Abstract
Custom-designed nucleases are a promising technology for genome editing through the catalysis of double-strand DNA breaks within target loci and subsequent repair by the host cell, which can result in targeted mutagenesis or gene replacement. Implementing this new technology requires a rapid means to determine the cleavage efficiency of these custom-designed proteins in planta. Here we present such an assay that is based on cleavage-dependent luciferase gene correction as part of a transient dual-luciferase® reporter (Promega) expression system. This assay consists of co-infiltrating Nicotiana benthamiana leaves with two Agrobacterium tumefaciens strains: one contains the target sequence embedded within a luciferase reporter gene and the second strain contains the custom-designed nuclease gene(s). We compared repair following site-specific nuclease digestion through non-homologous DNA end-joining, as opposed to single strand DNA annealing, as a means to restore an out-of-frame luciferase gene cleavage-reporter construct. We show, using luminometer measurements and bioluminescence imaging, that the assay for non-homologous end-joining is sensitive, quantitative, reproducible and rapid in estimating custom-designed nucleases’ cleavage efficiency. We detected cleavage by two out of three transcription activator-like effector nucleases that we custom-designed for targets in the Arabidopsis CRUCIFERIN3 gene, and we compared with the well-established ‘QQR’ zinc-finger nuclease. The assay we report requires only standard equipment and basic plant molecular biology techniques, and it can be carried out within a few days. Different types of custom-designed nucleases can be preliminarily tested in our assay system before their downstream application in plant genome editing.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Background
Our ability to engineer plant genomes is limited by the essentially random integration of introduced DNA molecules into the plant genome as well as by the random occurrence of mutations caused by various mutagenic agents. Custom-designed nucleases can promote targeted double-strand DNA breaks (DSBs) for precise genome editing, either by stimulating targeted gene replacement (Shukla et al. 2009; Townsend et al. 2009; Zhang et al. 2012; Fauser et al. 2012), or targeted mutagenesis [for review, see Tzfira et al. (2012)]. Custom-designed nucleases include zinc-finger nuclease (ZFN) enzymes, engineered-meganucleases, as well as Xanthomonas-derived transcription activator-like effector nuclease (TALEN) enzymes, which are a more-recent alternative that expand the number of genomic target sites available for cleavage (Cermak et al. 2011). Custom-designed nucleases are comprised of target site-specific DNA-recognition domains, consisting of triplet-binding zinc-fingers in the case of ZFNs (Tzfira et al. 2012), and central repeat domains that can bind individual DNA bases with specificity owing to particular repeat-variable diresidues in the case of TALENs (Moscou and Bogdanove 2009). A gene fusion can be made between the sequence encoding these target-recognizing domains, and the sequence encoding a Flavobacterium okeanokoites I (FokI) endonuclease DNA-cleavage domain, in order to form a nuclease monomer. For cleavage of the target DNA sequence to occur, two such monomers are required to bind their respective recognition sites on opposite DNA strands in orientations that can cause their respective nuclease domains to dimerize in the intervening ‘spacer’ region between their respective recognition sites (Zhang et al. 2012; Tzfira et al. 2012). Despite considerable progress in the field of custom-designed nucleases, there is still a need for an effective, rapid, preliminary means to validate candidate nucleases’ cleavage efficiency in planta. This preliminary testing is necessary because of: the multiple possibilities of design; the uncertainties related to the efficiency of nuclease custom design, with respect to DNA-recognition specificity and accuracy of FokI dimerization, and; the considerable time and effort involved in a whole organism gene targeting or mutagenesis experiment.
Several assays have been reported to obtain initial measurements for the efficiency of custom-designed nuclease cleavage in planta. These assays are based on reactivation of a defective reporter gene following DSB repair, but differ with respect to the DSB repair pathway. The first type of assay measures DSB repair via non-homologous end-joining (NHEJ), a process that tends to be error-prone (Gorbunova and Levy 1997; Lloyd et al. 2005; de Pater et al. 2009). For example, an assay reported by Tovkach et al. (2009), was based on activating a premature stop codon-disrupted uidA [β-glucuronidase (GUS)] reporter gene following cleavage with the previously-reported ‘QQR’ ZFN (Kim et al. 1997; Lloyd et al. 2005). With this assay, the target was genomically-embedded in a transgenic plant; the target-specific endonuclease was delivered via Agrobacterium tumefaciens-infiltration of tobacco (Nicotiana tabacum) leaves and; the GUS reading frame was restored through stop-codon removal as a result of error prone NHEJ repair. Variations of this assay were also reported (Tovkach et al. 2009), which included expressing the nuclease through a Virus-Aided Gene Expression system (Marton et al. 2010), or in a tissue specific manner, e.g. using an egg apparatus-specific enhancer sequence (Even-Faitelson et al. 2011). These assays are time-consuming due to their dependence on stably transformed plants, which limits their use towards rapidly screening a large number of nucleases. This time constraint was solved through the development of transient assays (Tovkach et al. 2009; Weinthal et al. 2010; Tzfira et al. 2012). One such assay, described by Mahfouz et al. (2011), involves reactivation of a GUS gene following A. tumefaciens-mediated co-transformation of a TALEN enzyme, and of its target within the defective GUS reporter, into Nicotiana benthamiana leaves. Quantitative assessments of nuclease activity were hindered by the varying transformation frequencies possible in the transient infiltrated-leaf system. Another type of assay measured the efficiency of custom-designed nucleases in tobacco (N. tabacum) protoplasts through restoration of the gene encoding the yellow fluorescent protein (YFP) following DSB induction and subsequent repair via the single-strand DNA annealing (SSA) pathway (Zhang et al. 2012). This assay is not limited by any translational reading frame (TGA, TAA, and TAG) stop codons within the nuclease target site as it is dependent on precise homology-based SSA recombination. In this assay, reporter vectors had a candidate nuclease target site flanked by direct repeats of a 255 bp portion of the YFP-encoding gene’s codon sequence. Twenty-four-hours after co-delivery of the reporter and TALEN expression-cassettes, TALENs’ cleavage efficiency was reported using fluorescence-activated cell sorting to detect the proportion of protoplasts with acquired YFP fluorescence. Some studies have described a means to assay for the DNA-recognition activity (but not cleavage efficiency) of a particular TALE protein based on transient transcriptional activation of a target promoter fused to a GUS reporter gene in planta (Boch et al. 2009; Li et al. 2012); however, the relevance of these assays for determining nuclease cleavage is limited by the absence of surveying FokI dimerization as is required to catalyze the formation of double-strand DNA breaks.
We describe an assay to measure the cleavage efficiency of various candidate custom-designed nucleases using a dual-luciferase® reporter gene system. In addition to the firefly Luciferase gene (LUC), which is activated following DSB repair, a novel feature of this assay was the use of a Renilla luciferase (REN) gene, as part of the dual-luciferase® reporter assay system (Promega), to normalize against the varying transformation frequencies (Hellens et al. 2005; Johnson et al. 2011). We show that this normalization allows for a quantitative cross-comparison between multiple custom-designed nucleases. The high degree of reproducibility of the assay is probably due to the buffering of experimental noise, such as variation in transformation frequencies, physiological state of leaves, Agrobacterium titer, etc. Furthermore, the assay is sensitive, rapid and simple, using standard luminometry equipment, which should facilitate its broad use in the assessment of custom-designed nucleases in genome-editing experiments.
Materials and methods
Reporter vectors
The pGreenII Tqqr-sc::LUC vector was derived from an inverse PCR of the pGreenII 0579-1 (35S::LUC) vector (material and information is available from www.AddGene.org with AddGene ID 44468) that was itself used to show constitutive LUC activity (Johnson et al. 2011). This PCR used LA-TAQ™ polymerase (TAKARA) according to the manufacturer’s instructions, and the oligonucleotide primers, RAJ-461 and 462 (described, along with all other oligonucleotide primers in Online Resource 1), which implemented XbaI and XhoI sites, respectively. These sites were used to clone the Tqqr-sc target site (see Table 1) as a T4 Polynucleotide Kinase-treated (New England BioLabs) primer-dimer of RAJ-493 & RAJ-494. The primers were kinase-treated individually, then annealed by mixing the pair together, heating the reaction to 94 °C for 30 seconds, holding at 65 °C for 20 minutes to denature the kinase, and then cooling to room temperature. The resulting clone was verified by sequencing with the oligonucleotide primers RAJ-182, RAJ-198, RAJ-199, RAJ-216, RAJ-365, RAJ-395, RPH-079 and RPH-248. The same sites of this pGreenII Tqqr-sc::LUC vector were then used to clone subsequent targets (Table 1): Tqqr-fs (RAJ-493 & RAJ-494); Tfs-qqr (RAJ-520 & RAJ-521); Tfs-494 (RAJ-524 & RAJ-525); Tfs-852 (RAJ-526 & RAJ-527; Tfs-1461 (RAJ-528 & RAJ-529); Tfs-494-5′ (RAJ-571 & RAJ-572), and; Tfs-494-3′ (RAJ-569 & RAJ-570). These vectors were partially-sequenced using RAJ-365 and RAJ-417. The pGreenII Tfs-qqr::LUC and pGreenII Tfs-494::LUC vectors (AddGene ID 44466 and 44467, respectively) can be used to clone additional nuclease targets to assay for their cleavage in planta. Gibbs Energy values were calculated for these target sites using the default parameters as part of the ‘Mfold’ web server (Zuker 2003). The pGreenII 0000 (No LUC) vector (AddGene ID 44465) was used to quantify background LUC activity in the Agro-infiltrated leaves (Johnson et al. 2011). These plant transformation vectors were delivered into A. tumefaciens strain LBA4404 cells along with the pSoup 0800 (35S::REN, AddGene ID 44469) vector (Johnson et al. 2011) using electroporation.
Intron-containing derivatives of the pGreenII Tqqr-sc::LUC, pGreenII Tqqr-fs::LUC, and pGreenII 35S::LUC vectors were constructed by insertion of a partial LUC gene sequence as a ScaI fragment that contained an intron of sequence 5′-GTGACTTCTTCTATTCAAGTAAGGTTTTTTAAGTCAAATTGGAGTGGTTTTAAATTGACTTTGGAATACTTCGTGTCATGAGAAATCTCAG-3′ starting from position 669 downstream of the ATG in the LUC gene, which was obtained from the pGreenII 0800 (I3@C) vector, kindly provided by Roger P. Hellens from the New Zealand Institute for Plant and Food Research Limited. The intron containing portion of the latter vector was isolated using a ScaI digestion, and ligated into the same sites pGreenII Tqqr-sc::LUC, pGreenII Tqqr-fs::LUC, and pGreenII 35S::LUC. To make the vectors consistent with their intron-less forms, a HincII NruI fragment was re-inserted from their original vectors.
The pGreenII LU::Tfs-qqr::UC vector was developed from the amplification of 5′Δ-LUC (or ‘UC’) and 3′Δ-LUC (or ‘LU’) gene fragments from pGreenII 35S::LUC using Phusion® DNA polymerase (New England BioLabs), as was used for subsequently-described amplifications. These products were amplified using the oligonucleotide primer pairs RAJ-198 & RAJ-523 and RAJ-522 and RAJ-182, respectively. The products were sequentially cloned as NotI XbaI and XhoI HincII fragments into pGreenII Tfs-qqr::LUC. The pGreenII LU::Tfs-qqr::UC plasmid was sequenced using RAJ-197, RAJ-182, RAJ-387, RAJ-417, RAJ-335, and RAJ-196.
Nuclease expression vectors
The QQR ZFN gene was amplified using RAJ-497 & RAJ-498 and RAJ-499 & RAJ-500 in a nested amplification from pART7 QQR (Even-Faitelson et al. 2011). The QQR ZFN gene was cloned into pDONR 221 using Gateway® recombination technology (Life Technologies, Inc.), sequence-verified, then sub-cloned into the pHEX2 (AddGene ID 44559) destination vector (Hellens et al. 2005; Johnson et al. 2011), again using Gateway® recombination technology. The resulting plasmid vector clone (AddGene ID 44463), along with other pHEX2-derivative vectors that were developed, was introduced into A. tumefaciens strain LBA4404 cells using electroporation.
The pHEX2 GUS vector (AddGene ID 44462) was used as a control (Johnson et al. 2011) against nuclease expression vectors in the transient expression assays for dual-luciferase® activity reported here.
A pair of TALEN heterodimers was designed using TALE-NT (Cermak et al. 2011; Doyle et al. 2012) to bind each of the following sites in the AtCRU3 gene: 5′-GCAAGGACAACAACAAGGCCAACCATGGCAAGGACGACAGGGACAACAAGGCCAACCATGGGAAGG-3′ (T494), 5′-AGAGTAATCTCATACTATATACAAAGACGCACATCCGCTATTAGAAGATCTACC-3′ (T852) and 5′-AGTCATATCCTAAATTATTTAAGAACGTATACAAATGTTTTAAAAATGAATTTGATAGAT-3′ (T1461). These sites respectively began 494, 852 and 1461 bp downstream of the translational reading frame (ATG) start codon and were chosen based on their low nucleosomal occupancies (http://epigenomics.mcdb.ucla.edu/Nuc-Seq/). TALE genes were custom made according to the previously reported ‘Golden Gate’ TALEN synthesis method, the materials for which were kindly provided by Daniel F. Voytas and Colby G. Starker from the University of Minnesota (Cermak et al. 2011; Zhang et al. 2012). The TALE genes were cloned into the AatII and StuI sites of the pTAL3 and pTAL4 yeast expression vectors, with each vector containing one monomer. The T494, T852 and T1461 targets where cloned into the BglII and SpeI sites of pCP5 (lacZ-based cleavage-reporter) using the annealing and T4 Polynucleotide Kinase-treatment (New England BioLabs) of the CRU3-494targerS & CRU3-494targerAS, CRU3-852targerS & CRU3-852targerAS, and CRU3-1461targerS & CRU3-1461targerAS primer pairs, respectively. The activity of these TALENs was checked using the assay for cleavage-based activation of the lacZ reporter in yeast (Cermak et al. 2011) with the exception that β-gal activity was estimated directly on plates containing X-gal (Fig. 5). The TAL effecter domains were transferred to pZHY500/pZHY501 plasmids as AatII StuI fragments, and then sub-cloned into the pCGS636 vector. The latter vector is a derivative of pZHY013 (AddGene ID 36185), a Gateway®-compatible entry vector for TALENs (Zhang et al. 2012), which contains, in addition, a ccdB negative-selection gene and a lacZ expression cassette to assist the cloning of the TAL effecter domains into the upstream and downstream cloning positions, respectively (C.G. Starker, personal communication). The gene encoding the TALE monomer designed to bind the 5′-recognition domain of T494 was cloned into the upstream TALEN expression site of the pCGS636 vector (before cloning the second TALEN monomer downstream). The construct containing the single TALEN recognizing the 5′ part of T494 was used for assessments with Tfs-494-5′, whereas for testing with Tfs-494-3′, the TALE monomer recognizing the 3′ part of T494 was cloned into the upstream TALEN expression site of pCGS636. These TALEN constructs were then cloned into the pHEX2 vector using an LR reaction as part of Gateway® recombination technology (Life Technologies, Inc.) according to the manufacturer’s instructions (the pHEX2 T494 TALENs vector has AddGene ID 44464).
Assays for transient dual-luciferase® activity
Transient expression assays for dual-luciferase® activity were conducted according to a previous study (Johnson et al. 2011). A. tumefaciens strain LBA4404 cells were cultured at 28 °C on Luria–Bertani media (Life Technologies, Inc.) with spectinomycin (100 μg ml−1) selection for strains carrying pHEX2 vectors, and kanamycin-selection (50 μg ml−1) for strains carrying both pGreenII and pSoup vectors. Only kanamycin was used to select for the pGreenII vector in A. tumefaciens strains also containing pSoup vectors as these cells require the presence of the pSoup vector (confers tetracycline-resistance) to provide factors for the replication of the pGreenII vector (Hellens et al. 2000). In order to verify the integrity of all A. tumefaciens strains that were used in the assays we report, plasmid DNA was extracted, re-transformed into E. coli cells, and then the DNA extracted from E. coli cells was analyzed by restriction enzyme digestion. The pGreenII-derivative plasmids were checked using BglII & XhoI digestion, and the pSoup-derivative plasmids were checked using StuI & XhoI digestion. Furthermore, the vectors reporting nuclease cleavage were subjected to DNA sequencing using oligonucleotide primers RAJ-365 and RAJ-417 as a further means of plasmid verification. Generally the pHEX2-derivative plasmids were checked using StuI digestion, although the pHEX2 GUS vector was checked using StuI & XhoI digestion, and the pHEX2 QQR ZFN vector was checked using BamHI & HindIII digestion. The pHEX2 plasmid was unsuited to reside in the Agrobacterium strain housing the LUC and REN reporters due to its origin of replication having the same incompatibility group (pRK2, IncP) as the pSoup plasmid (Hellens et al. 2000, 2005; Gleave 1992). Confluent Agrobacterium cells were grown overnight, then re-suspended in infiltration media (10 mM MgCl2, 1 μM Acetosyringone) to an Optical Density (in light with 600 nm wavelength) of 1.2 ± 0.05 then incubated at room temperature for 2 h without shaking. Reporter- and nuclease-containing vector strains were kept separate until immediately before ‘Agro-infiltration’, where they were mixed together in a 1:1 ratio, before these A. tumefaciens cells were introduced into the three youngest leaves per N. benthamiana plant using sterile needle-less syringes. For each sample infiltration, a minimum of two plants were used, which had been grown to the 8–16 leaf stage in glasshouse conditions at 22 °C. Care was taken to use only plants of a healthy appearance, with a consistent age and size across an experiment. The Agro-infiltrated plants were incubated in the aforementioned glasshouse conditions throughout the three-day time-course of the experiments (Hellens et al. 2005). The infiltrated leaf tissue was then harvested into three small plastic zip-lock bags per sample, with each sub-sample being ground in 500 μl of ‘passive lysis buffer’, with a 1/8 dilution being then used to provide 4 μl of diluted extract for analysis in each sample run. Aggregates of these sub-sample extracts were also used to prepare 6 to 7 replicate runs, with all measurements using a 96-well solid white flat bottom plate. Using the dual-luciferase® reporter assay system (Promega), 40 μl of ‘luciferase assay reagent II’ was added to each sample to measure LUC, then 40 μl of ‘stop and glow’ reagent was added to measure REN, in a Turner Modulus plate reader luminometer, with a 2-second delay and a 10-second measurement. Data of LUC and REN values were collected individually then converted to LUC:REN ratios. The cleavage efficiency of a nuclease was obtained by dividing the average LUC:REN ratio for the reporter strain in the presence of the nuclease(s), by the same reporter’s LUC:REN ratio when in the presence of a negative control gene (GUS) that was not anticipated to affect the LUC:REN ratio. These respective average LUC:REN ratios were checked for a significant statistical difference using a two-tailed Student’s t test (with an assumption unequal variance). The p-values quoted were derived using a mega-analysis of the LUC:REN ratios across two separate experiments. As a means to control against potential day-to-day and plant-to-plant effects (Hellens et al. 2005), only cleavage efficiency values that had p-values of less than 0.01 in 2 replicate experiments are discussed as significant.
Assays for bioluminescent luciferase activity
Leaves that had been Agro-infiltrated using the above procedure were subjected to imaging of their firefly luciferase activity after they were sprayed with 1 mM d-Luciferin, sodium salt (Gold Biotechnology, LUCNA-300). Images were taken with a Berthold LB 985 NightSHADE Camera, using a 45 s exposure time, 5 × 5 binning, high gain, and a slow readout. Quantification of luciferase activity was performed using IndiGO (version 2.0.3.0) software across the uniformly sized regions displayed in Fig. 8 using average luminescence counts per second, cps.
Results
The three vectors associated with the assay that we report here are represented in Fig. 1.
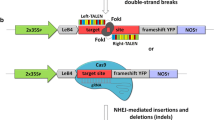
The vectors used in transient assays for testing nuclease cleavage in planta. a Shows a firefly-derived luciferase (LUC)-based reporter for nuclease cleavage-induced non-homologous end-joining (NHEJ). The sequence immediately downstream of the ATG start codon is expanded above the construct to show the presence of a frame-shift mutation (F/shift) that prematurely disrupts LUC, as well as the nuclease target (5′ Recog. site—Spacer—3′ Recog. site). The binding of both recognition domains by nuclease monomers (shown in green with a scissor symbol) can cause cleavage in the spacer region, leading to restoration of the LUC translational reading-frame as part of error-prone repair by NHEJ. b Shows a co-delivered Renilla reniformis-derived luciferase (REN)-based vector for normalizing the firefly LUC activity, and; c a vector for nuclease expression, either one QQR ZFN monomer, or two TALEN monomers. ‘35S’ refers to a constitutively-active cauliflower-mosaic virus promoter sequence and ‘T’ refers to a terminator/polyadenylation sequence
The first component, a constitutively-expressed firefly-derived luciferase (LUC)-based cleavage reporter (Fig. 1a), contains both a disruptive frame-shift mutation to abolish background LUC reporter expression as well as the target site of a candidate nuclease, contained in the pGreenII 0579-1 (35S::LUC) plant transformation vector (Johnson et al. 2011; Hellens et al. 2005). LUC expression can be activated as a result of target-site specific cleavage and subsequent error-prone NHEJ repair, which can restore the LUC translational reading frame in some of the DNA repair events. LUC activity is thus used as an indicator of the relative level of cleavage by the tested nuclease.
The second component, a constitutively-expressed Renilla reniformis-derived luciferase (REN) gene construct (Fig. 1b), was used to normalize the variable transformation frequency in LUC-based measurements. The REN gene construct was delivered using the pSoup 0800 (35S::REN) plant transformation vector (Johnson et al. 2011; Hellens et al. 2005). Infiltrations were carried out using an Agrobacterium strain that contained both the pSoup 0800 vector and a pGreenII-based vector, with both vectors being able to have their T-DNAs delivered separately.
The third component, a nuclease expression construct (Fig. 1c) is in the pHEX2 plant transformation vector (Johnson et al. 2011; Hellens et al. 2005), which was delivered into plant cells using a separate Agrobacterium strain. This vector has Gateway® recombination sites (Life Technologies, Inc.) compatible with the Golden Gate-based TALEN assembly vectors (Cermak et al. 2011) used for polycistronic TALEN expression with their architectures optimized for efficient cleavage in planta (Zhang et al. 2012). We describe the testing of TALENs designed to cleave target sites within the A. thaliana CRUCIFERIN3 gene (AtCRU3), which has been previously described in assays for targeted gene replacement (Even-Faitelson et al. 2011; Shaked et al. 2005). Leaves of N. benthamiana were infiltrated with A. tumefaciens (‘Agro-Infiltrated’) that contained the LUC-based cleavage reporter and the REN gene transformation-reporter, both with and without a nuclease. After three-day incubation in planta, tandem measurements of LUC and REN luminescence were made from crushed leaf-extracts using a luminometer, after sequentially adding the unique substrates of these enzymes.
Assay development
In order to develop and optimize the assay for nuclease cleavage efficiency, we used the established QQR ZFN (Kim et al. 1997; Lloyd et al. 2005) together with reporter vectors that were also delivered into plants without any nuclease to serve as a negative control. A no LUC-containing vector served as an additional negative control, whereas a constitutively-expressed LUC gene served as a positive control. In addition we tested whether a premature stop codon (Tovkach et al. 2009), or a newly-tested frame-shift mutation, would best report cleavage efficiency. These target sites were referred to as Tqqr-sc and Tqqr-fs, respectively, with ‘Tqqr’ meaning the target for the QQR ZFN, ‘sc’ referring to the premature stop codon, and ‘fs’ meaning frame-shift mutation. In these particular reporter vectors, the stop codon and frame-shift mutation were located in the spacer region of the QQR ZFN target site. The sequences of these targets and the other custom-designed nuclease targets tested in the work we report are described in Table 1. The results from testing the above-mentioned vectors are presented in Fig. 2 for two replicate experiments, with further details being described in the “Materials and methods”.
Assays for site-specific cleavage-induced NHEJ in response to QQR ZFN expression. Luciferase activity was measured from the firefly-derived cleavage reporter gene (LUC) after QQR target site-specific cleavage-induced NHEJ by the QQR ZFN in Agro-infiltrated N. benthamiana leaves, which was normalized by the activity of the Renilla transformation reporter gene (REN). Average normalized luciferase (LUC:REN) values were generated in a series of experiments using the following constructs: a an empty vector (No LUC), as well as the QQR ZFN with the empty vector (No LUC + ZFN), served as negative controls; b a cleavage reporter construct that contained a premature stop codon in the LUC ORF and was expressed either without (Tqqr-sc::LUC) or together with the QQR ZFN (Tqqr-sc::LUC + ZFN); c a cleavage reporter construct, with a single base-pair addition to cause a translational reading frame-shift mutation in the LUC ORF, was expressed either without (Tqqr-fs::LUC) or together with the QQR ZFN (Tqqr-fs::LUC + ZFN), and; d an intact, constitutive LUC gene expressed under the cauliflower mosaic virus 35S promoter, either without (35S::LUC) or together with the QQR ZFN (35S::LUC + ZFN), served as positive controls. All data are derived from two separate experiments (open circle, filled circle); in some cases, symbols overlap. There were 6–7 replicas in each experiment. Error bars, corresponding to the standard error, are not always visible due to their small size. Data is provided in Online Resource 3
A low basal activity was obtained with all the negative controls (No LUC, No LUC + ZFN, Tqqr-sc::LUC and Tqqr-fs::LUC) as shown in Fig. 2a–c. On the other hand, in the presence of the QQR ZFN, the reporter with a premature stop codon (Tqqr-sc::LUC) was found to have a significantly increased LUC:REN ratio (Tqqr-sc::LUC + ZFN, Fig. 2b). An even greater increase in the LUC:REN ratio was found for the measurement of a reporter with a single base-pair translational reading frame-shift mutation (Tqqr-fs::LUC) in the presence of the QQR ZFN (Tqqr-fs::LUC + ZFN, Fig. 2c). These data suggest that both means to assay for NHEJ, either based on repair of the premature stop codon or on restoration of the frame-shift, are suitable for estimating nuclease cleavage. A slightly (1.6-fold) higher sensitivity was measured using the frame-shifted reporter, even though it contained a spacer region that was one base-pair longer than the recommended size (Weinthal et al. 2010), so these frame-shifted constructs were explored further in subsequent experiments. We also tested the effect of including an intron in the LUC-based reporter constructs. These constructs where found to be less sensitive than their intron-lacking equivalents, showing 11-fold less normalized LUC activity (Online Resource 2). Intron-lacking constructs were therefore used in subsequent assessments of nuclease cleavage in planta due to their higher sensitivity in reporting cleavage events. One of the important features of the assay for NHEJ was the high reproducibility obtained in two separate experiments, which was supported by the normalization of the transformation frequency that we conducted using the REN reporter.
We further compared two reporters for alternative types of recombination in order to assay for site-specific nuclease cleavage activity by LUC reactivation: the first reporter was also used to test for NHEJ, whereas the second reporter was used to assay for single strand DNA annealing (SSA) repair at overlapping repeats (Fig. 3). The next reporter used to assay for NHEJ (Tfs-qqr::LUC) was a variant of the reporter referred to in Fig. 2 (Tqqr-fs::LUC), with the following difference: a translational reading frame-shift mutation was included upstream of the target cleavage site as shown in Fig. 1, rather than within the spacer region of the target site. The position of ‘fs’ in the name ‘Tfs-qqr’ refers to the frame-shift mutation being located before the QQR ZFN target site. This experiment was sought to determine whether the frame-shift mutation, positioned outside of the nuclease target site, would offer more sensitive cleavage-reporting than if this frame-shift mutation was located in spacer region, enlarging it over the recommended spacer size (Weinthal et al. 2010). The additionally-tested SSA-reporter (LU::Tfs-qqr::UC) was surveyed as a possible means to detect cleavage of nuclease target sites that contained many stop codons. Both the SSA-reporter and the NHEJ reporter included the Tfs-qqr site in order to identify which of these constructs facilitated the highest measurements of cleavage-induced LUC activity. The Tfs-qqr site in the SSA-reporter was flanked by direct repeats of 550 bp LUC gene portions, with the frame-shift mutation not anticipated to hinder LUC reconstitution upon SSA due to the predicted removal of the nuclease target site. The results obtained from testing the vectors reporting SSA and NHEJ, which were made using the QQR ZFN, are presented in Fig. 3.
Comparison of an assay for non-homologous end-joining (NHEJ), versus an assay for single-strand DNA annealing (SSA), in order to detect site-specific cleavage by the QQR ZFN. Average normalized LUC activity (LUC:REN ratio) is shown for various reporter constructs in Agro-infiltrations of N. benthamiana leaves that were delivered either with or without the QQR ZFN expression construct. The following reporter constructs were used: a a negative control (No LUC); b a cleavage reporter construct detecting SSA repair (LU::Tfs-qqr::UC); c a cleavage reporter construct detecting NHEJ repair (Tfs-qqr::LUC), as well as; d a positive control (35S::LUC). All data are derived from two separate experiments (open circle, filled circle); in some cases, the symbols overlap. There were 7 replicas in each experiment. Error bars, corresponding to the standard error, are not always visible due to their small size. Data is provided in Online Resource 4
Normalized LUC activity from the vector reporting NHEJ was significantly induced by 90-fold (p = 9.8 × 10−14) when co-delivered with the QQR ZFN (Tfs-qqr::LUC vs Tfs-qqr::LUC + ZFN as shown in Fig. 3). This induction was found to be 2.5-fold greater when using the reporter with a frame-shift mutation located outside the target site, meaning it had a standard spacer region size, compared with measurements from the reporter with this mutation enlarging the spacer region (Fig. 3c compared with Fig. 2c). Additionally, this induction was fourfold greater than was obtained using the reporter with a premature stop codon (Fig. 3c compared with Fig. 2b). On the other hand, assessments that were made using our reporter to assay for SSA did not detect a significant enhancement in normalized LUC activity (LU::Tfs-qqr::UC vs LU::Tfs-qqr::UC + ZFN, as shown in Fig. 3b). On the basis of these results, subsequent experiments to detect cleavage from custom-designed TALENs were carried out with derivatives of the construct detecting NHEJ, rather than the construct detecting SSA. Subsequent experiments also assayed for restoration of a frame-shift mutation that was located outside of the nuclease target site, meaning that the spacer regions of surveyed TALEN sites were in their native, unmodified form.
Testing custom-designed TALENs
Sites desired for cleavage by TALENs were chosen in the AtCRU3 gene (Even-Faitelson et al. 2011; Shaked et al. 2005), as shown in Fig. 4.
The location of target sites for the custom-designed TALENs in the Arabidopsis thaliana CRUCIFERIN3 gene (AtCRU3). The locations of the TALEN target sites are shown in green within the AtCRU3 gene (AT4G28520). Exons are shown as red arrows and introns are the red lines in between exons. The target site numbers T494, T852 and T1461 refer to the coordinates within the AtCRU3 gene, starting from the translation start site (START)
Three target sites for cleavage within the AtCRU3 gene were chosen and named according to the number of bp downstream of the gene’s (ATG) start codon. Targets 494, 852 and 1461 are subsequently referred to as T494, T852 and T1461 (respectively). These targets were used for the custom-development of three TALEN pairs that were capable of recognizing these targets (see “Materials and methods”). The developed TALENs were then tested for functional activity in yeast (Saccharomyces cerevisiae) based on an established assay system (Townsend et al. 2009; Cermak et al. 2011), where accurate target site cleavage by a TALEN pair was reported as blue-colored colony phenotypes based on lacZ gene reconstitution by SSA. The mating combinations used to test for TALEN cleavage activity are shown in Fig. 5.
Testing the activity of custom-designed TALENs in yeast. (A) The components of a yeast-based assay for TALEN cleavage activity adapted from Cermak, Doyle et al. (2012) included: a parental ‘α’-strain with two TALEN monomer-expressing plasmids, and; a parental ‘a’-strain, which contained a reporter for cleavage-induced SSA. This SSA-reporter vector contained the target for a TALEN pair and a URA3 auxotrophic marker (to select against spontaneous reporter reversion), which were flanked by direct repeats of the lacZ gene’s codon sequence (Cermak et al. 2011). The target site-specific cleavage by a TALEN pair (shown in green) was reported by a LacZ-positive (blue) colony phenotype, as was determined in the mating grid, (B) the haploid parental strains and relevant control matings are also shown in the grid
The observation of blue-colored (LacZ-positive) yeast colony phenotypes for the T494, T852, and T1461 sites with their corresponding TALEN pairs (Fig. 5) shows functional target-specific cleavage by each TALEN heterodimer. No background LacZ activity or off-target TALEN cleavage was observed in this assay. Development was undertaken to create plant vectors that contained the target sites for these TALENs (preceded by a disruptive frame-shift mutation) to report cleavage by LUC activity, in addition to vectors for the polycistronic expression of these TALEN pairs as conducted by Zhang et al. (2012)—see “Materials and methods”. Comparative testing of these custom-designed nucleases’ activity at their target sites was then undertaken in planta, using the assay for NHEJ in Agro-infiltrated N. benthamiana that was described above (Fig. 1). The results for these custom-designed TALENs are shown in Fig. 6, in comparison with the QQR ZFN that served as a positive control reference.
Assaying for the nuclease activity of custom-designed TALENs. Average normalized LUC activity (LUC:REN ratio) assayed from a series of constructs designed to test cleavage by custom-designed TALENs and the QQR ZFN reference, following NHEJ DSB repair in Agro-infiltrated N. benthamiana leaves. The following reporter constructs were used: a a negative control (No LUC); nuclease cleavage reporter constructs embedded with target sites; b Tfs-494, c Tfs-852, d Tfs-1461, and e Tfs-qqr sites, which were delivered either without or together with their respective nuclease. f Shows a positive control (35S::LUC) reporter construct. All data are derived from two separate experiments (open circle, filled circle); in some cases, the symbols overlap. There were 7 replicas in each experiment. Error bars, corresponding to the standard error, are not always visible due to their small size. Data is provided in Online Resource 5
The TALEN pair recognizing T494 was found to induce a significant (p = 6.1 × 10−7) 6.6-fold greater average LUC:REN ratio than its negative control measurement made without the nuclease pair (Fig. 6b). The T852 TALEN pair elicited a lower, but also significant (p = 1.0 × 10−13), 2.2-fold increase in normalized average LUC activity relative to the nuclease-omitting control (Fig. 6c). The TALEN pair recognizing T1461 did not significantly activate LUC signal (Fig. 6d). This observation was either due to the low activity of the nuclease or to the presence of stop codons in its target site, which may have reduced the chances for NHEJ-mediated LUC-reactivation. These findings can be compared with the 78-fold LUC induction from cleavage by the QQR ZFN (Fig. 6e) over its negative control.
As the candidate TALENs were tested as heterodimers, whereas the QQR ZFN was tested as a homodimer (Fig. 6), it was then sought to assay for the cleavage efficiency of TALEN homodimers, assuming that a less efficient TALEN monomer might be a ‘bottleneck’ for heterodimer formation. The TALEN pair recognizing T494 was selected for further assessment due to its greatest cleavage efficiency (Fig. 6b), with the sites that were homodimeric for the T494 site’s 5′ and 3′ recognition domains, named Tfs-494-5′ and Tfs-494-3′, respectively. The results of the heterodimeric and homodimeric TALENs are shown in Fig. 7 in comparison with the homodimeric QQR ZFN.
Assay for cleavage by homodimeric versus heterodimeric TALENs. Average normalized LUC-activity (LUC:REN ratio) from various constructs, designed to test cleavage by homodimeric and heterodimeric T494 TALENs, or the QQR ZFN, in Agro-infiltrated N. benthamiana leaves. The following reporter constructs were used with and without their respective nuclease(s): a a negative control (No LUC); b a reporter for homodimeric cleavage by the TALEN binding the 5′ DNA recognition domain of T494 (Tfs-494-5′::LUC); c a reporter for heterodimeric cleavage by the T494 TALEN (Tfs-494::LUC); d a reporter for homodimeric cleavage by the TALEN binding the 3′ DNA recognition domain of T494 (Tfs-494-3′::LUC), and; e a reporter for cleavage by the QQR ZFN homodimer (Tfs-qqr::LUC). f shows a positive control (35S::LUC) reporter construct. All data are derived from two separate experiments (open circle, filled circle); in some cases, the symbols overlap. There were 7 replicas in each experiment. Error bars, corresponding to the standard error, are not always visible due to their small size. Data is provided in Online Resource 6
We observed that homodimeric TALEN pairs recognizing T494 in inverted repeats of the 5′ and 3′ recognition sites (Tfs-494-5′ and Tfs-494-3′, respectively) did not show statistically significant differences in normalized LUC activity compared to their respective controls, suggesting that they were not effective at catalyzing the cleavage of their DNA target sites (Fig. 7b, d). In contrast, the average LUC:REN ratio for the heterodimeric Tfs-494 site-containing reporter was 6.3-fold greater in the presence of its heterodimeric TALEN pair than without them (Fig. 7c). This measurement for the Tfs-494 site was closely comparable to the previously observed data presented in Fig. 6, as was the measurement for the QQR ZFN (Fig. 7e). These data suggest that TALEN target sequences containing inverted repeats of TALE-recognition sites are not effectively cleaved by homodimerized TALEN monomers.
Luminescence imaging of NHEJ events
The cleavage efficiencies of the T494 TALEN and the QQR ZFN were assayed by imaging firefly-derived luciferase activity in Agro-infiltrated N. benthamiana leaves, using the experimental conditions from the LUC:REN luminometer-based assays shown in Fig. 6. Leaves’ bioluminescence was imaged after they were sprayed with the LUC substrate, d-Luciferin, using a Berthold LB 985 NightSHADE Camera for ultra-low light detection (see “Materials and methods”). The findings are displayed in Fig. 8.
Imaging of luciferase activity in assays for NHEJ using the QQR ZFN and a custom-designed TALEN pair. Representative images of bioluminescence in leaves of N. benthamiana that were Agro-Infiltrated with: (A) A no LUC-containing vector as a negative control; (B) the Tfs-494 site-containing reporter without nucleases; (C) the Tfs-494 reporter with its TALEN pair; (D) the Tfs-qqr::LUC (NHEJ) reporter without nucleases; (E) the Tfs-qqr::LUC (NHEJ) reporter with the QQR ZFN, and; (F) a constitutively expressed LUC gene (35S::LUC) as a positive control. The photograph of the leaf is shown in greyscale, with the measured light intensity (in luminescence counts per second) being mapped using a color-code shown on the right-hand side of the figure
The spotted pattern of LUC activity determined for the T494 TALEN pair (Fig. 8C) and the QQR ZFN (Fig. 8E) was consistent with the notion that LUC-gene restoration relied on leaf cells receiving both the reporter and nuclease-expression constructs, then having cleavage occur, followed by the frame-shift mutation being removed as part of error-prone NHEJ repair. This spotted pattern is most evident for the T494 TALEN pair (Fig. 8C), with LUC activity visualized as discrete ‘spots’ in a leaf despite conducting the infiltration of Agrobacterium strains in a uniform manner across the entire leaf surfaces. This observation suggests that cleavage-induced LUC-restoration was less frequent in response to this TALEN pair than for the QQR ZFN; however, these data, unlike the luminometer-based measurements presented above, do not account for possible variation in the transformation frequency between samples. By quantification of luminescence, using IndiGO software, the findings of total LUC activity were found to broadly correlate with the LUC:REN measurements shown in Fig. 6, despite some saturation in the signal observed for the control measurement of a constitutively expressed LUC gene (Fig. 8F). The average LUC values that were quantified for the strains shown in Fig. 8 are shown plotted in Online Resource 7.
Discussion
We have developed a new assay for estimating the relative cleavage efficiency of custom-designed nucleases. The assay is based on the normalized transient activity of a firefly luciferase reporter in Agro-infiltrated leaves of N. benthamiana leaves following correction of the inactive reporter upon error-prone DSB repair by NHEJ.
In this work, the highest normalized LUC expression was obtained by assaying for NHEJ to correct a frame-shift mutation in the 5′ extremity of the LUC gene, upstream of the nuclease target site (as shown in Figs. 1a, 3c). In earlier reports of NHEJ-based assays for nuclease cleavage, a premature stop codon was embedded within the spacer region (Tovkach et al. 2009; Mahfouz et al. 2011). The slightly higher sensitivity that was measured using the frame-shifted reporter compared with the stop codon-halted reporter (Fig. 2b vs Fig. 2c) may have been due to a greater flexibility of base changes being capable of correcting a frame-shift mutation, compared with removing a stop codon. Our finding that a translational reading frame-shift mutation positioned outside the nuclease recognition site (Figs. 1a, 3c) could reliably report nuclease cleavage efficiency is an improvement upon other NHEJ-based assays as it allows us to survey diverse genomic target sites while keeping a fixed spacer sequence, thus introducing fewer unknown parameters into the system.
The SSA-based assay for nuclease cleavage that we trialed was less sensitive than our alternative assay using NHEJ (Fig. 3), which contrasted with other reports showing that SSA was an efficient means to assay for nuclease cleavage efficiency (Zhang et al. 2012). The reasons for this discrepancy are not clear: factors such as the nature of delivered DNA (linear vs circular), the homology length and the cell type might underlie this difference. In principle, an SSA-based assay for nuclease cleavage efficiency is more precise than an NHEJ-based alternative as every SSA repair event is expected to restore the interrupted gene’s activity; however, it seemed to be impractical or not sensitive enough when our reporter construct was transiently delivered via Agro-infiltration in the assay described here.
The NHEJ-based nuclease cleavage assay we report provides an underestimate of the actual repair efficiency as approximately one quarter of cleavage-mutagenized molecules were expected to correct the single base-pair insertion to the translational reading frame. This proportion was estimated by the incidence of sequences that were recovered from site-specific nuclease mutagenesis with a net reduction of one base-pair to the translational reading frame: 27 % (Lloyd et al. 2005); 21 % (de Pater et al. 2009); 10 % (Shukla et al. 2009), and; 50 % (Zhang et al. 2012) of the total product molecules. Nevertheless, the assay for frame-shift restoration by NHEJ was sensitive enough to measure DSB induction by nucleases.
Another limitation in the use of the NHEJ-based assay is when the recognition site contains stop codons in the three (5′–3′) reading frames as was the case for the T1461 TALEN, which we designed from within an intron sequence (Table 1). While this limitation is not expected for most sites, especially when the recognition site is within an exon, this issue may occur for sites in un-translated regions or introns, therefore, it should be taken into consideration when selecting a recognition site or designing a cleavage reporter. The positioning of additional base-pairs outside of the TALEN target sites was also found to be a way to shift additional (ATG) start codons out-of-frame, such that un-cleaved reporter constructs would not show constitutive LUC expression. This approach was conducted for the Tfs-494-5′ site, where a frame-initiating start codon was shifted by positioning additional base-pairs in 5′ and 3′ positions relative to the nuclease target site. It was found that the problem of stop codons in the T1461 site could, in theory, be mitigated by assaying for its cleavage in a reverse complementary orientation.
One important feature of the assay we describe, which was not conducted in earlier reports (Mahfouz et al. 2011; Tovkach et al. 2009; Zhang et al. 2012), is the normalization of the transformation frequency in each experiment. This feature allowed us to obtain quantitative and reproducible findings for the cleavage efficiency of nucleases, and can be seen from the small standard error between replicas in all LUC assays shown here, as well as from the small variation between experiments. Interestingly, the QQR ZFN that we used as a positive control had a higher nuclease activity than all three custom-designed TALENs that we tested. This finding might be due to the fact that the QQR ZFN had an artificial target optimized for zinc-finger protein binding, causing it to be a ‘gold standard’ for custom-designed nucleases for many years in a broad range of species (Shi and Berg 1995; Desjarlais and Berg 1993; Kim et al. 1997; Lloyd et al. 2005; Marton et al. 2010). Additionally, the homodimeric QQR ZFN targets what is essentially, aside from the spacer sequence, a palindrome formed by two recognition sites as inverted repeats. These palindrome-like targets differ from the targets of the heterodimeric TALENs we surveyed. We hypothesized that the combination of palindrome-like targets with homodimeric nucleases could allow for more efficient cleavage than non-palindromic targets with heterodimeric nucleases. Contrary to this expectation, we found little or no activity for homodimeric TALENs (Fig. 7). The reason for that finding might be that the recognition sites (22-26 bp), in the homodimeric TALEN target sites, formed long stem-loop structures that provided a poor substrate for cleavage. Indeed, we calculated Gibbs Energy values (Zuker 2003) that showed the stem-loop structures possible between the 22–26 bp repeats in the homodimeric TALEN targets were threefold more thermodynamically stable than between the 9 bp repeats in the QQR ZFN target. Another relevant aspect of the ZFN versus TALENs comparison, which supports the greater versatility of TALENs, is that we have not been able to design a valid ZFN to our AtCRU3 gene target (data not shown) while we have succeeded, with no earlier experience, to obtain 2 (out of the 3 we tested) active TALENs (Fig. 6).
In conclusion, the assay described here enabled us to test the cleavage efficiency of a known nuclease (QQR ZFN) as well as of three custom-made TALENs that were targeted to the AtCRU3 gene. This assay requires only basic plant molecular biology research skills (e.g. Agrobacterium culture and N. benthamiana leaf injection) as well as standard equipment (e.g. a luminometer), and can be carried out within a few days. Assuming that the activity of TALENs in a preliminary transient screen correlates well with their activity against genomically-embedded target sites in planta, as shown by Zhang et al. (2012), our results support the view of TALENs as a versatile platform for genome editing. Other target and nuclease combinations can be surveyed in our vectors owing to the unique cloning sites that are compatible with those previously reported for TALEN generation (Zhang et al. 2012; Cermak et al. 2011). This assay can also be adapted to the next generation of site-specific DNA cleavage technologies, such as the Streptococcus pyogenes-derived type II clustered regularly interspaced short palindromic repeats (CRISPR) system, where the cleavage of target DNA can be directed by a guide RNA construct (Cong et al. 2013; Mali et al. 2013).
Abbreviations
- ΔG:
-
Gibbs energy values for DNA secondary structures
- 35S:
-
Cauliflower mosaic virus-derived 35S promoter
- AtCRU3 :
-
Arabidopsis thaliana CRUCIFERIN3 gene
- Cps:
-
Counts per second
- F/shift, fs:
-
A translational reading frame-shift mutation
- Kb:
-
Kilo base pair(s)
- LUC :
-
Gene encoding a luciferase enzyme derived from Photinus pyralis
- NHEJ:
-
Non-homologous end-joining
- QQR:
-
The name given to a previously-reported zinc-finger nuclease
- ORF:
-
Translational open reading frame
- Recog. site:
-
The DNA site recognized by a custom-designed nuclease
- REN :
-
The gene encoding a luciferase enzyme derived from Renilla reniformis
- SSA:
-
Single-strand annealing
- Sc:
-
A premature stop codon
- T:
-
A transcription ‘terminator’ sequence
- T-DNA:
-
A. tumefaciens transfer-DNA
- TALEN:
-
Transcription activator-like effector nuclease
- YFP:
-
Yellow fluorescent protein
- ZFN:
-
Zinc-finger nuclease
References
Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U (2009) Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326(5959):1509–1512. doi:10.1126/science.1178811
Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF (2011) Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res 39(12):e82. doi:10.1093/nar/gkr218
Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339(6121):819–823. doi:10.1126/science.1231143
de Pater S, Neuteboom LW, Pinas JE, Hooykaas PJ, van der Zaal BJ (2009) ZFN-induced mutagenesis and gene-targeting in Arabidopsis through Agrobacterium-mediated floral dip transformation. Plant Biotechnol J 7(8):821–835. doi:10.1111/j.1467-7652.2009.00446.x
Desjarlais JR, Berg JM (1993) Use of a zinc-finger consensus sequence framework and specificity rules to design specific DNA binding proteins. Proc Natl Acad Sci U S A 90(6):2256–2260
Doyle EL, Booher NJ, Standage DS, Voytas DF, Brendel VP, VanDyk JK, Bogdanove AJ (2012) TAL effector-nucleotide targeter (TALE-NT) 2.0: tools for TAL effector design and target prediction. Nucleic Acids Res 40(W1):W117–W122. doi:10.1093/nar/gks608
Even-Faitelson L, Samach A, Melamed-Bessudo C, Avivi-Ragolsky N, Levy AA (2011) Localized egg-cell expression of effector proteins for targeted modification of the Arabidopsis genome. Plant J 68(5):929–937. doi:10.1111/j.1365-313X.2011.04741.x
Fauser F, Roth N, Pacher M, Ilg G, Sanchez-Fernandez R, Biesgen C, Puchta H (2012) In planta gene targeting. Proc Natl Acad Sci. doi:10.1073/pnas.1202191109
Gleave AP (1992) A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol Biol 20:1203–1207
Gorbunova V, Levy AA (1997) Non-homologous DNA end joining in plant cells is associated with deletions and filler DNA insertions. Nucleic Acids Res 25(22):4650–4657. doi:10.1093/nar/25.22.4650
Hellens RP, Anne Edwards E, Leyland NR, Bean S, Mullineaux PM (2000) pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol 42(6):819–832
Hellens R, Allan A, Friel E, Bolitho K, Grafton K, Templeton M, Karunairetnam S, Gleave A, Laing W (2005) Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1(1):13. doi:10.1186/1746-4811-1-13
Johnson RA, Hellens RP, Love DR (2011) A transient assay for recombination demonstrates that Arabidopsis SNM1 and XRCC3 enhance non-homologous recombination. Genet Mol Res 10(3):2104–2132. doi:10.4238/vol10-3gmr1347
Kim Y-G, Shi Y, Berg JM, Chandrasegaran S (1997) Site-specific cleavage of DNA–RNA hybrids by zinc finger/FokI cleavage domain fusions. Gene 203(1):43–49. doi:10.1016/S0378-1119(97)00489-7
Li L, Piatek M, Atef A, Piatek A, Wibowo A, Fang X, Sabir J, Zhu J-K, Mahfouz M (2012) Rapid and highly efficient construction of TALE-based transcriptional regulators and nucleases for genome modification. Plant Mol Biol 78(4):407–416. doi:10.1007/s11103-012-9875-4
Lloyd A, Plaisier CL, Drews GN, Carroll D (2005) Targeted mutagenesis using zinc-finger nucleases in Arabidopsis. Proc Natl Acad Sci USA 102(6):2232–2237
Mahfouz MM, Li L, Shamimuzzaman M, Wibowo A, Fang X, Zhu J-K (2011) De novo-engineered transcription activator-like effector (TALE) hybrid nuclease with novel DNA binding specificity creates double-strand breaks. Proc Natl Acad Sci. doi:10.1073/pnas.1019533108
Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM (2013) RNA-guided human genome engineering via Cas9. Science 339(6121):823–826. doi:10.1126/science.1232033
Marton I, Zuker A, Shklarman E, Zeevi V, Tovkach A, Roffe S, Ovadis M, Tzfira T, Vainstein A (2010) Nontransgenic genome modification in plant cells. Plant Physio 154(3):1079–1087. doi:10.1104/pp.110.164806
Moscou MJ, Bogdanove AJ (2009) A simple cipher governs DNA recognition by TAL effectors. Science 326(5959):1501. doi:10.1126/science.1178817
Shaked H, Melamed-Bessudo C, Levy AA (2005) High-frequency gene targeting in Arabidopsis plants expressing the yeast RAD54 gene. PNAS 102(34):12265–12269
Shi Y, Berg JM (1995) Specific DNA–RNA hybrid binding by zinc finger proteins. Science 268(5208):282–284
Shukla VK, Doyon Y, Miller JC, DeKelver RC, Moehle EA, Worden SE, Mitchell JC, Arnold NL, Gopalan S, Meng X, Choi VM, Rock JM, Wu YY, Katibah GE, Zhifang G, McCaskill D, Simpson MA, Blakeslee B, Greenwalt SA, Butler HJ, Hinkley SJ, Zhang L, Rebar EJ, Gregory PD, Urnov FD (2009) Precise genome modification in the crop species Zea mays using zinc-finger nucleases. Nature 459(7245):437–441. doi:10.1038/nature07992
Tovkach A, Zeevi V, Tzfira T (2009) A toolbox and procedural notes for characterizing novel zinc finger nucleases for genome editing in plant cells. Plant J 57(4):747–757. doi:10.1111/j.1365-313X.2008.03718.x
Townsend JA, Wright DA, Winfrey RJ, Fu F, Maeder ML, Joung JK, Voytas DF (2009) High-frequency modification of plant genes using engineered zinc-finger nucleases. Nature 459(7245):442–445. doi:10.1038/nature07845
Tzfira T, Weinthal D, Marton I, Zeevi V, Zuker A, Vainstein A (2012) Genome modifications in plant cells by custom-made restriction enzymes. Plant Biotechnol J 10(4):373–389. doi:10.1111/j.1467-7652.2011.00672.x
Weinthal D, Tovkach A, Zeevi V, Tzfira T (2010) Genome editing in plant cells by zinc finger nucleases. Trends Plant Sci 15(6):308–321
Zhang Y, Zhang F, Li X, Baller JA, Qi Y, Starker CG, Bogdanove AJ, Voytas DF (2012) TALENs enable efficient plant genome engineering. Plant Physiol. doi:10.1104/pp.112.205179
Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31(13):3406–3415. doi:10.1093/nar/gkg595
Acknowledgments
We thank Avishai Mor from the laboratory of Robert Fluhr in the Department of Plant Sciences at the Weizmann Institute of Science for his help with imaging bioluminescent luciferase activity. We also thank all members of Avraham A. Levy’s laboratory in the Department of Plant Sciences at the Weizmann Institute of Science, especially Assaf Gavish for his help infiltrating N. benthamiana plants with Agrobacterium, followed by assaying for LUC and REN activities. This work was funded by an EU-FP7 TRACTAR grant from the European Research Council.
Conflict of Interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
11103_2013_52_MOESM1_ESM.pdf
Online Resource 1 The sequences and applications of the oligonucleotide primer sequences that were used in the study. Supplementary material 1 (PDF 36 kb)
11103_2013_52_MOESM2_ESM.pdf
Online Resource 2 Assays for site-specific cleavage-induced NHEJ in response to QQR ZFN expression, which complements Fig. 2. Supplementary material 2 (PDF 136 kb)
11103_2013_52_MOESM3_ESM.pdf
Online Resource 3 The luciferase data from assays for NHEJ with the QQR ZFN, which complements Fig. 2 and Online Resource 2.Supplementary material 3 (PDF 22 kb)
11103_2013_52_MOESM4_ESM.pdf
Online Resource 4 The luciferase data from testing reporter signal levels arising from SSA, versus NHEJ, for the QQR ZFN, which complements Fig. 3.Supplementary material 4 (PDF 94 kb)
11103_2013_52_MOESM5_ESM.pdf
Online Resource 5 The luciferase data from comparing the cleavage efficiency of TALENs recognizing the AtCRU3 gene at T494, T852 and T1461 sites, which complements Fig. 6.Supplementary material 5 (PDF 28 kb)
11103_2013_52_MOESM6_ESM.pdf
Online Resource 6 The luciferase data from comparing two TALEN homodimers and their heterodimer that was custom-designed for T494 within the AtCRU3 gene, which complements Fig. 7.Supplementary material 6 (PDF 34 kb)
11103_2013_52_MOESM7_ESM.pdf
Online Resource 7 Quantification of firefly-derived luciferase activity in imaged leaves, which complements Fig. 8.Supplementary material 7 (PDF 141 kb)
Rights and permissions
About this article
Cite this article
Johnson, R.A., Gurevich, V. & Levy, A.A. A rapid assay to quantify the cleavage efficiency of custom-designed nucleases in planta . Plant Mol Biol 82, 207–221 (2013). https://doi.org/10.1007/s11103-013-0052-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-013-0052-1