Abstract
System identification has been applied in diverse areas over past decades. In particular, parametric modelling approaches such as linear and nonlinear autoregressive with exogenous inputs models have been extensively used due to the transparency of the model structure. Model structure detection aims to identify parsimonious models by ranking a set of candidate model terms using some dependency metrics, which evaluate how the inclusion of an individual candidate model term affects the prediction of the desired output signal. The commonly used dependency metrics such as correlation function and mutual information may not work well in some cases, and therefore, there are always uncertainties in model parameter estimates. Thus, there is a need to introduce a new model structure detection scheme to deal with uncertainties in parameter estimation. In this work, a distance correlation metric is implemented and incorporated with a bagging method. The combination of these two implementations enhances the performance of existing forward selection approaches in that it provides the interpretability of nonlinear dependency and an insightful uncertainty analysis for model parameter estimates. The new scheme is referred as bagging forward orthogonal regression using distance correlation (BFOR-dCor) algorithm. A comparison of the performance of the new BFOR-dCor algorithm with benchmark algorithms using metrics like error reduction ratio, mutual information, or the Reversible Jump Markov Chain Monte Carlo method has been carried out in dealing with several numerical case studies. For ease of analysis, the discussion is restricted to polynomial models that can be expressed in a linear-in-the-parameters form.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
System identification is a challenging and interesting engineering problem that has been extensively studied for decades. It consists in identifying a mathematical model that describes the behaviour of a system based on recorded input–output data [1]. In general, most of the real-life systems of interest are nonlinear [2]. Extensive research has been developed in the nonlinear realm for system identification since 1980s [1, 3, 4]. In particular, one of the most popular approaches is the Nonlinear AutoRegressive with eXogenous inputs (NARX) methodology, which has proved to be a well-suited scheme for nonlinear system identification problems [1, 5]. Such approach ranks a set of candidate terms based on their contribution to the output data and identifies parsimonious models that generalise well on new data. The commonly used criterion to measure the dependency between candidate model terms and the desired output is linear correlation; however, it can only identify linear dependency. Therefore, new metrics have been implemented recently to identify nonlinear dependencies. Some of these new metrics are entropy [6] and mutual information [7–10]. In particular, mutual information has been extensively used because it captures both linear and nonlinear correlations and has no assumption on the distribution of the data [11]. Although most of the research is promising, the mutual information is hard to interpret because its maximum value is not fixed and depends on the entropy of the variables involved.
Another important issue is the need to extend the deterministic notion of the NARX model to accommodate uncertainties in the parameter estimates, as well as the identified model and the computed predictions. Some authors have worked towards the incorporation of the Bayesian approach within the NARX methodology. An interesting example is the work by Baldacchino et al [12] which developed a computational Bayesian framework for Nonlinear AutoRegressive Moving Average with eXogenous inputs (NARMAX) models using the Reversible Jump Markov Chain Monte Carlo (RJMCMC) procedure, an iterative sampling technique for performing inference in the context of model selection [13]. In [12], Bayesian inference is a key element to estimate not only the parameters but also the model. The results obtained are interesting; however, the main drawback is that there are many assumptions in the probability distributions of the parameters involved, and the likelihood and prior distributions are selected carefully to be conjugate priors, an assumption that may not always be accurate.
In this work, we address both the use of a novel metric to detect nonlinearities within the data set, and the extension of the deterministic notion of the NARX model. For the first case, the distance correlation metric is implemented, which is a measure that belongs to a new class of functions of distances between statistical observations and is able to detect all types of nonlinear or non-monotone dependencies between random vectors with finite first moment, but not necessarily with equal dimension [14, 15]. This is the first time that the distance correlation is introduced and implemented to the well-known orthogonal forward regression [16]. For the second case, the bagging method is used. Bagging consists of running an algorithm several times on different bootstrap realisations, and the results obtained are combined to predict a numerical value via averaging (for regression problems) or via voting (for classification problems). The combination of these two implementations enhances the performance of a NARX model and provides interpretability of nonlinear dependencies together with an insightful uncertainty analysis. For simplicity, the discussion is restricted to polynomial models that can be expressed in a linear-in-the-parameters form.
This work is organised as follows. In Sect. 2, a brief summary of nonlinear system identification, that includes the Orthogonal Forward Regression algorithm, is discussed. Section 3 reviews the bootstrap and bagging method. In Sect. 4, the distance correlation metric is described. Our new Bagging Forward Orthogonal Regression using distance Correlation (BFOR-dCor) algorithm is proposed in Sect. 5. Three case studies that show the effectiveness of the new algorithm are presented in Sect. 6. The work is concluded in Sect. 7.
2 Nonlinear system identification
System identification is an experimental approach that aims to identify and fit a mathematical model of a system based on experimental data that record the system inputs and outputs behaviour [1, 17]. Linear system identification has been extensively used in past years; however, its applicability is limited since the linearity assumption is strict, and in real life most of the systems of interest are nonlinear [2]. One of the most popular approaches used to deal with nonlinear systems is the Nonlinear AutoRegressive with eXogenous inputs (NARX) methodology, which has been extensively used in different case studies and interesting results have been obtained [1, 5, 18–21].
In general, system identification consists of three steps [17, 22]:
-
1.
Model structure detection
-
2.
Parameter estimation
-
3.
Model validation
Model structure detection has been extensively studied, and there is considerable amount of information in the literature [3]. It consists of determining the model order and selecting model terms that contribute to explaining the variation of the system output [1]. In general, most of the candidate model terms in an initially predetermined model are redundant or spurious; therefore, their contribution to the system output is negligible [23]. Furthermore, a model that includes a large number of terms tends to generalise poorly on unseen data [24]. Because of this, different methods have been developed to search and select the significant model terms that play a major role in the identification process. Some of these methods include clustering [24, 25], the Least Absolute Shrinkage and Selection Operator (LASSO) [26, 27], elastic nets [28, 29], genetic programming [30, 31], the Orthogonal Forward Regression (OFR) using the Error Reduction Ratio (ERR) approach [19], and a recently developed multiobjective extension known as the Multiobjective ERR (MERR) [32]. Once the structure has been identified, the parameter of each model term needs to be estimated for testing the term’s significance [33, 34]. Finally, a fundamental part of system identification is model validation. It consists in testing the identified model to check whether the parameter estimates are biased and if the final model is an adequate representation of the recorded data set [1, 22]. For the latter, Billings and Voon [35] developed a set of statistical correlation tests that can be used for nonlinear input–output model testing and validation. In summary, system identification has to consider a trade-off between model parsimony, accuracy, and validity [36].
2.1 Orthogonal forward regression algorithm
The NARX model is a nonlinear recursive difference equation with the following general form:
where \(f\left( \cdot \right) \) represents an unknown nonlinear mapping, \(y\left( k\right) \), \(u\left( k\right) \), and \(\xi \left( k\right) \) are the output, input, and prediction error sequences with \(k=1,2,\ldots ,N\), where \(N\) is the total number of observations, and the maximum lags for the output and input sequences are \(n_{y}\) and \(n_{u}\) [9]. For simplicity, in this work we assume that the function \(f\left( \cdot \right) \) is a polynomial model of nonlinear degree \(\ell \).
One of the most popular algorithms to work with the NARX identification approach is the Orthogonal Forward Regression (OFR) algorithm, which is also known as the Forward Orthogonal Regression (FOR) algorithm [1, 37]. This was developed in the late 1980s by Billings et al. [1]. It is a greedy algorithm [38] that belongs to the class of recursive-partitioning procedures [39]. It identifies and fits a deterministic parsimonious NARX model that can be expressed in a generalised linear regression form [4, 9]. The original OFR algorithm used the Error Reduction Ratio (ERR) index as dependency metric [1]. The ERR of a term represents the percentage reduction in the total mean square error that is obtained if such term is included in the final model [6], and it is defined as the non-centralised squared correlation coefficient \(C\left( \mathbf {x},\mathbf {y}\right) \) between two associated vectors \(\mathbf {x}\), and \(\mathbf {y}\) [8]
The non-centralised squared correlation only detects linear dependencies; therefore, new metrics have been implemented recently to identify nonlinear dependencies [6, 8, 9]. Some of these new metrics are entropy [6] and mutual information [7–10]. In particular, mutual information \(I\left( \mathbf {x},\mathbf {y}\right) \) provides a measure of the amount of information that two variables share with each other [8]. It is defined as
Although most of the research is promising, the mutual information is hard to interpret. Furthermore, the conventional OFR method may incorrectly select some spurious model terms due to the effect of noise, and there is still a need to extend the deterministic notion of the NARX methodology to deal with uncertainties in the parameter estimates, the identified model and the computed predictions.
In the remaining of this work, we refer to the original OFR algorithm as OFR-ERR (Orthogonal Forward Regression using Error Reduction Ratio) [1], and if the mutual information is used as dependency metric, then it is referred as FOR-MI (Forward Orthogonal Regression using Mutual Information) [8]. These will be used later in Sect. 6 for comparison with our new developed algorithm.
3 The bootstrap and bagging methods
The bootstrap method was developed by Bradley Efron [40]. It is a computer-based method that computes measures of accuracy to statistical estimates. Bootstrapping consists of randomly sampling \(R\) times, with replacement, from a given data set where it is assumed that the observations are independent of each other. Each of the resamples is called a bootstrap realisation and has the same length as the original data set. The bootstrap realisations can be treated as unique data sets that produce their own results when used in a specific algorithm, method, or technique. Such results contain information that can be used to make inferences from the original data set [41, 42].
The bootstrap method has been previously used for system identification of NARX models. In [43, 44], bootstrapping was used for structure detection where a backward elimination scheme was implemented to find the significant model terms. Such methodology is computationally expensive, as the bootstrap method must be applied every time a model term is eliminated. Furthermore, the methodology may not work when the lag order of the system is large. In [45], the bootstrap was used for parameter estimation of a fixed model. Although the parameter estimation is improved, by fixing the model there is no guarantee that the bootstrapped data come from the true model. The main drawback of these previous works is that the model structure needs to be correct for bootstrap to work [45].
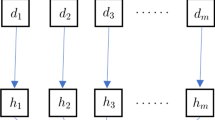
In this work, the bootstrap method is applied in a different way based on [41]. Considering that observations at a given time may depend on previously measured observations, the data set is split into overlapping blocks of fixed length \(B\). The first and last observations appear in fewer blocks than the rest; therefore, the data set is wrapped around a circle to make all data points participate equally [42]. Then the blocks are sampled with replacement until a new data set is created with the same length as the original one. This methodology is known as moving blocks bootstrap for time series [41], and it is illustrated in Fig. 1. By sampling the blocks, the correlation present in observations less than \(B\) units apart is preserved. This methodology is less “model dependent” than the bootstrapping of the residuals approach [41]. It is important to notice that the choice of \(B\) is quite important. If it is too small, the correlation within the observations may be lost. If it is too big, there would be no distinction between the original data set and the bootstrap realisations. Effective methods for choosing \(B\) are still been investigated. In the remaining of this work, we assume that \(B\) is known beforehand.
The bootstrap technique can be extended to a very popular approach nowadays. Assume that a total of \(R\) bootstrap realisations have been carried out and each of them has been used in a specific algorithm to duplicate a result of its own. Therefore, \(R\) outputs are generated and all of them can be used to predict a numerical value via averaging (for regression problems) or via voting (for classification problems). This procedure is known as bagging (that stands for bootstrap aggregating) and was proposed by Leo Breiman [46].
4 The distance correlation
The distance correlation was recently developed by Székely, et al. [14]. It is a measure that belongs to a new class of functions of distances between statistical observations [15]. Distance correlation, denoted as \(dCor\left( \mathbf {x},\mathbf {y}\right) \), provides a new approach to measure all types of nonlinear or non-monotone dependencies between two random vectors with finite first moment, but not necessarily with equal dimension.
The distance correlation has the following properties [14, 15]:
-
(i)
\(0\le dCor\left( \mathbf {x},\mathbf {y}\right) \le 1\)
-
(ii)
If \(dCor\left( \mathbf {x},\mathbf {y}\right) =1\), then the dimensions of the linear subspaces spanned by \(\mathbf {x}\) and \(\mathbf {y}\) are almost surely equal. Therefore, there exists a vector \(\mathbf {a}\), a nonzero real number \(b\) and an orthogonal matrix \(\mathbf {C}\) such that \(\mathbf {y}=\mathbf {a}+b\mathbf {C}\mathbf {x}\).
The distance correlation is analogous to Pearson product-moment correlation coefficient \(\rho \). However, Pearson’s coefficient only characterises linear dependency between two variables while distance correlation is a more general measure that characterises independence of random variables [15]. The procedure to compute this metric is shown in [14, 15].
As a simple comparison, Fig. 2 displays three distinct noisy data sets. These have been created using a linear (\(y=x\)), sinusoidal (\(y=\sin \left( x+\frac{\pi }{2}\right) \)), and circular (\(x^{2}+y^{2}=1\)) relationship with additive white noise. Each of the figures shows the respective values for the Pearson product-moment correlation coefficient, mutual information, and distance correlation. The Pearson coefficient is able to detect a linear dependency in the first data set, but finds no such dependency in the other cases, as expected. The mutual information provides a better insight in each of the data sets, but its value is difficult to interpret because the maximum value of the mutual information is not fixed and depends on the entropy of each of the variables involved. Finally, the distance correlation is able to detect dependencies in all cases. Also, the distance correlation is not as strict as the Pearson coefficient, and the fixed range between 0 and 1 for possible values of the distance correlation is an important characteristic that plays a key role in our new algorithm when determining significant terms. It is important to mention that one drawback of the distance correlation metric is its computation time, since it can take three times longer to compute it compared with the Pearson coefficient or the mutual information.
5 The new BFOR-dCor algorithm
The bagging method and distance correlation are combined with the OFR algorithm to produce the Bagging Forward Orthogonal Regression using distance Correlation (BFOR-dCor) algorithm. This is the first time that the distance correlation metric is introduced and incorporated to the well-known orthogonal forward regression [16]. This algorithm is divided into two parts. In Algorithm 1, the Forward Orthogonal Regression algorithm using the distance correlation dependency metric is described. It is important to mention that in contrast to the original algorithm developed by Billings et al. [1], that requires a threshold in the Error-to-Signal Ratio (ESR), the user needs to specify the maximum number of terms \(n_{max}\) that the algorithm will look for [23]. In this algorithm, lines 1–4 search for the candidate term that has the most significant influence on the system output based on the distance correlation metric. Once found, lines 5–8 create an orthogonal projection of \(\mathbf {y}\) with respect to \(\mathbf {q}_{1}\) using the modified Gram–Schmidt process. This orthogonalisation sequence is repeated in lines 11–25 until the maximum number of models \(n_{max}\) specified by the user is achieved. To avoid redundant candidate terms, we introduced lines 14–16, which check the squared norm-2 of a candidate term, and if it is less than \(10^{-10}\), it is simply removed. Following [23], we introduced the concept of Leave-One-Out Cross Validation (LOOCV) in order to prevent under- and over-fitting. Every time a new model term is added, the LOOCV statistic is computed with its standard error (SE) using the following equations:
where \(e_{i}\) is the residual obtained from fitting the model to all \(N\) observations using the selected candidate terms at each iteration \(s\), and \(h_{i}\) are the diagonal values of the influence matrix for the fitted model [47]. Once the maximum number of terms \(n_{max}\) is achieved, the most parsimonious model with \(n\le n_{max}\) terms is selected in line 26 using the 1 SE rule [48], i.e. select the simplest model for which the LOOCV is within 1 SE from the minimum LOOCV. Finally, the parameters \(\varvec{\theta }\) are computed in line 27, and the algorithm returns them together with the significant terms selected. The parameter \(n_{max}\) can be selected heuristically, by running Algorithm 1 a couple of times and inspecting the resulting LOOCV curve.
Algorithm 2 describes the new BFOR-dCor algorithm. Here, Algorithm 1 is repeated \(R\) times, each with a different bootstrap realisation taken from the original input and output signals. Every time a bootstrap realisation is used, the identified model is recorded in a table. After all the \(R\) bootstrap realisations are taken, the table is summarised to identify the different models that were found, and each of them is assigned a value that is equal to the number of times it was selected within the \(R\) bootstrap realisations.
The BFOR-dCor algorithm is a new method that has been applied for the first time to nonlinear model selection. The proposed algorithm outperforms the conventional OFR algorithm in that the new method aims to find correct model terms within noisy data by introducing a voting mechanism in the algorithm.


6 Case studies
In this section, several examples are provided to illustrate the effectiveness of the BFOR-dCor algorithm. First, a comparison of the new method with both the traditional OFR-ERR and the recent FOR-MI algorithms is performed. Second, the BFOR-dCor technique is applied to a testing model in [12] where the RJMCMC algorithm is applied. Finally, the BFOR-dCor algorithm is applied to the sunspot data provided by the Solar Influences Data Center (SIDC), RWC Belgium, World Data Center for the Sunspot Index, Royal Observatory of Belgium [49]. The data consist of annual number of sunspots from 1700 to 2013.
6.1 Comparison with OFR-ERR and FOR-MI
The following model was taken from [9]:
where the input \(u\left( t\right) \sim \mathcal {U}\left( -1,1\right) \), that is \(u\left( t\right) \) is evenly distributed over \(\left[ -1,1\right] \), and the error \(e\left( t\right) \sim \mathcal {N}\left( 0,0.02^{2}\right) \). Following [9], the maximum lags for the input and output are chosen to be \(n_{u}=n_{y}=4\) and the nonlinear degree is \(\ell =3\). The stop criterion for the OFR-ERR and FOR-MI algorithms is when the ESR is less than 0.05. A total of 500 input–output data points were generated, and the same random seed is used to ensure a fair comparison. The results for the OFR-ERR algorithm are shown in Table 1 and Fig. 3. It can be seen that all the model terms selected are correct except for the first one. Likewise, the results for the FOR-MI algorithm are displayed in Table 2 and Fig. 4. The four model terms selected are correct, still the algorithm failed to find one of the five terms required. From Tables 1 and 2, both models failed to select all the true model terms in (6). It is interesting to notice that, except by the spurious term found by the OFR-ERR algorithm, the union set of the model terms found by the OFR-ERR and FOR-MI algorithms is equivalent to the true model terms set. As explained in [9], both the OFR-ERR and FOR-MI algorithms can be used at the same time to select the model terms based on the t-tests; however, this example shows that the selection is still hard to perform as all the terms selected by both methods are statistically significant.
Model terms selected for (6) by the OFR-ERR algorithm with their corresponding ERR and the updated sum of ERR (SERR)
Model terms selected for (6) by the FOR-MI algorithm with the updated ESR
The BFOR-dCor algorithm is applied to model (6) using a total of \(R=1000\) bootstrap realisations and a block length \(B=5\). The maximum number of terms to look for is \(n_{max}=10\). On Table 3, the 3 top model structures obtained by the BFOR-dCor algorithm are shown. These 3 model structures correspond to 96.5 % of the bootstrap realisations. The most-voted model structure has a structure that coincides with the true model (6), something that was not obtained with the OFR-ERR and FOR-MI algorithms.
For the 924 realisations that have the most-voted model structure, Fig. 5 shows the beanplots [50] for each of the parameter estimates, which clearly suggest that each parameter bootstrap distribution is not Gaussian. Furthermore, Table 4 shows a statistical summary of the parameter estimates. It is interesting to notice that all but one of the true values are within 2 standard deviation (SD) from the mean. The exception is the \(y^{3}\left( t-1\right) \) term. A frequency analysis may reveal an insightful understanding of the contribution of this term.
Beanplots for the parameter estimates of the model terms identified in the most-voted model structure using the BFOR-dCor algorithm for identification of (6), where the red dotted line represents the parameter true value while the black solid line represents the parameter mean estimated value. (Colour figure online)
The results presented here show that the BFOR-dCor algorithm is able to identify 924 realisations with the true model structure together with a bootstrap distribution of the parameter estimates. Furthermore, having different equal-structure models is beneficial for the forecasting task since all the models or a sample from them can be used to compute an average prediction with the corresponding SD.
6.2 Comparison with RJMCMC algorithm
The following model was taken from [12]:
In [12], the authors developed a computational Bayesian identification framework for NARMAX models that uses the RJMCMC algorithm to perform structure detection and parameter estimation together with a characterisation of the probability distribution over models. The algorithm is stochastic in nature, which encourages a global search over the model term space while at the same time ensuring that the identified model is parsimonious [12, 13]. In their work, the algorithm is executed 10 times on the same input–output data. From the 10 runs, the algorithm is able to get the true model structure 7 times. The main drawbacks of this method are that it is computationally expensive, and it needs to define different probability distributions for the parameters involved. Most of these distributions are chosen to be conjugate prior to ease the computations, but of course this does not mean that such distributions are faithful to the real unknown distributions.
The BFOR-dCor algorithm requires no assumptions about probability distributions, and it can work extremely well once the basic parameters are defined. Here again the maximum lags for the input and output are \(n_{u}=n_{y}=4\) and the nonlinear degree is \(\ell =3\), exactly the same values as in [12]. A total of 500 input–output data points were generated. The BFOR-dCor algorithm is applied to (7) using a total of \(R=1000\) bootstrap realisations, a block length \(B=5\), and the maximum number of terms is \(n_{max}=10\). On Table 5, the 3 top model structures obtained by the BFOR-dCor algorithm are shown. These 3 model structures correspond to 88.1 % of the bootstrap realisations. The most-voted model structure has a structure that coincides with the true model (7).
Figure 6 shows the beanplots for each of the parameter estimates, which suggest that each parameter may be treated as a Gaussian random variable. Likewise, Table 6 shows a statistical summary of the parameter estimates. It is interesting to notice that all the true values are within 2 SD from the mean.
Beanplots for the parameter estimates of the model terms identified in the most-voted model structure using the BFOR-dCor algorithm for identification of (7), where the red dotted line represents the parameter true value while the black solid line represents the parameter mean estimated value. (Colour figure online)
These results show that the BFOR-dCor algorithm is extremely efficient and works well without the need of assumptions of probability distributions.
6.3 Forecasting the annual sunspot number
The sunspot time series provided by the Solar Influences Data Center (SIDC), RWC Belgium, World Data Center for the Sunspot Index, Royal Observatory of Belgium [49] consists of 314 observations of the annual number of sunspots from 1700 to 2013. The data from 1700 to 1950 are used for structure detection and parameter estimation while the data from 1951 to 2013 is used for model performance testing and validation. It is assumed that the annual number of sunspots depends only on previous annual observations, i.e. \(n_{u}=0\). Furthermore, it is well known that the sun’s north and south poles reverse around every 11 years which corresponds to a period of great solar activity known as the solar max [51]. Therefore, we choose \(n_{y}=12\), and employ a Nonlinear AutoRegressive (NAR) model with nonlinear degree \(\ell =3\) to test the performance of the proposed BFOR-dCor algorithm.
The BFOR-dCor algorithm is applied using a total of \(R=1000\) bootstrap realisations, a block length \(B=15\), and the maximum number of terms is \(n_{max}=15\). The five top model structures obtained by the BFOR-dCor algorithm are shown in Table 7, which correspond to 7.2 % of the bootstrap realisations.
For the 30 realisations that have the most-voted model structure, Fig. 7 shows the beanplots for each of the parameter estimates, which clearly suggest that most of the bootstrap parameter distributions are not Gaussian. Furthermore, Table 8 shows a statistical summary of the parameter estimates. Figures 8 and 9 show the one-step ahead and model predicted outputs together with the 2 SD region, respectively. In both cases, from these two graphs we can see that a simple NAR model has successfully captured the general trend of the sunspots behaviour. The root-mean-square error (RMSE) for the one-step ahead predicted output is 19.39716, while the RMSE for the model predicted output is 28.77858.
One-step ahead predicted output for the sunspot time series using the most-voted model structure identified by the BFOR-dCor algorithm, where the black solid line with circles indicates the true measurements, the empty blue circles represent the one-step ahead predicted output, and the blue shadow represents the 2 SD region. (Colour figure online)
Model predicted output for the sunspot time series using the most-voted model structure identified by the BFOR-dCor algorithm, where the black solid line with circles indicates the true measurements, the green diamonds represent the model predicted output, and the green shadow represents the 2 SD region. (Colour figure online)
In [52], Billings and Tao developed a set of tests that are effective for time series model validation:
where \(\xi \left( k\right) =\xi _{k}\) is the prediction error sequence with \(k=1,2,\ldots ,N\), \(\xi '_{k}=\xi _{k}-\overline{\xi }\) and \(\left( \xi ^{2}\right) '_{k}=\xi _{k}^{2}-\overline{\xi ^{2}}\). Fig. 10 shows the statistical correlation tests for the one-step ahead predicted output of the most-voted NAR model identified by the BFOR-dCor algorithm. It can be seen that the second and third tests, i.e. \(\phi _{\xi '\left( \xi ^{2}\right) '}\left( \tau \right) =0\) and \(\phi _{\left( \xi ^{2}\right) '\left( \xi ^{2}\right) '} \left( \tau \right) =\delta \left( \tau \right) \;\forall \,\tau \), are not ideally satisfied, suggesting that autoregressive models may not be sufficient to fully characterise the entire dynamics of the process. Nevertheless, the results obtained by the BFOR-dCor algorithm are still remarkable given the complexity of the system.
Statistical correlation tests (8), with 95 % confidence limits, for the one-step ahead predicted output of the most-voted NAR model identified for the sunspot time series using the BFOR-dCor algorithm
7 Conclusion
A new algorithm for model structure detection and parameter estimation has been developed. This new algorithm combines two different concepts that enhance the performance of the original OFR algorithm. First, the distance correlation metric is used, which measures all types of nonlinear or non-monotone dependencies between random vectors. Second, the bagging method is implemented, which produces different models for each resample from the original data set. Identified models, or a subset of them, can be used together to generate improved predictions via averaging (for regression problems) or via voting (for classification problems). A main advantage of these concepts in the new BFOR-dCor algorithm is that it provides the interpretability of nonlinear dependencies and an insightful uncertainty analysis. The algorithm can be slow since the distance correlation is a complex computation compared with other metrics; nevertheless, it produces results that outperform its counterparts and requires no assumptions of probability distributions like the RJMCMC algorithm. All these have been demonstrated through numerical case studies.
References
Billings, S.A.: Nonlinear System Identification: NARMAX Methods in the Time, Frequency, and Spatio-Temporal Domains. Wiley, London (2013)
Pope, K.J., Rayner, P.J.W.: Non-linear system identification using Bayesian inference. In: IEEE International Conference on Acoustics, Speech, and Signal Processing, ICASSP-94, 1994 vol. IV, pp. 457–460 (1994)
Haber, R., Unbehauen, H.: Structure identification of nonlinear dynamic systems–a survey on input/output approaches. Automatica 26(4), 651–677 (1990)
Aguirre, L.A., Letellier, C.: Modeling nonlinear dynamics and chaos: a review. Mathe. Prob. Eng. 2009, 35 (2009). doi:10.1155/2009/238960
Billings, S.A., Coca, D.: Identification of NARMAX and related models. tech. rep., Department of Automatic Control and Systems Engineering, The University of Sheffield, UK, (2001)
Guo, L.Z., Billings, S.A., Zhu, D.Q.: An extended orthogonal forward regression algorithm for system identification using entropy. Int. J. Control 81(4), 690–699 (2008)
Koller, D., Sahami, M.: Toward optimal feature selection. In: In 13th International Conference on Machine Learning (1995)
Billings, S.A., Wei, H.-L.: Sparse model identification using a forward orthogonal regression algorithm aided by mutual information. IEEE Trans. Neural Netw. 18(1), 306–310 (2007)
Wei, H.-L., Billings, S.A.: Model structure selection using an integrated forward orthogonal search algorithm assisted by squared correlation and mutual information. Int. J. Model. Identif. Control 3(4), 341–356 (2008)
Wang, S., Wei, H.-L., Coca, D., Billings, S.A.: Model term selection for spatio-temporal system identification using mutual information. Int. J. Syst. Sci. 44(2), 223–231 (2013)
Han, M., Liu, X.: Forward Feature Selection Based on Approximate Markov Blanket. In: Advances in Neural Networks-ISNN 2012, pp. 64–72, Springer, Berlin (2012)
Baldacchino, T., Anderson, S.R., Kadirkamanathan, V.: Computational system identification for Bayesian NARMAX modelling. Automatica 49, 2641–2651 (2013)
Ninness, B., Brinsmead, T.: A Bayesian Approach to System Identification using Markov Chain Methods. Tech. Rep. EE02009, University of Newcastle, Australia, NSW (2003)
Székely, G.J., Rizzo, M.L., Bakirov, N.K.: Measuring and testing dependence by correlation of distances. Annals Stat. 35(6), 2769–2794 (2007)
Székely, G.J., Rizzo, M.L.: Energy statistics: a class of statistics based on distances. J. Stat. Plan. Inf. 143(8), 1249–1272 (2013)
Chen, S., Billings, S., Luo, W.: Orthogonal least squares methods and their application to non-linear system identification. Int. J. Control 50(5), 1873–1896 (1989)
Söderström, T., Stoica, P.: System Identification. Prentice Hall, New Jersey (1989)
Wei, H.-L., Balikhin, M.A., Billings, S.A.: Nonlinear time-varying system identification using the NARMAX model and multiresolution wavelet expansions. Tech. Rep. 829, The University of Sheffield, United Kingdom (2003)
Wei, H.-L., Billings, S.A., Liu, J.: Term and variable selection for non-linear system identification. Int. J. Control 77(1), 86–110 (2004)
Wei, H.-L., Billings, S.A., Zhao, Y., Guo, L.: Lattice dynamical wavelet neural networks implemented using particle swarm optimization for spatio-temporal system identification. IEEE Trans. Neural Netw. 20(1), 181–185 (2009)
Rashid, M.T., Frasca, M., Ali, A.A., Ali, R.S., Fortuna, L., Xibilia, M.G.: Nonlinear model identification for Artemia population motion. Nonlinear Dyn. 69(4), 2237–2243 (2012)
Haynes, B.R., Billings, S.A.: Global analysis and model validation in nonlinear system identification. Nonlinear Dyn. 5(1), 93–130 (1994)
Billings, S.A., Wei, H.-L.: An adaptive orthogonal search algorithm for model subset selection and non-linear system identification. Int. J. Control 81(5), 714–724 (2008)
Aguirre, L.A., Jácôme, C.: Cluster analysis of NARMAX models for signal-dependent systems. In: IEE Proceedings Control Theory and Applications, vol. 145, pp. 409–414, IET, July (1998)
Feil, B., Abonyi, J., Szeifert, F.: Model order selection of nonlinear input-output models–a clustering based approach. J. Process Control 14(6), 593–602 (2004)
Kukreja, S.L., Lofberg, J., Brenner, M.J.: A least absolute shrinkage and selection operator (LASSO) for nonlinear system identification. Syst. Identif. 14, 814–819 (2006)
Qin, P., Nishii, R., Yang, Z.-J.: Selection of NARX models estimated using weighted least squares method via GIC-based method and l 1-norm regularization methods. Nonlinear Dyn. 70(3), 1831–1846 (2012)
Zou, H., Hastie, T.: Regularization and variable selection via the elastic net. J. R. Stat. Soc. B 67(2), 301–320 (2005). (Statistical Methodology)
Hong, X., Chen, S.: An elastic net orthogonal forward regression algorithm. In: 16th IFAC Symposium on System Identification, pp. 1814–1819, July (2012)
Sette, S., Boullart, L.: Genetic programming: principles and applications. Eng. Appl. Artif. Intell. 14(6), 727–736 (2001)
Madár, J., Abonyi, J., Szeifert, F.: Genetic programming for the identification of nonlinear input-output models. Ind. Eng. Chem. Res. 44(9), 3178–3186 (2005)
Martins, S.A.M., Nepomuceno, E.G., Barroso, M.F.S.: Improved structure detection for polynomial NARX models using a multiobjective error reduction ratio. J. Control Autom. Electr. Syst. 24(6), 764–772 (2013)
Baldacchino, T., Anderson, S.R., Kadirkamanathan, V.: Structure detection and parameter estimation for NARX models in a unified EM framework. Automatica 48(5), 857–865 (2012)
Teixeira, B.O., Aguirre, L.A.: Using uncertain prior knowledge to improve identified nonlinear dynamic models. J. Process Control 21(1), 82–91 (2011)
Billings, S.A., Voon, W.S.F.: Correlation based model validity tests for nonlinear models. Tech. Rep. 285, The University of Sheffield, United Kingdom, October (1985)
Billings, S.A., Wei, H.-L.: The wavelet-NARMAX representation: a hybrid model structure combining polynomial models with multiresolution wavelet decompositions. Int. J. Syst. Sci. 36(3), 137–152 (2005)
Guo, Y., Guo, L., Billings, S., Wei, H.-L.: An iterative orthogonal forward regression algorithm. Int. J. Syst. Sci. 46(5), 776–789 (2015)
Billings, S.A., Chen, S., Backhouse, R.J.: The identification of linear and non-linear models of a turbocharged automotive diesel engine. Mech. Syst. Signal Process. 3(2), 123–142 (1989)
Dietterich, T.G.: Machine Learning for Sequential Data: A Review. In: Structural, Syntactic, and Statistical Pattern Recognition, pp. 15–30, Springer, Berlin (2002)
Efron, B.: Computers and the theory of statistics: thinking the unthinkable. SIAM Rev. 21, 460–480 (1979)
Efron, B., Tibshirani, R.J.: An Introduction to the Bootstrap, vol. 57 of Monographs on Statistics and Applied Probability. Chapman & Hall, London (1993)
Davison, A.C.: Bootstrap Methods and their Application. Cambridge University Press, Cambridge (1997)
Kukreja, S.L., Galiana, H., Kearney, R.: Structure detection of NARMAX models using bootstrap methods. In: Proceedings of the 38th IEEE Conference on Decision and Control, 1999. vol. 1, pp. 1071–1076 (1999)
Kukreja,: A suboptimal bootstrap method for structure detection of NARMAX models. Tech. Rep. LiTH-ISY-R-2452, Linköpings universitet, Linköping, Sweden (2002)
Wei, H.-L., Billings, S.A.: Improved parameter estimates for non-linear dynamical models using a bootstrap method. Int. J. Control 82(11), 1991–2001 (2009)
Breiman, L.: Bagging predictors. Tech. Rep. 421, University of California, Berkeley, California, USA, September (1994)
Hyndman, R.J., Athanasopoulos, G.: Forecasting: Principles and Practice. OTexts (2014). https://www.otexts.org/book/fpp
James, G., Witten, D., Hastie, T., Tibshirani, R.: An Introduction to Statistical Learning with Application in R, vol. 103 of Springer Texts in Statistics. Springer, Berlin (2013)
Sunspot Data, (2003)
Kampstra, P.: Beanplot: A boxplot alternative for visual comparison of distributions. J. Stati. Softw. 28, 1–9 (2008)
Lin, H., Varsik, J., Zirin, H.: High-resolution observations of the polar magnetic fields of the Sun. Solar Phys. 155(2), 243–256 (1994)
Billings, S.A., Tao, Q.H.: Model validity tests for non-linear signal processing applications. Int. J. Control 54(1), 157–194 (1991)
Acknowledgments
The authors acknowledge the support for J. R. Ayala Solares from a University of Sheffield Full Departmental Fee Scholarship and a scholarship from the Mexican National Council of Science and Technology (CONACYT). The authors also gratefully acknowledge that this work was partly supported by EPSRC.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ayala Solares, J.R., Wei, HL. Nonlinear model structure detection and parameter estimation using a novel bagging method based on distance correlation metric. Nonlinear Dyn 82, 201–215 (2015). https://doi.org/10.1007/s11071-015-2149-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11071-015-2149-3