEW, or alternatively S249 mutated to Ala.
Discussion
cDNA of TabZIP2 was isolated from a cDNA library prepared from developing spikes and leaf tissues of the drought tolerant wheat genotype RAC875, subjected to drought and heat stresses (Eini et al. 2013). The analysis of TabZIP2 expression in a variety of wheat tissues revealed high levels of transcripts in all tested tissues. Levels of expression in leaves, parts of flowers and the endosperm of mature grains were two- to threefold higher than in other tissues. This strong and ubiquitous expression in wheat organs may suggest that the activity of TabZIP2 is predominantly regulated by post-translational modification rather than by changes in transcript and protein levels. The analysis of influence of cold and slowly (during several weeks) developing drought on transcription of TabZIP2 revealed no response to cold and a weak response to drought (Figure S2). However, the rapid dehydration of wheat seedlings (T. aestivum cv. Bobwhite, low tolerance to drought) within 2–3 days revealed a five- to sixfold increase of TabZIP2 transcripts in leaves of seedlings at wilting point, suggesting that transcriptional regulation of this TF took place only in rapidly developing and critical-for-plant stress situations. Similar, but weaker activation of TabZIP2 transcription under the same conditions was observed in a drought-tolerant wheat genotype (cv. Gladius) (Figure S3).
To investigate the practical significance of our research, we determined the influence of overexpression of TabZIP2 on grain yield of transgenic wheat under drought conditions. Transgenic wheat (T. aestivum cv. Gladius) plants were generated, single copy transgenic lines were selected and the T2 generation plants were analysed as previously described (Yadav et al. 2015; Amalraj et al. 2016). The drought inducible maize Rab17 promoter was chosen because it is strongly induced by relatively mild drought and shows low or no basal levels of activity in wheat (Morran et al. 2011). The use of this promoter allowed us to obtain about a five- to eightfold increase in TabZIP2 transgene transcripts under stress over the level of TabZIP2 endogene transcripts; this could not be achieved in wheat using the currently available constitutive promoters. Drought-inducible expression of TabZIP2 prevented any significant changes in phenotype or yield characteristics of transgenic wheat lines under well-watered conditions of cultivation. Under drought, however, the T2 plants of two of four analysed transgenic lines were smaller than those of control (WT) plants. The plants of all four T2 transgenic lines produced on average one spike-less and 30–50% fewer seeds than those of control (WT) plants. Single seed weight was, however, increased in three of four lines, which to some extent compensated for the seed number-dependent yield loss. Grain yield per plant was maintained in two of the lines and was reduced by 25–40% in the other three lines. We suggest that these changes were the result of rearrangements in carbohydrate and nutrient partitioning, because of plant response to drought during spike and grain development. This response was stronger in transgenic than in wild type plants because of wrong information was received by transgenic plants about the strength of the drought stress. This wrong information was triggered by an increased number of TabZIP2 transcripts in the transgenic plants, which in WT plants happens only with strong stress. The overexpression of TabZIP2 seemed to re-program plants to sacrifice late developing spikes and some seeds in each spike, to ensure the best possible development of remaining seeds under stress conditions. Additional mechanisms of a positive influence of TabZIP2 on grain development and particularly on grain filling because of preferable transcription of TabZIP2 in flowers and endosperm, cannot be excluded. However, these possibilities were not further examined in this study.
Initially, seven wheat promoters of stress-responsive genes from our in-house collection that might be potential targets for bZIP TFs were tested, to select a promoter able to be activated by TabZIP2. One of the tested promoters, the promoter of TdCor39, was significantly activated by TabZIP2 in a transient expression assay performed in cultured wheat cells, whereby the activation was further enhanced by exogenously added ABA (Fig. 4b). The TdCor39 gene is a late stress-responsive gene, and was originally identified as a gene encoding a group II LEA protein that accumulates in root, leaf and crown tissues of wheat during cold acclimation (Guo et al. 1992). It is strongly activated by elevated concentrations of ABA, drought, cold and mechanical wounding (Kovalchuk et al. 2013).
Mapping of the functional cis-elements in the TdCor39 promoter was performed in a transient expression assay using constitutive overexpression of TabZIP2 in the presence of additional ABA. Two functional ABRE elements and one functional CRT element in the TdCor39 promoter were revealed. The interaction of TabZIP2 with both mapped ABREs was confirmed in vitro by a pull-down assay. The mapped CRT element was likely to operate through endogenous ABA-inducible DREB/CBF or ERF types of TFs, since neither the CRT element nor its flanking nucleotide sequences were able to interact with TabZIP2 in the pull-down assay (Fig. 5b). In addition to being able to quantify the genuine physical interaction and activation of the TdCor39 promoter by TabZIP2, the transient expression assay now gave us a testing system for analysis of the influence of interacting/modifying TabZIP2 proteins on the TabZIP2-mediated activation of a stress-responsive promoter.
We next identified interacting partners of TabZIP2 using a Y2H screen of the same cDNA library which was initially used for the isolation of TabZIP2 cDNA. The full-length TabZIP2 protein behaved as a transcriptional activator in both wheat and yeast cells and therefore could not be used as a bait in the Y2H screen (Figs. 4, 6). Therefore, the N-terminal half of the protein containing the activation domain (AD) was removed. A screen with truncated TabZIP2 protein (D2) resulted in isolation of two cDNAs: the first encoded a bZIP protein of group A, designated TaABI5L, and the second cDNA encoded a 14-3-3 protein which was earlier identified as TaWIN1 (Ikeda et al. 2000). The analysis of 96 colonies revealed no other bZIP TF or 14-3-3 proteins, so the protein–protein interactions we identified, are considered as fairly specific. We also showed by Y2H assay that hetero-dimerisation of TabZIP2 and TaABI5L did not depend on the presence of the 14-3-3 binding motif at the C-terminus of TabZIP2 (D2), and therefore occurs only through the leucine zippers of both proteins. In contrast, binding of TaWIN1 to TabZIP2 took place through four amino acid residues (LRR--S) of the predicted 14-3-3 binding motif.
Analyses of TabZIP2 and TaABI5L homo- and hetero-dimerisation patterns using a pull-down assay revealed that in the case of simultaneous presence of both proteins in a plant cell, the formation of a hetero-dimer will be about twice as preferable to that of the TabZIP2 homo-dimer. Since post-transcriptionally modified TabZIP2 can already efficiently activate promoters as a homo-dimer, the formation of a hetero-dimer may change the strength of target promoter activation and increase the complexity of the TabZIP2 activity modulation by increasing the number of regulatory pathways affecting this protein.
Ikeda et al. (2000) used a wheat protein kinase 4, WPK4, as a bait for the identification of proteins involved in signal transduction through WPK4, and isolated two cDNA clones encoding very closely related 14-3-3 proteins designated as TaWIN1 and TaWIN2. It was shown that both proteins, upon auto-phosphorylation of the kinase, bind the C-terminal regulatory domain of WPK4. Mutational analysis through amino acid residue substitution revealed that TaWIN1 and TaWIN2 primarily bind WPK4 through C-terminal phosphoserines at positions 388 and 418 (Ikeda et al. 2000). Amino acid residues of TaWIN1 responsible for binding to WPK4 kinase and bZIP TFs are demonstrated in the molecular model of the TaWIN1 dimer (Figure S6). It can clearly be seen that specific residues are required for binding to either a bZIP or a kinase. Further modelling and experimental analysis is required to compare the strengths of binding to each moiety, and to investigate if the presence of one affects TaWIN1 binding to the other.
WPK4 belongs to the SNF1 related protein kinase 3 (SnRK3) class of protein kinases from wheat, later classified as calcineurin B-like protein (CBL) interacting protein kinase (CIPK) (Sano and Youssefian 1994; Ikeda et al. 1999; Nozawa et al. 2003; Ruuska et al. 2006; Mao et al. 2016). SnRK3/CIPK regulate signal transduction under abiotic stresses, nutrient deprivation and hormonal changes and operate as regulators of sodium (Na+), potassium (K+), magnesium (Mg2+), nitrate (NO3−) and proton (H+) homeostasis (Das and Pandey 2010; Sun et al. 2015; Mao et al. 2016). Four Arabidopsis CIPK proteins were found to group in the same clade with WPK4 (Figure S8, Table S5). Two of the members of this clade, CIPK12 and CIPK19, play fundamental roles during pollen germination and pollen tube growth (Steinhorst et al. 2015; Zhou et al. 2015). A similar role of WPK4 has not been reported. However, the involvement of this kinase in the regulation of wheat metabolism has been demonstrated. It was shown that WPK4 transcripts are upregulated in various wheat tissues by cytokinins, nutrient deprivation and changes in sugar concentrations (Sano and Youssefian 1994; Ikeda et al. 1999; Ruuska et al. 2006). In addition, WPK4 phosphorylates the essential nitrogen metabolism enzyme, nitrate reductase, to which TaWIN1 binds. These findings prompted the suggestion that TaWIN1 and WPK4 are involved in plant nitrogen metabolism (Ikeda et al. 2000). A precise role for WPK4 was later proposed to be in carbon storage metabolism during reproductive wheat stem growth under low nitrogen (Ruuska et al. 2006).
Our Y2H screen with WPK4 as a bait confirmed earlier findings by Ikeda et al. (2000) that the main interacting partner of this kinase is the TaWIN1 protein, as found in a majority of clones. We also demonstrated by Y2H assay that WPK4 does not interact directly with full-length TabZIP2.
The influence of TaWIN1 and WPK4 on the activation of TabZIP2 was tested in a transient expression assay in the absence and presence of ABA. In the absence of ABA, both TaWIN1 and WPK4, and in combination, inhibited the activation of the TdCor39 promoter by TabZIP2. In contrast, promoter activation was significantly enhanced by the addition of ABA, and further increased by the co-expression of TabZIP2 and WPK4. However, if TabZIP2 was co-expressed with TaWIN1 alone or with both TaWIN1 and WPK4 simultaneously, its activity was significantly inhibited, although it was not totally abolished. Hence, TaWIN1 behaves as a negative regulator of TabZIP2, and WPK4 as a positive regulator of TabZIP2.
Deletion of the 14-3-3 recognition motif or substitution of a single amino acid residue inside of this motif (S249A) led to a similar outcome to the activation of the promoter in absence of ABA, inhibiting the activation of TabZIP2. This suggested that S249 was most likely phosphorylated by the WPK4 kinase and that this phosphorylation is very important for the activation of bZIP TF. Activation can result in a release of TabZIP2 from the TabZIP2-TaWIN1 inactive complex and the deactivation of WPK4 by binding to a released TaWIN1 protein.
Notably, TaWIN1 was identified as one of the most constantly expressed genes among 24 tested candidate genes from Triticum aestivum cv. Cubus flag leaves, grown under organic and conventional farming systems. The expression levels of these genes were evaluated with the aim to select suitable genes for normalisation of quantitative real-time PCR (Q-PCR) reactions (Tenea et al. 2011). Taking into consideration a near-constitutive expression of TabZIP2, one can imagine that most of the TabZIP2 protein under favourable plant growth conditions exists in an inactive form as a complex of non-phosphorylated TabZIP2 with TaWIN1, which could be disrupted under stress due to an ABA-dependent S249 phosphorylation by the nutrient starvation-dependent WPK4, where WPK4 is a part of signal pathway activated during a phase of strong drought. This may lead to TabZIP2 activation through its release from the complex with TaWIN1, whereby TaWIN1 binds to WPK4. The formation of the TabZIP2-TaABI5L dimer could further increase the complexity of control of transcriptional activity of downstream stress-related genes. This in turn induces enrichments of drought tolerance protein transcripts and/or proteins involved in carbohydrate rearrangements in plant tissues in favour of grain.
In summary, TabZIP2 molecular characteristics and phenotypes of transgenic plants suggested that the TabZIP2 gene may play roles in signalling pathways controlling carbohydrate rearrangements and a nutrient flow between plant organs in response to drought-induced starvation.
Abbreviations
- 3D:
-
Three-dimensional
- ABA:
-
Abscisic acid
- COR:
-
Cold responsive
- DOPE:
-
Discrete optimised protein energy
- DHN:
-
Dehydrin
- GFP:
-
Green fluorescent protein
- HD-Zip I:
-
Homeodomain-leucine zipper class I
- IWGSC:
-
International Wheat Genome Sequencing Consortium
- LEA:
-
Late embryogenesis abundant
- MOF:
-
Modeller objective function
- Q-PCR:
-
Quantitative RT-PCR
- Ta:
-
Triticum aestivum
- TF(s):
-
Transcription factor(s)
- Y1H:
-
Yeast-1-hybrid
- Y2H:
-
Yeast-2-hybrid
References
Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T (2005) FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309:1052–1056
Alonso R, Oñate-Sánchez L, Weltmeier F, Ehlert A, Diaz I, Dietrich K, Vicente-Carbajosa J, Dröge-Laser W (2009) A pivotal role of the basic leucine zipper transcription factor bZIP53 in the regulation of Arabidopsis seed maturation gene expression based on heterodimerization and protein complex formation. Plant Cell 21:1747–1761
Amalraj A, Luang S, Kumar MY, Sornaraj P, Eini O, Kovalchuk N, Bazanova N, Li Y, Yang N, Eliby S, Langride P, Hrmova M, Lopato S (2016) Change of function of the wheat stress-responsive transcriptional repressor TaRAP2.1L by repressor motif modification. Plant Biotechnol J 14:820–832
Chae MJ, Lee JS, Nam MH, Cho K, Hong JY, Yi SA, Suh SC, Yoon IS (2007) A rice dehydration-inducible SNF1-related protein kinase 2 phosphorylates an abscisic acid responsive element-binding factor and associates with ABA signaling. Plant Mol Biol 63:151–169
Choi H, Hong J, Ha J, Kang J, Kim SY (2000) ABFs, a family of ABA-responsive element binding factors. J Biol Chem 275:1723–1730
Chuang CF, Running MP, Williams RW, Meyerowitz EM (1999) The PERIANTHIA gene encodes a bZIP protein involved in the determination of floral organ number in Arabidopsis thaliana. Genes Dev 13:334–344
Colcombet J, Hirt H (2008) Arabidopsis MAPKs: a complex signalling network involved in multiple biological processes. Biochem J 413:217–226
Curtis MD, Grossniklaus U (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133:462–469
Danquah A, de Zelicourt A, Colcombet J, Hirt H (2014) The role of ABA and MAPK signaling pathways in plant abiotic stress responses. Biotechnol Adv 32:40–52
Das R, Pandey GK (2010) Expressional analysis and role of calcium regulated kinases in abiotic stress signaling. Curr Genomics 11:2–13
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Eini O, Yang N, Pyvovarenko T, Pillman K, Bazanova N, Tikhomirov N, Eliby S, Shirley N, Sivasankar S, Tingey S, Langridge P, Hrmova M, Lopato S (2013) Complex regulation by Apetala2 domain-containing transcription factors revealed through analysis of the stress-responsive TdCor410b promoter from durum wheat. PLoS ONE 8:e58713
Ellenberger TE, Brandl CJ, Struhl K, Harrison SC (1992) The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted alpha helices: crystal structure of the protein-DNA complex. Cell 71:1223–1237
Fletcher SJ (2014) qPCR for quantification of transgene expression and determination of transgene copy number. Methods Mol Biol 1145:213–237
Foster R, Izawa T, Chua NH (1994) Plant bZIP proteins gather at ACGT elements. FASEB J 8:192–200
Fujii H, Zhu JK (2009) Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc Nat Acad Sci USA 106:8380–8385
Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY, Cutler SR, Sheen J, Rodriguez PL, Zhu JK (2009) In vitro reconstitution of an abscisic acid signalling pathway. Nature 462:660–664
Fujita Y, Nakashima K, Yoshida T, Katagiri T, Kidokoro S, Kanamori N, Umezawa T, Fujita M, Maruyama K, Ishiyama K, Kobayashi M, Nakasone S, Yamada K, Ito T, Shinozaki K, Yamaguchi-Shinozaki K (2009) Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol 50:2123–2132
Fujita Y, Fujita M, Shinozaki K, Yamaguchi-Shinozaki K (2011) ABA-mediated transcriptional regulation in response to osmotic stress in plants. J Plant Res 124:509–525
Furihata T, Maruyama K, Fujita Y, Umezawa T, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K (2006) Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc Natl Acad Sci USA 103:1988–1993
Gahlaut V, Jaiswal V, Kumar A, Gupta PK (2016) Transcription factors involved in drought tolerance and their possible role in developing drought tolerant cultivars with emphasis on wheat (Triticum aestivum L.). Theor Appl Genet 129:2019–2042
Gangappa SN, Crocco CD, Johansson H, Datta S, Hettiarachchi C, Holm M, Botto JF (2013) The Arabidopsis B-BOX protein BBX25 interacts with HY5, negatively regulating BBX22 expression to suppress seedling photomorphogenesis. Plant Cell 25:1243–1257
Gonzalez-Guzman M, Pizzio GA, Antoni R, Vera-Sirera F, Merilo E, Bassel GW, Fernández MA, Holdsworth MJ, Perez-Amador MA, Kollist H, Rodriguez PL (2012) Arabidopsis PYR/PYL/RCAR receptors play a major role in quantitative regulation of stomatal aperture and transcriptional response to abscisic acid. Plant Cell 24:2483–2496
Göransson O, Deak M, Wullschleger S, Morrice NA, Prescott AR, Alessi DR (2006) Regulation of the polarity kinases PAR-1/MARK by 14-3-3 interaction and phosphorylation. J Cell Sci 119:4059–4070
Guo W, Ward RW, Thomashow MF (1992) Characterization of a cold-regulated wheat gene related to a Arabidopsis cor47. Plant Physiol 100:915–922
Harris JC, Sornaraj P, Taylor M, Bazanova N, Baumann U, Lovell B, Langridge P, Lopato S, Hrmova M (2016) Molecular interactions of the γ-clade homeodomain-leucine zipper class I transcription factors during the wheat response to water deficit. Plant Mol Biol 90:435–452
Hrmova M, Lopato S (2014) Enhancing abiotic stress tolerance in plants by modulating properties of stress responsive transcription factors. In: Tuberosa R, Graner A, Frison E (eds) Genomics of plant genetic resources, vol 2, Part II: Crop productivity, food security and nutritional quality pp 291–316. Springer Verlag, Netherlands, ISBN 978-94-007-7574-9
Hurst HC (1994) Transcription factors. 1: bZIP proteins. Protein Profile 1:123–168
Ikeda Y, Koizumi N, Kusano T, Sano H (1999) Sucrose and cytokinin modulation of WPK4, a gene encoding a SNF1-related protein kinase from wheat. Plant Physiol 121:813–820
Ikeda Y, Koizumi N, Kusano T, Sano H (2000) Specific binding of a 14-3-3 protein to autophosphorylated WPK4, an SNF1-related wheat protein kinase, and to WPK4-phosphorylated nitrate reductase. J Biol Chem 275:31695–31700
Ismagul A, Iskakova G, Harris JC, Eliby S (2014) Biolistic transformation of wheat with centrophenoxine as a synthetic auxin. Methods Mol Biol 1145:191–202
Izawa T, Foster R, Chua NH (1993) Plant bZIP protein DNA binding specificity. J Mol Biol 230:1131–1144
Jakoby M, Weisshaar B, Dröge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F (2002) bZIP transcription factors in Arabidopsis. Trends Plant Sci 7:106–111
Kang J, Choi H, Im M, Kim SY (2002) Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell 14:343–357
Kang SG, Price J, Lin PC, Hong JC, Jang JC (2010) The Arabidopsis bZIP1 transcription factor is involved in sugar signaling, protein networking, and DNA binding. Mol Plant 3:361–373
Kim S, Kang JY, Cho DI, Park JH, Kim SY (2004) ABF2, an ABRE-binding bZIP factor, is an essential component of glucose signaling and its overexpression affects multiple stress tolerance. Plant J 40:75–87
Kobayashi Y, Murata M, Minami H, Yamamoto S, Kagaya Y, Hobo T, Yamamoto A, Hattori T (2005) Abscisic acid-activated SNRK2 protein kinases function in the gene-regulation pathway of ABA signal transduction by phosphorylating ABA response element-binding factors. Plant J 44:939–949
Kobayashi F, Maeta E, Terashima A, Kawaura K, Ogihara Y, Takumi S (2008a) Development of abiotic stress tolerance via bZIP-type transcription factor LIP19 in common wheat. J Exp Bot 59:891–905
Kobayashi F, Maeta E, Terashima A, Takumi S (2008b) Positive role of a wheat HvABI5 ortholog in abiotic stress response of seedlings. Physiol Plant 134:74–86
Kovalchuk N, Jia W, Eini O, Morran S, Pyvovarenko T, Fletcher S, Bazanova N, Harris J, Beck-Oldach K, Shavrukov Y, Langridge P, Lopato S (2013) Optimization of TaDREB3 gene expression in transgenic barley using cold-inducible promoters. Plant Biotechnol J 11:659–670
Landschulz WH, Johnson PF, McKnight SL (1988) The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science 240:1759–1764
Li ZF, Zhang LX, Yu YW, Quan YD, Zhang ZJ, Zhang HW, Huang RF (2011) The ethylene response factor AtERF11 that is transcriptionally modulated by the bZIP transcription factor HY5 is a crucial repressor for ethylene biosynthesis in Arabidopsis. Plant J 68:88–99
Lopez-Molina L, Chua NH (2000) A null mutation in a bZIP factor confers ABA-insensitivity in Arabidopsis thaliana. Plant Cell Physiol 41:541–547
Lopez-Molina L, Mongrand S, McLachlin DT, Chait BT, Chua NH (2002) ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J 32:317–328
Lozano-Durán R, Robatzek S (2015) 14-3-3 proteins in plant pathogen interactions. Mol Plant Microbe Interact 28:511–518
Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E (2009) Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324:1064–1068
Mao J, Manik SM, Shi S, Chao J, Jin Y, Wang Q, Liu H (2016) Mechanisms and physiological roles of the CBL-CIPK networking system in Arabidopsis thaliana. Genes (Basel) 7:62
Molzan M, Weyand M, Rose R, Ottmann C (2012) Structural insights of the MLF1/14-3-3 interaction. FEBS J 279:563–571
Morran S, Eini O, Pyvovarenko T, Parent B, Singh R, Ismagul A, Eliby S, Shirley N, Langridge P, Lopato S (2011) Improvement of stress tolerance of wheat and barley by modulation of expression of DREB/CBF factors. Plant Biotechnol J 9:230–249
Nozawa A, Sawada Y, Akiyama T, Koizumi N, Sano H (2003) Variable interactions between sucrose non-fermented 1-related protein kinases and regulatory proteins in higher plants. Biosci Biotechnol Biochem 67:2533–2540
Ohnishi N, Himi E, Yamasaki Y, Noda K (2008) Differential expression of three ABA-insensitive five binding protein (AFP)-like genes in wheat. Genes Genet Syst 83:167–177
Pei J, Kim B-H, Grishin NV (2008) PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res 36:2295–2300
Prasad VB, Gupta N, Nandi A, Chattopadhyay S (2012) HY1 genetically interacts with GBF1 and regulates the activity of the Z-box containing promoters in light signaling pathways in Arabidopsis thaliana. Mech Dev 129:298–307
Pyvovarenko T, Lopato S (2011) Isolation of plant transcription factors using a yeast one-hybrid system. Methods Mol Biol 754:45–66
Ruuska SA, Rebetzke GJ, van Herwaarden AF, Richards RA, Fettell NA, Tabe L, Jenkins CLD (2006) Genotypic variation in water-soluble carbohydrate accumulation in wheat. Funct Plant Biol 33:799–809
Saavedra X, Modrego A, Rodríguez D, González-García MP, Sanz L, Nicolás G, Lorenzo O (2010) The nuclear interactor PYL8/RCAR3 of Fagus sylvatica FsPP2C1 is a positive regulator of abscisic acid signaling in seeds and stress. Plant Physiol 152:133–150
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sano H, Youssefian S (1994) Light and nutritional regulation of transcripts encoding a wheat protein kinase homolog is mediated by cytokinins. Proc Natl Acad Sci USA 91:2582–2586
Schweighofer A, Hirt H, Meskiene I (2004) Plant PP2C phosphatases: emerging functions in stress signaling. Trends Plant Sci 9:236–243
Shavrukov Y, Gupta NK, Miyazaki J, Baho MN, Chalmers KJ, Tester M, Langridge P, Collins NC (2010) HvNax3–a locus controlling shoot sodium exclusion derived from wild barley (Hordeum vulgare ssp. spontaneum). Funct Integrat Gen 10:277–291
Shen H, Cao K, Wang X (2007) A conserved proline residue in the leucine zipper region of AtbZIP34 and AtbZIP61 in Arabidopsis thaliana interferes with the formation of homodimer. Biochem Biophys Res Commun 362:425–430
Silveira AB, Gauer L, Tomaz JP, Cardoso PR, Carmello-Guerreiro S, Vincentz M (2007) The Arabidopsis AtbZIP9 protein fused to the VP16 transcriptional activation domain alters leaf and vascular development. Plant Sci 172:1148–1156
Sirichandra C, Davanture M, Turk BE, Zivy M, Valot B, Leung J, Merlot S (2010) The Arabidopsis ABA-activated kinase OST1 phosphorylates the bZIP transcription factor ABF3 and creates a 14-3-3 binding site involved in its turnover. PLoS ONE 5:e13935
Sornaraj P, Luang S, Lopato S, Hrmova M (2016) Basic leucine zipper (bZIP) transcription factors involved in abiotic stresses: a molecular model of a wheat bZIP factor and implications of its structure in function. Biochim Biophys Acta 1860:46–56
Steinhorst L, Mähs A, Ischebeck T, Zhang C, Zhang X, Arendt S, Schültke S, Heilmann I, Kudla J (2015) Vacuolar CBL-CIPK12 Ca(2+)-sensor-kinase complexes are required for polarized pollen tube growth. Curr Biol 25:1475–1482
Sun T, Wang Y, Wang M, Li T, Zhou Y, Wang X, Wei S, He G, Yang G (2015) Identification and comprehensive analyses of the CBL and CIPK gene families in wheat (Triticum aestivum L.). BMC Plant Biol 15:269
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Tang N, Zhang H, Li X, Xiao J, Xiong L (2012) Constitutive activation of transcription factor OsbZIP46 improves drought tolerance in rice. Plant Physiol 158:1755–1768
Taoka K, Ohki I, Tsuji H, Furuita K, Hayashi K, Yanase T, Yamaguchi M, Nakashima C, Purwestri YA, Tamaki S, Ogaki Y, Shimada C, Nakagawa A, Kojima C, Shimamoto K (2011) 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature 476:332–335
Tenea GN, Peres-Bota A, Cordeiro-Raposo F, Maquet A (2011) Reference genes for gene expression studies in wheat flag leaves grown under different farming condition. BMC Res Notes 4:373
Thalor SK, Berberich T, Lee SS, Yang SH, Zhu X, Imai R, Takahashi Y, Kusano T (2012) Deregulation of sucrose-controlled translation of a bZIP-Type transcription factor results in sucrose accumulation in leaves. PLoS ONE 7:e33111
Umezawa T, Nakashima K, Miyakawa T, Kuromori T, Tanokura M, Shinozaki K, Yamaguchi-Shinozaki K (2010) Molecular basis of the core regulatory network in ABA responses: sensing, signaling and transport. Plant Cell Physiol 51:1821–1839
Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K (2000) Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci USA 97:11632–11637
Walsh J, Waters CA, Freeling M (1998) The maize gene liguleless2 encodes a basic leucine zipper protein involved in the establishment of the leaf blade sheath boundary. Genes Dev 12:208–218
Wang Y, Li L, Ye T, Lu Y, Chen X, Wu Y (2013) The inhibitory effect of ABA on floral transition is mediated by ABI5 in Arabidopsis. J Exp Bot 64:675–684
Wang J, Li Q, Mao X, Li A, Jing R (2016) Wheat transcription factor TaAREB3 participates in drought and freezing tolerances in Arabidopsis. Int J Biol Sci 12:257–269
Würtele M, Jelich-Ottmann C, Wittinghofer A, Oecking C (2003) Structural view of a fungal toxin acting on a 14-3-3 regulatory complex. EMBO J 22:987–994
Xu DB, Gao SQ, Ma YZ, Xu ZS, Zhao CP, Tang YM, Li XY, Li LC, Chen YF, Chen M (2014) ABI-like transcription factor gene TaABL1 from wheat improves multiple abiotic stress tolerances in transgenic plants. Funct Integr Genomics 14:717–730
Xue GP (2005) A CELD-fusion method for rapid determination of the DNA-binding sequence specificity of novel plant DNA-binding proteins. Plant J 41:638–649
Xue GP, Bower NI, McIntyre CL, Riding GA, Kazan K, Shorter R (2006) TaNAC69 from the NAC superfamily of transcription factors is up-regulated by abiotic stresses in wheat and recognises two consensus DNA-binding sequences. Funct Plant Biol 33:43–57
Yadav D, Shavrukov Y, Bazanova N, Chirkova L, Borisjuk N, Kovalchuk N, Ismagul A, Parent B, Langridge P, Hrmova M, Lopato S (2015) Constitutive overexpression of the TaNF-YB4 gene in transgenic wheat significantly improves grain yield. J Exp Bot 66:6635–6650
Yamamoto MP, Onodera Y, Touno SM, Takaiwa F (2006) Synergism between RPBF Dof and RISBZ1 bZIP activators in the regulation of rice seed expression genes. Plant Physiol 141:1694–1707
Yang Y, Luang S, Harris J, Riboni M, Li Y, Bazanova N, Hrmova M, Haefele S, Kovalchuk N, Lopato S (2017) Overexpression of the class I homeodomain transcription factor TaHDZipI-5 increases drought and frost tolerance in transgenic wheat. Plant Biotechnol J. https://doi.org/10.1111/pbi.12865
Yoshida T, Fujita Y, Sayama H, Kidokoro S, Maruyama K, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2010) AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J 61:672–685
Yoshida T, Fujita Y, Maruyama K, Mogami J, Todaka D, Shinozaki K, Yamaguchi-Shinozaki K (2015) Four Arabidopsis AREB/ABF transcription factors function predominantly in gene expression downstream of SnRK2 kinases in abscisic acid signalling in response to osmotic stress. Plant Cell Environ 38:35–49
Zhang L, Wang H, Liu D, Liddington R, Fu H (1997) Raf-1 kinase and exoenzyme S interact with 14-3-3zeta through a common site involving lysine 49. J Biol Chem 272:13717–13724
Zhang L, Zhang L, Xia C, Zhao G, Liu J, Jia J, Kong X (2015) A novel wheat bZIP transcription factor, TabZIP60, confers multiple abiotic stress tolerances in transgenic Arabidopsis. Physiol Plant 153:538–554
Zhou L, Lan W, Chen B, Fang W, Luan S (2015) A calcium sensor-regulated protein kinase, CALCINEURIN B-LIKE PROTEIN-INTERACTING PROTEIN KINASE19, is required for pollen tube growth and polarity. Plant Physiol 167:1351–1360
Zhu W, Zhang L, Lv H, Zhang H, Zhang D, Wang X, Chen J (2014) The dehydrin wzy2 promoter from wheat defines its contribution to stress tolerance. Funct Integr Genomics 14:111–125
Zou M, Guan Y, Ren H, Zhang F, Chen F (2008) A bZIP transcription factor, OsABI5, is involved in rice fertility and stress tolerance. Plant Mol Biol 66:675–683
Acknowledgements
The plant transformation contribution of Ainur Ismagul is acknowledged. We also thank Ursula Langridge, Larissa Chirkova and Yagnesh Nagarajan for technical assistance, and Julie Hayes and Carl Simmons for critically reading the manuscript. Pradeep K. Agarwal is grateful to the Council of Scientific and Industrial Research of India (Raman Research Fellowship) and Syed Sarfraz Hussain to DuPont Pioneer for funding their fellowships. This work was supported by the Australian Research Council (LP120100201 to MH and SL), the Australian Grains Research and Development Corporation, the Government of South Australia, and DuPont Pioneer, as part of LP120100201.
Author information
Authors and Affiliations
Contributions
Conceived, designed experiments and analysed data: SLu, PS, MH and SLo. Plant growth and transformation: PS, NK, OE and PKA. Cloning, Y1H, pull-down and transient expression assays, Q-PCR experiments: WJ, NBa and SLo. Protein–protein interactions: SH and MH. 3D molecular modelling: SLu and MH. Discussed the data and contributed to writing: SLu and PS. Writing of the manuscript: SLo and MH.
Corresponding author
Ethics declarations
Conflict of interest
Authors declare that they have no conflict of interest.
Additional information
GeneBank accession numbers
TabZIP2 (KT224373), WPK4 (AB011670), TaABI5L (MF034144), and TaWIN1 (MF034145).
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Luang, S., Sornaraj, P., Bazanova, N. et al. The wheat TabZIP2 transcription factor is activated by the nutrient starvation-responsive SnRK3/CIPK protein kinase. Plant Mol Biol 96, 543–561 (2018). https://doi.org/10.1007/s11103-018-0713-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-018-0713-1