Abstract
Chemokines are a superfamily of small heparin-binding cytokines that induce leukocytes to migrate to sites of inflammation or injury through interacting with specific transmembrane G protein-coupled receptors. Currently, attention is focused on chemokine/chemokine receptor pairs and their ability to promote tumor cell migration and angiogenesis. The chemokine receptor CXCR3 is involved in tumor metastasis and is used as a prognostic biomarker. However, its relationship with the clinicopathological features of primary glioblastoma multiforme (pGBM) and its potential prognostic value have yet to be investigated. Here, we report that high CXCR3 expression conferred poor survival in pGBM patients. Further analysis showed that CXCR3 served as an independent prognostic biomarker for pGBM patients. In addition, functional assays indicated that CXCR3 induced glioma cell invasion. Therefore, this evidence indicates CXCR3 is an independent prognostic factor for pGBM patients and promotes an invasive phenotype, which suggests a new potential biotarget for glioblastoma multiforme therapy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Glioblastoma multiforme (GBM), the most malignant type of glioma, remains one of the most lethal tumors because of its highly invasive behavior and recurrence rate. Furthermore, primary glioblastoma multiforme (pGBM) contributes to the majority of GBM (~90 %), which develops rapidly and de novo in older patients, and in the absence of clinical or histologic evidence of a less malignant precursor lesion. pGBM also indicates a significantly poorer prognosis [1]. A major challenge of GBM therapy is the propensity of cancer cells to invade into adjacent normal brain tissues and/or spread to the contralateral cerebral hemisphere [2, 3]. Tumor cell migration/invasion is a complicated multi-step process, and an improved understanding of the biochemical and molecular mechanisms of glioma cell migration/invasion might provide valuable insights into the underlying biological features of GBM and highlight potential targets for anti-GBM therapies [4]. In recent years, multiple factors that contribute to tumor invasion have been reported [5–9]. Among these mediators, increased attention has focused on chemokine receptors expressed on the surface of cancer cells. The migration and metastasis of cancer cells shows similarities with leukocyte trafficking [10, 11].
CXCR3, a CXC chemokine receptor subfamily member, is expressed by most cancer cells. It regulates cell invasion and migration as well as facilitating tumor metastasis to lymph nodes [12–14], and antagonism of CXCR3 can suppress tumor metastasis in tumor models [15–17]. Furthermore, CXCR3 expression is associated with poor survival in breast cancer [14], colorectal carcinoma [18], and renal cell carcinoma patients [19]. CXCR3 and its ligands are elevated in high-grade glioma cells [20], and can promote glioma cell growth [21] and migration [22]. However, the expression level of CXCR3 and its relationship with the pathological features and overall survival in human pGBM is still unclear, although studies indicate the prognostic value of CXCR3. Therefore, we investigated the functional role and prognostic value of CXCR3 in human pGBM.
Materials and methods
Samples and patients
A total of 65 pGBM samples were confirmed by pathological diagnosis according to the 2007 WHO classification. After confirmation on HE stained slides, only samples with tumor cells >80 % were remained for further studies. Normal brain tissues were obtained from patients with severe traumatic brain injury who required post-trauma surgery. Extent of resection was graded as gross total resection (GTR) or non-GTR using MRIs obtained within 72 h after surgical resection by two independent radiologists. This study was approved by the institutional review boards of Nanjing Medical University and Capital Medical University. Written informed consent was obtained from all patients.
Cell culture and oligonucleotide transfection
Human glioma cell line U87, U251, U373, LN229 were purchased from the Chinese Academy of Sciences Cell Bank. All cell lines were maintained in a 37 °C, 5 % CO2 incubator in DMEM supplemented with 10 % fetal bovine serum (FBS). CXCR3 small interference RNA (siRNA) were chemically synthesized by Shanghai GenePharma Company (Shanghai, China). The siRNA sequence was 5′-GCUGCUCUAUGCCUUUGUATT-3′, 5′-UACAAAGGCAUAGAGCAGCTT-3′. And the negative control (NC) sequence was 5′-UUCUCCGAACGUGUCACGUTT-3′, 5′-ACGUGACACGUUCGGAGAATT-3′. Cells of 70–80 % confluence were transfected with the siRNA and NC by using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, USA), transfection was performed according to the manufacturer’s instructions. CXCR3 over-expressing vectors and NC vectors were constructed by Wuhan Genesil (Genesil, Wuhan, China). Vectors were transfected into human glioma cell lines with FuGENE HD6 (Roche, Basel, Switzerland), according to the manufacturer’s instructions, and screened by the aminoglycoside G418.
RNA extraction and PCR
Total RNA was extracted from cells by using TRIzol reagent (Invitrogen, Carlsbad, USA). RNA was quantified by absorbance reading at A260/A280 = 1.6–1.8 and stored at −80 °C until use. Total 20 μl mixture containing 1 μg RNA was used for reverse transcription reaction (TaKaRa Biotechnology, China). For PCR reaction, 1 μl of RT product was used. Primers for CXCR3 and GAPDH were as follows, CXCR3 forward primer: 5′-CCTTCCTGCCAGCCCTCTACAG-3′, reverse primer: 5′-TGGGCATAGCAGTAGGCCATGA-3′. GAPDH forward primer: 5′-CCACCCATGGCAAATTCCATGGCA-3′, reverse primer: 5′-TCTAGACGGCAGGTCAGGTCCACC-3′. PCR condition was 94 °C, 5 min; 35 cycles of 94 °C, 30 s; 56 °C, 30 s; 72 °C, 30 s; 72 °C, 5 min.
Western blot
Total protein was collected using a Total Protein Extraction Kit (KeyGen, China) and quantified using a BCA Protein Assay Kit (KeyGen, China). 20 μg of protein were added and subjected to SDS-PAGE on (SDS)-polyacrylamide gel electrophoresis using the discontinuous buffer system of Laemmli (Bio-Rad Laboratories, USA). The electrophoresed proteins were transferred to a polyvinylidene difluoride (PVDF) membrane and subjected to immunoblot analysis with a mouse primary antibody against CXCR3 (Abcam, 1:1,000), followed by HRP-conjugated goat anti-mouse secondary antibodies (Bioworld Technology, USA, 1:5,000). GAPDH (KangCheng, China, 1:2,000) was used as control.
Immunohistochemistry
Immunohistochemical staining with streptavidin–biotin immunoperoxidase assay was performed on 65 formalin-fixed, paraffin-embedded pGBM tissues to detect CXCR3 expression by using mouse-anti human CXCR3 (Abcam, 1:800) primary antibody. Slides were individually reviewed and scored by two independent observers. The proportion of positively-stained tumor cells was graded as follows: 0 (<5 % positive tumor cells); 1 (6–30 % positive tumor cells); 2 (31–50 % positive tumor cells); and 3 (>50 % positive tumor cells). The intensity of staining was graded as follows: 0 (no staining), 1 (weak staining, light yellow), 2 (moderate staining, yellowish brown), and 3 (strong staining, brown). The staining score = staining intensity × proportion of positively stained tumor cells. High CXCR3 expression was defined as a staining score >4, while low expression was defined as a staining score ≤4. The final immunostaining score was the average of the two observers.
Invasion assay in vitro
To investigate the cells’ invasion ability after CXCR3 inhibition or overexpressing, transwell invasion assay was done as previous report [23]. Cells were plated into 24-well Boyden chambers (Coring costar, MA, USA) with an 8-μm pore polycarbonate membrane, which was coated with 20 μg of Matrigel (BD Biosciences, San Jose, CA). Cells were in the upper chamber with 200 μl of serum-free medium, and medium containing 20 % FBS was added to the lower chamber to serve as a chemoattractant. After 24 h, the cells were washed three times with PBS, cells were removed from the upper well by cotton swabs (non invasive cells), and the invasive cells were then fixed with 4 % paraformaldehyde, stained with 0.1 % crystal violet, and photographed (×200) in six independent fields for each well. The results were averaged through three independent experiments.
Immunofluorescence
Glioma cells were fixed with 4 % paraformaldehyde for 20 min, incubated overnight at 4 °C with mouse-anti human CXCR3 (Abcam, 1:400), incubated for 1 h at 37 °C with Dylight 488 goat anti-mouse secondary antibody (Sigma, 1:200), then counterstained with DAPI (Sigma, USA) to reveal nuclei. Cells were detected by laser confocal scanning microscopy.
Statistical analysis
Statistical analysis was performed using SPSS Graduate Pack 16.0 and GraphPad Prism 5.0 statistical software. Descriptive statistics included mean ± SE. Student’s t test, one-way ANOVAs and Exact Sig (2-sided) χ2 test were used to analyze significant differences. Kaplan–Meier survival analysis was used to estimate the survival distributions, and the log-rank test was used to assess the statistical significance between stratified survival groups. Karnofsky performance status (KPS), extent of resection, and temozolomide treatment were incorporated into a Cox multivariate analysis. A two-sided P value of <0.05 was considered significant.
Results
CXCR3 expression in glioma cell lines and pGBM samples
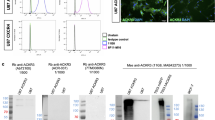
To detect the expression of CXCR3 mRNA and protein in glioma cell lines, RT-PCR, western blot, and immunofluorescence were employed. Both CXCR3 mRNA and protein were positive in glioma cell lines U87, U251, U373, and LN229 (Fig. 1, one normal brain sample was used as a control). Furthermore, immunochemistry was performed to analyze the CXCR3 protein expression pattern in 65 pGBM tissue samples. As shown in Fig. 2a, we observed CXCR3 protein positive staining to be mainly located in the cytoplasm of cells. According to the CXCR3 staining score, 38 (58.46 %) patients were classified as the low-CXCR3 group and 27 (41.54 %) patients were classified as the high-CXCR3 group.
a CXCR3 mRNA was detected by RT-PCR in a normal brain sample, U87, U251, LN229, and U373 glioma cell lines. b Western blot showed CXCR3 protein was positive in the samples in a. c CXCR3 protein (red) was detected in four cell lines by immunofluorescence (magnification ×200). The cell nucleus (blue) was stained by DAPI. (N normal brain sample)
High expression of CXCR3 confers a poor prognosis
To investigate whether CXCR3 protein expression levels were linked to the clinical parameters of pGBM patients, a correlation between CXCR3 expression values and various clinical pathological features was analyzed. However, there was no significant correlation between CXCR3 expression and patient sex (P = 0.591), age (P = 0.816), and KPS score (P = 0.692). Next, the relationship between CXCR3 expression and survival was determined by Kaplan–Meier survival curve analysis with a log-rank comparison of the 65 pGBM patients. pGBM patients with higher levels of CXCR3 expression had poorer survival compared with those with lower levels of CXCR3 expression (Fig. 2b). Thus, CXCR3 might affect both progression-free survival and overall survival of pGBM patients.
CXCR3 is an independent prognostic biomarker for pGBM
To determine the prognostic value of CXCR3 in pGBM, we used univariate Cox regression analysis. As shown in Table 1, high CXCR3 expression was shown to be a risk factor for pGBM [P < 0.01, HR 2.336, 95 % confidence interval (CI) 1.341–4.071]. Additionally, some factors (KPS score, extent of resection, and temozolomide treatment) were significantly associated with the overall survival of pGBM patients. To evaluate further whether CXCR3 could act as an independent prognostic biomarker for pGBM patients, we performed a stepwise, multivariate Cox regression analysis incorporating CXCR3 expression, KPS score, extent of resection, and temozolomide treatment status [24]. This analysis revealed that CXCR3 expression level was an independent prognostic factor for the overall survival of pGBM patients. Taken together, these results indicated that CXCR3 might be a useful independent prognostic biomarker for pGBM.
CXCR3 promotes glioma cell invasion
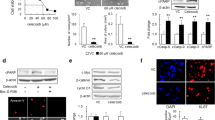
To assess the functional role of CXCR3 in glioma, siRNA was used to silence CXCR3 expression, and a CXCR3 over-expressing vector was used to elevate CXCR3 levels. The efficiency of siRNA and vectors were examined by RT-PCR and western blot (Fig. 3). Both CXCR3 mRNA expression and protein levels were decreased by CXCR3 siRNA transfection in U87 and U251 cell lines. CXCR3 over-expressing vectors up-regulated CXCR3 mRNA and protein levels in the LN229 cell line. A transwell assay was performed to investigate whether CXCR3 could affect glioma cell invasion. First, loss-of-function analysis was used. Antagonism of CXCR3 expression reduced migrating cell numbers by 48.27 ± 3.09 % compared with controls in U87 cells (Fig. 4a, d) and by 53.61 ± 4.26 % in U251 cells (Fig. 4b, e). A gain-of-function assay in LN229 cells with over-expression of CXCR3 showed a 1.68 ± 0.12-fold increase in invasion compared with controls (Fig. 4c, f).
CXCR3 mRNA and protein were decreased by CXCR3 siRNA in U87 and U251 cells, and were over-expressed by CXCR3-expressing Vector in LN229 cells. Transfection efficient was measured by RT-PCR (a) and western blot (b). (NC negative control, siRNA CXCR3 small interference RNA, Vector CXCR3 expressing Vector)
CXCR3 affects cell invasion ability in glioma cell lines (magnification ×200). a and d U87 cells transfected with CXCR3 siRNA have a lower invasive potential (reduction of cell numbers was 48.27 ± 3.09 %) compared with the blank control. b and e U251 cells transfected with CXCR3 siRNA showed a 53.61 ± 4.26 % reduction in the number of invasive cells compared with the blank control. c and f LN229 cells transfected with the CXCR3-expressing Vector showed a higher invasive potential (cell numbers were increased 1.68 ± 0.12-fold) compared with the blank control. *P < 0.05 compared with the blank control
Discussion
GBM has a highly invasive behavior and high recurrence rate of cells infiltrating to adjacent tissues during the early stage of disease. Currently, little is known about the mechanisms of cell migration and infiltration into adjacent tissues. The chemokine superfamily contains more than 50 cytokines that were initially characterized through their ability to bind to 18–22 G protein-coupled receptors to direct the migration of leukocytes to sites of inflammation or injury [25]. Furthermore, chemokine/chemokine receptor pairs affect cell migration, survival, adhesion, invasion, and proliferation responses associated with tumor growth [7, 26]. CXCR3 belongs to the CXC chemokine receptor subfamily and has three endogenous ligands, CXCL9, CXCL10, and CXCL11. CXCR3 has been detected in many malignant tumors and is associated with patient outcomes. However, the functional role of CXCR3 is controversial when considered for various malignant cancers. In melanoma, CXCR3 contributed to cell motility during invasion as well as the regulation of cell proliferation and survival [27]. Moreover, CXCR3 expression was positively correlated with tumor thickness and the presence of distant metastases. The study also indicated that CXCR3 could mediate melanoma metastasis to draining lymph nodes [28]. In breast cancer, high CXCR3 is an independent predictor of poor prognosis and worse long-term survival, and which can promote metastasis in mice [14]. Antagonism of CXCR3 inhibited lung metastasis in a murine model of metastatic breast cancer [15]. The functional role of CXCR3 gene silencing was further confirmed in osteosarcoma [16]. Kawada found that CXCR3 promoted colon cancer metastasis to lymph nodes and that CXCR3-positive patients had a worse prognosis [13]. They also showed the prognostic value of CXCR3 in colorectal carcinoma patients [18]. The aberrant expression of CXCR3 was also found to promote prostate tumor metastasis via stimulating cell migration and invasion [29]. However, other studies have shown the opposite results. In renal cell carcinoma, patients with low CXCR3 expression had a significantly worse prognosis than patients with high CXCR3 expression, and CXCR3 was an independent predictor of improved disease-free survival [19]. In chronic lymphocytic leukemia (CLL), low CXCR3 levels also predicted a shorter survival time [30].
These studies reported opposite conclusions in different tumors; however, CXCR3 can promote the growth and migration of glioma [21, 22] and CXCR3 and its paired ligands were detected in glioma samples and cell lines [20, 31, 32]. Thus, it is valuable to confirm the functional role and prognostic value of CXCR3 expression in human pGBM. In this study, we found that CXCR3 mRNA and protein were both positively expressed in U87, U251, U373, and LN229 cell lines. CXCR3 protein was also detected in pGBM samples and was mainly located in the cytoplasm. However, there was no significant correlation between CXCR3 expression and pGBM patient clinical features. Kaplan–Meier survival analysis of pGBM samples revealed that high expression levels of CXCR3 conferred a poor prognosis. Furthermore, the prognostic value of CXCR3 expression was analyzed by univariate Cox regression analysis. IDH1 mutation, KPS, extent of resection, TMZ treatment, and CXCR3 expression were then subjected to multivariate Cox regression analysis, which showed that CXCR3 could be used as an independent prognostic biomarker for pGBM. In addition, in vitro functional analysis showed that CXCR3 promoted glioma cell invasion. Thus, CXCR3 might affect malignant progression and function as an oncogene via the induction of GBM cell invasion and migration.
Previous studies have shown that the role of chemokine/chemokine receptor pairs in malignant tumors is complex. Anti-tumor activity was induced by stimulating immune cells, whereas other studies showed CXCR3 increased tumor growth and metastasis by directly promoting growth, enhancing cell motility and angiogenesis, and inducing cell invasion. Interestingly, Liu et al. found that glioma-bearing CXCR3-deficient mice had a significantly shorter median survival time and reduced numbers of tumor-infiltrated natural killer and natural killer T cells as compared with tumor-bearing wild-type (WT) mice. However, pharmacological antagonism of CXCR3 with NBI-74330 prolonged the median survival times of both tumor-bearing WT and CXCR3-deficient mice when compared with vehicle-treated groups [21]. This study also indicated the converse functional role of CXCR3 in tumors. Although CXCR3 can promote glioma growth, migration, and angiogenesis, it also induced immune cells to migrate to the tumor site and kill tumor cells. Our studies show that pGBM patients with higher than median levels of CXCR expression have a poor survival rate. In our opinion, in addition to CXCR3, the blood–brain barrier plays a vital role in glioma cell invasion, by preventing the trafficking of immune cells to the intra-calvarium.
In conclusion, we showed that CXCR3 was detected in pGBM and might be a useful independent prognostic biomarker for pGBM patients. Inhibiting CXCR3 expression in GBM cells could significantly suppress cell invasion. Together, these results indicate that CXCR3 might be a potential target for anti-invasive GBM therapy. However, further studies are needed to analyze how CXCR3 participates in the carcinogenesis and progression of glioma.
References
Okumus NO, Gursel B, Meydan D, Ozdemir O, Odabas E, Gonullu G (2012) Prognostic significance of concomitant radiotherapy in newly diagnosed glioblastoma multiforme: a multivariate analysis of 116 patients. Ann Saudi Med 32(3):250–255. doi:10.5144/0256-4947.2012.250
Drappatz J, Norden AD, Wen PY (2009) Therapeutic strategies for inhibiting invasion in glioblastoma. Expert Rev Neurother 9(4):519–534. doi:10.1586/ern.09.10
Demuth T, Berens ME (2004) Molecular mechanisms of glioma cell migration and invasion. J Neurooncol 70(2):217–228. doi:10.1007/s11060-004-2751-6
Nakada M, Nakada S, Demuth T, Tran NL, Hoelzinger DB, Berens ME (2007) Molecular targets of glioma invasion. Cell Mol Life Sci 64(4):458–478. doi:10.1007/s00018-007-6342-5
Mikheeva SA, Mikheev AM, Petit A, Beyer R, Oxford RG, Khorasani L, Maxwell JP, Glackin CA, Wakimoto H, Gonzalez-Herrero I, Sanchez-Garcia I, Silber JR, Horner PJ, Rostomily RC (2010) TWIST1 promotes invasion through mesenchymal change in human glioblastoma. Mol Cancer 9:194. doi:10.1186/1476-4598-9-194
Thomas SL, Alam R, Lemke N, Schultz LR, Gutierrez JA, Rempel SA (2010) PTEN augments SPARC suppression of proliferation and inhibits SPARC-induced migration by suppressing SHC-RAF-ERK and AKT signaling. Neuro Oncology 12(9):941–955. doi:10.1093/neuonc/noq048
Fulton AM (2009) The chemokine receptors CXCR4 and CXCR3 in cancer. Curr Oncol Rep 11(2):125–131
Asadi-Moghaddam K, Chiocca EA, Lawler SE (2010) Potential role of miRNAs and their inhibitors in glioma treatment. Expert Rev Anticancer Ther 10(11):1753–1762. doi:10.1586/era.10.168
Yan W, Zhang W, Sun L, Liu Y, You G, Wang Y, Kang C, You Y, Jiang T (2011) Identification of MMP-9 specific microRNA expression profile as potential targets of anti-invasion therapy in glioblastoma multiforme. Brain Res 1411:108–115. doi:10.1016/j.brainres.2011.07.002
Tanaka T, Bai Z, Srinoulprasert Y, Yang BG, Hayasaka H, Miyasaka M (2005) Chemokines in tumor progression and metastasis. Cancer Sci 96(6):317–322. doi:10.1111/j.1349-7006.2005.00059.x
Zlotnik A (2006) Chemokines and cancer. Int J Cancer 119(9):2026–2029. doi:10.1002/ijc.22024
Kawada K, Sonoshita M, Sakashita H, Takabayashi A, Yamaoka Y, Manabe T, Inaba K, Minato N, Oshima M, Taketo MM (2004) Pivotal role of CXCR3 in melanoma cell metastasis to lymph nodes. Cancer Res 64(11):4010–4017. doi:10.1158/0008-5472.CAN-03-1757
Kawada K, Hosogi H, Sonoshita M, Sakashita H, Manabe T, Shimahara Y, Sakai Y, Takabayashi A, Oshima M, Taketo MM (2007) Chemokine receptor CXCR3 promotes colon cancer metastasis to lymph nodes. Oncogene 26(32):4679–4688. doi:10.1038/sj.onc.1210267
Ma X, Norsworthy K, Kundu N, Rodgers WH, Gimotty PA, Goloubeva O, Lipsky M, Li Y, Holt D, Fulton A (2009) CXCR3 expression is associated with poor survival in breast cancer and promotes metastasis in a murine model. Mol Cancer Ther 8(3):490–498. doi:10.1158/1535-7163.MCT-08-0485
Walser TC, Rifat S, Ma X, Kundu N, Ward C, Goloubeva O, Johnson MG, Medina JC, Collins TL, Fulton AM (2006) Antagonism of CXCR3 inhibits lung metastasis in a murine model of metastatic breast cancer. Cancer Res 66(15):7701–7707. doi:10.1158/0008-5472.CAN-06-0709
Pradelli E, Karimdjee-Soilihi B, Michiels JF, Ricci JE, Millet MA, Vandenbos F, Sullivan TJ, Collins TL, Johnson MG, Medina JC, Kleinerman ES, Schmid-Alliana A, Schmid-Antomarchi H (2009) Antagonism of chemokine receptor CXCR3 inhibits osteosarcoma metastasis to lungs. Int J Cancer 125(11):2586–2594. doi:10.1002/ijc.24665
Cambien B, Karimdjee BF, Richard-Fiardo P, Bziouech H, Barthel R, Millet MA, Martini V, Birnbaum D, Scoazec JY, Abello J, Al Saati T, Johnson MG, Sullivan TJ, Medina JC, Collins TL, Schmid-Alliana A, Schmid-Antomarchi H (2009) Organ-specific inhibition of metastatic colon carcinoma by CXCR3 antagonism. Br J Cancer 100(11):1755–1764. doi:10.1038/sj.bjc.6605078
Wu Z, Han X, Yan J, Pan Y, Gong J, Di J, Cheng Z, Jin Z, Wang Z, Zheng Q, Wang Y (2012) The prognostic significance of chemokine receptor CXCR3 expression in colorectal carcinoma. Biomed Pharmacother 66(5):373–377. doi:10.1016/j.biopha.2011.12.003
Klatte T, Seligson DB, Leppert JT, Riggs SB, Yu H, Zomorodian N, Kabbinavar FF, Strieter RM, Belldegrun AS, Pantuck AJ (2008) The chemokine receptor CXCR3 is an independent prognostic factor in patients with localized clear cell renal cell carcinoma. J Urol 179(1):61–66. doi:10.1016/j.juro.2007.08.148
Maru SV, Holloway KA, Flynn G, Lancashire CL, Loughlin AJ, Male DK, Romero IA (2008) Chemokine production and chemokine receptor expression by human glioma cells: role of CXCL10 in tumour cell proliferation. J Neuroimmunol 199(1–2):35–45. doi:10.1016/j.jneuroim.2008.04.029
Liu C, Luo D, Reynolds BA, Meher G, Katritzky AR, Lu B, Gerard CJ, Bhadha CP, Harrison JK (2011) Chemokine receptor CXCR3 promotes growth of glioma. Carcinogenesis 32(2):129–137. doi:10.1093/carcin/bgq224
Honeth G, Staflin K, Kalliomaki S, Lindvall M, Kjellman C (2006) Chemokine-directed migration of tumor-inhibitory neural progenitor cells towards an intracranially growing glioma. Exp Cell Res 312(8):1265–1276. doi:10.1016/j.yexcr.2005.12.018
Sun L, Yan W, Wang Y, Sun G, Luo H, Zhang J, Wang X, You Y, Yang Z, Liu N (2011) MicroRNA-10b induces glioma cell invasion by modulating MMP-14 and uPAR expression via HOXD10. Brain Res 1389:9–18. doi:10.1016/j.brainres.2011.03.013
Zhang JP, Lu WG, Ye F, Chen HZ, Zhou CY, Xie X (2007) Study on CXCR4/SDF-1alpha axis in lymph node metastasis of cervical squamous cell carcinoma. Int J Gynecol Cancer 17(2):478–483. doi:10.1111/j.1525-1438.2007.00786.x
Koizumi K, Hojo S, Akashi T, Yasumoto K, Saiki I (2007) Chemokine receptors in cancer metastasis and cancer cell-derived chemokines in host immune response. Cancer Sci 98(11):1652–1658. doi:10.1111/j.1349-7006.2007.00606.x
Vandercappellen J, Van Damme J, Struyf S (2008) The role of CXC chemokines and their receptors in cancer. Cancer Lett 267(2):226–244. doi:10.1016/j.canlet.2008.04.050
Robledo MM, Bartolome RA, Longo N, Rodriguez-Frade JM, Mellado M, Longo I, van Muijen GN, Sanchez-Mateos P, Teixido J (2001) Expression of functional chemokine receptors CXCR3 and CXCR4 on human melanoma cells. J Biol Chem 276(48):45098–45105. doi:10.1074/jbc.M106912200
Monteagudo C, Martin JM, Jorda E, Llombart-Bosch A (2007) CXCR3 chemokine receptor immunoreactivity in primary cutaneous malignant melanoma: correlation with clinicopathological prognostic factors. J Clin Pathol 60(6):596–599. doi:10.1136/jcp.2005.032144
Wu Q, Dhir R, Wells A (2012) Altered CXCR3 isoform expression regulates prostate cancer cell migration and invasion. Mol Cancer 11:3. doi:10.1186/1476-4598-11-3
Jones D, Benjamin RJ, Shahsafaei A, Dorfman DM (2000) The chemokine receptor CXCR3 is expressed in a subset of B-cell lymphomas and is a marker of B-cell chronic lymphocytic leukemia. Blood 95(2):627–632
Flynn G, Maru S, Loughlin J, Romero IA, Male D (2003) Regulation of chemokine receptor expression in human microglia and astrocytes. J Neuroimmunol 136(1–2):84–93
Bajetto A, Barbieri F, Dorcaratto A, Barbero S, Daga A, Porcile C, Ravetti JL, Zona G, Spaziante R, Corte G, Schettini G, Florio T (2006) Expression of CXC chemokine receptors 1-5 and their ligands in human glioma tissues: role of CXCR4 and SDF1 in glioma cell proliferation and migration. Neurochem Int 49(5):423–432. doi:10.1016/j.neuint.2006.03.003
Acknowledgments
This work is supported by National Natural Science Foundation of China (Grant Nos. 81302183, 81302200). Natural Science Foundation of Jiangsu Province (Grant No. BK20141099).
Author information
Authors and Affiliations
Corresponding author
Additional information
Yi Pu and Shouwei Li have contributed equally to this work.
Rights and permissions
About this article
Cite this article
Pu, Y., Li, S., Zhang, C. et al. High expression of CXCR3 is an independent prognostic factor in glioblastoma patients that promotes an invasive phenotype. J Neurooncol 122, 43–51 (2015). https://doi.org/10.1007/s11060-014-1692-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11060-014-1692-y