Abstract
In this paper, we propose a large language models (LLMs) assisted algorithm that uses ChatGPT to summarize clinical notes and then concatenate these generated summaries with structured data to predict sepsis. We perform a human evaluation of the summaries generated by ChatGPT and evaluate our algorithm using an independent test set. Our algorithm achieves a high prediction AUC of 0.93 (95% CI 0.92-0.93), accuracy of 0.92 (95% CI 0.91-0.92), and specificity of 0.89 (95% CI 0.88-0.90) 4 hours before the onset of sepsis. The ablation study demonstrated a 2% improvement in predicted AUC score when utilizing ChatGPT for clinical notes summarization compared to traditional methods, 4 hours before the sepsis onset. The experiment results in turn revealed the remarkable performance of ChatGPT in the domain of clinical notes summarization.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Sepsis is a systemic disease characterized by an impaired immune response to a bloodstream infection, rendering patients vulnerable to organ damage and mortality [1]. Early prediction of sepsis is crucial in preventing mortality due to the high level of time sensitivity associated with sepsis management. Despite significant medical advances in recent decades, sepsis continues to be a prominent cause of in-hospital mortality. Sepsis imposes a substantial burden on hospitals and healthcare systems due to its alarmingly high mortality rate [2]. In the United States, sepsis accounts for over one-third of all inpatient deaths. Sepsis management in hospitals incurs annual costs of approximately $24 billion, with a significant portion of these costs attributed to patients who develop sepsis during their hospital stay. These costs exceed those associated with any other health condition. Furthermore, research has shown that delaying the administration of intravenous antibiotics by just one hour can result in a sepsis mortality increase ranging from 4% to 8% [3]. Hence, early and accurate diagnosis and treatment of sepsis can improve patient outcomes, reduce the mortality rate, and decrease the cost of care. The source code of our method can be found at https://github.com/TJU-MHB/ChatGPT-sepsis-prediction.
The majority of current methods for early sepsis detection solely rely on structured data from the electronic health records (EHR) system [4,5,6]. However, research has indicated that approximately 80% of clinical data in EHR systems comprises unstructured data, which refers to data stored without a predetermined or standardized format [7]. Free-form text (e.g., clinical notes) and images (e.g., radiological images) are common examples of this unstructured data. Unstructured clinical data contain valuable information, specifically, additional clinical details that are not captured in the structured data fields of the EHR. Physicians utilize unstructured clinical data fields to record "free-form" clinical notes since structured data is intended for storing predetermined discrete data, such as patient vital signs. Furthermore, physicians rely on unstructured data to review judgments and critical clinical information inputted by other clinicians, aiming to enhance their understanding of a patient’s condition and the effects of their treatment [8]. Previous studies have shown that incorporating both structured and unstructured data can significantly enhance the prediction accuracy of sepsis [8,9,10]. Nevertheless, their approach to handling clinical notes is either confined to removing special characters and stop words [9] or, in some cases [10, 11], lacks a detailed explanation of the preprocessing applied to clinical notes. Additionally, in some instances [8, 12, 13], they do not utilize pre-trained natural language processing (NLP) models to obtain a representation of clinical notes. These processed notes are then fed into clinical Bidirectional Encoder Representations from Transformers (ClinicalBERT) [14] to obtain document-level representations, which are later concatenated with structured features and used as input for sepsis prediction. However, clinical notes often contain various forms of noise, such as abbreviations, grammatical errors, misspellings, and irrelevant formatting [15].
The presence of such noise has the potential to adversely affect the model’s performance. The emergence of LLMs offers a solution to this problem. LLMs have a significantly larger scale in terms of model parameters and training data compared to previous pre-trained models, and they differ in that they do not require fine-tuning [16]. LLMs have demonstrated promising results in zero-shot and few-shot tasks spanning diverse domains [17], which has generated considerable interest in their potential for automated summarization. The emergence of LLMs, such as ChatGPT [18], has led to a substantial advancement in the accurate summarization of text, effectively tackling various noise-related challenges within the text. ChatGPT is a language model that employs reinforcement learning techniques [19, 20] and showcases robust capabilities in the domain of NLP. Goyal et al. [21] conducted an extensive series of experiments, showcasing that ChatGPT’s summarization ability yields results comparable to the current state-of-the-art models, BRIO [22] and T0 [23] in the field of news summarization. They also conducted a human evaluation, revealing that human evaluators preferred the summarization content generated by ChatGPT. This, in turn, has motivated us to explore ChatGPT’s performance in the realm of clinical notes summarization. Consequently, we employ ChatGPT to summarize clinical notes with the aim of eliminating noise. At the same time, to assess both the readability and faithfulness of the generated content to the original clinical note, we invited two professional doctors for human evaluation. To the best of our knowledge, we are the first to utilize ChatGPT for the summarization of clinical notes, and subsequently, employ ClinicalBERT to capture representations of these summaries for downstream tasks such as sepsis prediction. In this study, we developed an LLMs-assisted deep learning algorithm, which comprises a structured data preprocessing pipeline, a clinical notes summarization and representation module, and a sepsis predictive module. This integrated approach combines NLP analysis of clinical notes with structured data, enhancing our early predictive capability for sepsis. The contributions of this paper are as follows:
-
We employ ChatGPT for the purpose of eliminating non-diagnostic noise from clinical notes and summarizing the content of these notes. Subsequently, we utilized the resulting summaries in the early prediction of sepsis. The generated clinical notes summaries are subject to review by ICU doctors, revealing the performance of ChatGPT in clinical notes summarization.
-
We propose an LLMs-assisted deep learning model that integrates structured and unstructured EHR data, facilitating early sepsis prediction. We also elucidate several essential data processing intricacies related to MIMIC-III.
-
We performed a comprehensive analysis and comparison of the experimental results. The doctors expressed great satisfaction with the produced summary report and deemed it faithful to the original clinical notes. Extensive experiments demonstrate that the proposed method outperforms traditional methods in early sepsis prediction, specifically at 4 h, 6 h, and 12 h before the onset of sepsis.
The rest of this paper is structured as follows: Section 2 discusses related works, Section 3 describes the methods we use, Section 4 presents the details of data preprocessing and our approach, Section 5 presents the corresponding experimental results and analysis, and finally, Section 6 concludes this study.
2 Realted work
Sepsis is a time-sensitive disease, early identification of sepsis following rapid initiation of antibiotic treatment improves patient outcomes. Furthermore, a delay of 6 h in treatment has been shown to increase the mortality risk by 7.6% [2]. Unfortunately, sepsis is frequently subject to misdiagnosis and mistreatment due to the similarity of deterioration and organ failure in other diseases, which complicates the identification and treatment of sepsis [24].
2.1 Clinical score-based sepsis prediction
The concept of Systemic Inflammatory Response Syndrome (SIRS) was first introduced in 1991 as the initial definition of sepsis [25]. In 2001, the International Sepsis Definitions Conference revised the definition of sepsis (Sepsis-2), significantly enhancing physicians’ ability to diagnose sepsis directly at the patient’s bedside [26]. In 2016, the Third International Consensus Definitions for Sepsis (Sepsis-3) were published, introducing a new definition for sepsis and septic shock [1]. Within the framework of Sepsis-3, the Sequential Organ Failure Assessment (SOFA) score and the quick Sequential Organ Failure Assessment (qSOFA) score are recommended as assessment measures for a patient’s organ dysfunction [27]. Through the utilization of these clinical scoring systems, clinicians can objectively assess the severity of sepsis, identify patients at high risk, and make well-informed decisions regarding treatment strategies. However, it is important to note that these systems operate based on rules and the reliability of the results they provide is limited.
2.2 Structured data-based sepsis prediction
Machine learning offers a promising methodology to address the complexities related to interpreting high-dimensional, nonlinear, and longitudinal EHR data, surpassing the limitations of conventional clinical statistical methods. The InSight algorithm, introduced by Calvert et al. [28], is regarded as one of the pioneering machine learning models for early sepsis prediction. Subsequently, Desautels et al. [29] and Mao et al. [30] proposed prediction models based on the InSight algorithm. Yang et al. [31] secured unofficial first place in the PhysioNet Computing in Cardiology Challenge 2019 [6] and the 2019 DII National Data Science Challenge using the gradient boosting algorithm. Goh et al. [8] introduced the sepsis early risk assessment (SERA) algorithm, which combines structured and unstructured data for sepsis prediction.
Deep learning models have emerged as the predominant solutions for sepsis prediction. These models specifically utilize neural networks with multiple layers, allowing for the extraction of valuable insights from EHR through a hierarchical architecture. Michael et al. [32] employ a temporal convolutional network embedded within a multi-task Gaussian Process adapter framework (MGP-TCN), allowing for its direct application to irregularly-spaced time series data. Shashikumar et al. [33] utilized a recurrent neural survival model called DeepAISE to forecast sepsis onset. DeepAISE successfully decreased the false-positive rate by capturing predictive features linked to higher-order interactions and temporal patterns among clinical risk factors for sepsis. Traditional machine learning algorithms face challenges in effectively capturing the long-term dependencies within medical time series sequences, leading to suboptimal performance. Furthermore, current deep learning models primarily depend on structured data for sepsis prediction, failing to fully leverage the unstructured data, which constitutes 80% of the EHR data.
2.3 Unstructured data-based sepsis prediction
Apostolova et al. [12] identified notes indicating suspected or confirmed cases of septic infection to establish a system for the identification of signs and symptoms of infection within unstructured nursing notes. Liu et al. [34] anticipated the occurrence of septic shock in sepsis patients prior to the fulfillment of established septic shock criteria. This was done to showcase an approach utilizing features extracted from clinical notes for the prediction of septic shock. Goh et al. [8] developed an algorithm that uses structured and unstructured data to diagnose and predict sepsis. Amrollahi et al. [9] employed ClinicalBERT to represent clinical notes and concatenated these vectors with structured data. They subsequently fed this combined data into a long short-term memory network for sepsis prediction.
Although these works have yielded remarkable results, these approaches for integrating structured and unstructured data frequently overlook the presence of noise in clinical notes during the handling of unstructured data. The introduction of LLMs offers a solution to this issue. Goyal et al. [21] recently reported that, despite receiving lower Rouge scores compared to traditional fine-tuning techniques, the summaries generated by GPT-3 were favored by human evaluators. Zhang et al. [16] conducted a comprehensive comparative experiment and demonstrated that LLMs summaries are considered comparable to human-written summaries. ChatGPT has demonstrated remarkable summarizing capabilities in the news domain. Thus, we tried to leverage ChatGPT’s potential in the medical domain to mitigate noise in clinical notes and summarize clinical notes, then concatenate with structured data for sepsis prediction.
3 Methods
This study expands upon existing methods discussed for sepsis prediction based on physiological and clinical data by integrating features extracted from clinical notes through a neural language model. The proposed model offers sequential hourly predictions for sepsis by utilizing data available at or prior to the prediction time, enabling prospective deployment. Additionally, LLMs have demonstrated promising results in zero-shot and few-shot tasks across diverse domains, generating considerable interest for their potential in automatic summarization.
Schematic diagram of the proposed model, including structured data preprocessing pipeline, clinical notes summarization and representation module, and sepsis predictive module. The clinical notes summarization and representation module involves obtaining the contextual embeddings for one patient by averaging the ClinicalBERT embedding representations of notes, which were generated by ChatGPT. "C" denotes concatenate, "\(\left\{ m_i\right\} \)" and "\(\left\{ x_i\right\} \)" represent the feature vectors of unstructured data and structured data respectively. The resulting representations are then concatenated with the structured clinical data and fed into a BiLSTM-based model for early prediction of sepsis. The version of ChatGPT we are using is "gpt-3.5-turbo", and the prompt we use is "Summarize the following clinical notes"
Therefore, we employ the state-of-the-art model, GPT-3.5-Turbo, to polish clinical notes and eliminate noise commonly found in clinical notes. Figure 1 illustrates the schematic diagram of our feature extraction pipeline and model architecture.
3.1 Problem define
We consider the early detection of sepsis in the ICU as a multivariate time series binary classification task [35]. Specifically, our focus is to combine irregularly sampled multivariate time series of physiological measurements and corresponding clinical notes for early sepsis prediction. Figure 2 illustrates the issues. For prediction purposes, we solely utilize previous clinical data and clinical notes from each time interval. As depicted in Fig. 2, we selected data from 6 hours prior to the prediction time point as the reference dataset for the early sepsis prediction.
For each patient i, we can acquire a set of samples \(X^i=\left\{ \left( x_i, m_i\right) \right\} _{i=1}^t\), where \(X^i \in \mathbb {R}^{t \times d}\). Here, x denotes the patient’s structured data, and m denotes the patient’s Clinical notes, and t signifies the selection of clinical data 6 hours before the prediction time point. d represents the selection of d clinical test values per hour. A predictive model for binary classification tasks can be represented as a mapping that takes inputs and assigns them to one of two categories, i.e.\(f\left( X^i\right) : \mathbb {R}^{t \times d} \rightarrow \{0,1\}\), where 0 denotes non-sepsis and 1 denotes sepsis. The entire sample data set is then denoted as \(\Phi =\left\{ X^i, y_i\right\} _{i=1}^N\), where \(y_i\) is the target label and N is the number of patients. The predictive model is derived from the input data \(X^i\) to obtain the output \(y_i^*=f\left( X^i, \theta \right) \), where \(\theta \) is the parameters of the model. The primary objective of our work is to acquire the optimal parameter \(\theta \), aiming to minimize the disparity between the model’s output \(y_i^*\) and the true label \(y_i\).
3.2 Using GPT-3.5 for summarization
Clinical notes in EHR frequently contain issues like disease-independent formats and abbreviations from doctors’ personal habits. Such noise can potentially impact the model’s performance. LLMs have shown promising results in zero-shot and few-shot tasks across various domains, garnering significant interest due to their potential for automatic summarization. A previous study [21] has shown that GPT-3’s summarization ability in the news domain is slightly inferior to the best fine-tuned models based on automatic metrics but significantly better based on human evaluation. Consequently, we utilize the state-of-the-art LLM, GPT-3.5, to polish clinical notes, mitigate the common noise found within them, and summarize the clinical notes. Simultaneously, we investigated its performance to summarize clinical notes by doing human evaluation. GPT-3.5 introduces additional features and parameters to enhance the accuracy and performance of GPT-3 in various NLP tasks, and the prompt we use is "Summarize the following clinical notes".
The manual entry of each clinical note into ChatGPT individually is undeniably a time-consuming task. Consequently, we developed a custom code and employed the GPT-3.5 Application Programming Interface (API) to process the notes. It is an NLP tool developed by OpenAI, capable of performing various tasks such as text completion, summarization, and translation. Within our system, we utilize the "gpt-3.5-turbo" version of the GPT-3.5 API to generate medical summary reports using a predefined prompt. The developed API generates a summary by utilizing the given prompt and content. Notably, the length of the generated report is not limited, as clinical notes encompass varying lengths, ranging from lengthy physician notes to concise ECG notes. Allowing unrestricted length enables ChatGPT to effectively capture essential information from the original clinical notes. Subsequently, we performed a human evaluation of the generated summary reports with two experienced ICU doctors. The generated reports are subsequently inputted into ClinicalBERT to acquire contextual embedding representations.
In the area of clinical notes mining, researchers have used NLP mainly to identify and extract medical events, medication information, and clinical workflow from unstructured data stored in EHR systems. Among the many language models [36,37,38] proposed in NLP, BERT stands out [39]. It performs extremely well and achieves the best performance for virtually all NLP tasks. Conventional word-level vector representations, such as Word2Vec [40] and GloVe [41], encode all potential meanings of a word into a single vector representation, lacking the ability to disambiguate word senses within the surrounding context or account for negations. The BERT language model resolves this challenge by offering context-sensitive embeddings for individual words within a sentence, which are valuable for downstream tasks such as sepsis predictive modeling. Emily et al. [14] introduced ClinicalBERT trained on MIMIC-III notes and showed that ClinicalBERT can successfully outperform prior models in several clinical NLP tasks. For representing the note, we leverage the activation levels of neurons in the final hidden layers of the ClinicalBERT model. A patient-level representation is computed by inputting all patient notes into the ClinicalBERT model and averaging the resulting note-level representations for each patient.
3.3 Sepsis predictive module and loss function
Subsequently, we combine the ultimate patient-level clinical notes representations \(m_i \in \mathbb {R}^{t_0 \times d_0}\), with their corresponding structured data \(x_i \in \mathbb {R}^{t_0 \times d_1}\), to derive the ultimate patient features \(V_i \in \mathbb {R}^{t_0 \times (d_0 + d_1)}\), where i represents the i-th patient, \(t_0\) is the time window, and \(d_0\) and \(d_1\) are the length of feature. We chose the Bidirectional LSTM (BiLSTM) as our base network, which exhibits advantages in its ability to capture long-term dependencies and handle intricate patterns within sequential data, thereby enabling efficient and accurate predictions, classification, and analysis of time series data:
where \(\overrightarrow{U}\), \(\overleftarrow{U}\), \(\overrightarrow{W}\), and \(\overleftarrow{W}\) are the learnable parameters of our model, \(\overrightarrow{b}\) and \(\overleftarrow{b}\) represent the forward bias and backward bias of the model, respectively. N represents the total number of patients. Then \(\overrightarrow{k_i}\) and \(\overleftarrow{k_i}\) are concatenated and input into a point-wise Multilayer perceptron (MLP) network and get corresponding classification logits via a sigmoid function:
where \(h \in \mathbb {R}^{d \times c}\), c is the number of categories, \(\phi _c\left( \cdot \right) \) represents the classifier, z denotes logits. Then we use the binary cross-entropy loss as our target loss:
by optimizing \(\mathcal {L}_{cls}\), the classifier can achieve its optimal performance.
4 Experiments
We begin by outlining the criteria for selecting sepsis cases. Based on it, we introduce the preprocessing steps for structured and unstructured data of the study subjects. Finally, we expound on the baseline models used for comparison and provide detailed information regarding our model parameters.
4.1 Dataset and sepsis label definition
We utilized the MIMIC-III v1.4 dataset, sourced from the Beth Israel Deaconess Medical Center (BIDMC) in Boston, MA, spanning the period from 2001 to 2012 [42]. The MIMIC-III database is a freely accessible repository of clinical data that researchers worldwide can utilize. The database comprises de-identified clinical and physiological data from approximately 60,000 ICU admissions, along with clinical notes from over 50,000 ICU admissions. Like other EHR databases, it maintains comprehensive records of patients’ demographics, vital signs, laboratory tests, clinical notes, and additional data.
For each encounter with a suspicion of infection(SI), we extract a 72 h window around the first SI event (-48 h to 24 h) as the SI window. The SOFA score is then evaluated for every hour. Following the SOFA definition, to arrive at a SOFA score we considered the worst organ scores of the last 24 h. Figure is modified from [32]
We follow the latest sepsis-3 definition [1], which requires a co-occurrence of suspected infection (SI) and organ dysfunction. For SI, we implemented the SI cohort based on the recommendations of Seymour et al. [43]. Singer et al. [1] established that the organ dysfunction criterion is met when the SOFA score exhibits a minimum increase of 2 points. To ascertain this, we adopt the recommended approach by Singer et al., which involves a time window of -48 h to 24 h around the suspected infection. Figure 3 depicts the implementation of Sepsis-3 in our study. Early detection of sepsis is crucial, and accurately determining the onset time is vital. We followed the recommendation of Moor et al. [32] to establish the Sepsis-3 label on an hourly basis. If sepsis is determined solely by checking whether a patient meets the criteria upon admission, only patients who enter the ICU with sepsis would be considered cases, excluding the potentially more intriguing cases of patients developing the syndrome during their ICU stay.
4.2 Data filtering
4.2.1 Study cohort
We employed a set of patient inclusion criteria to determine the suitable study cohort. Patients below the age of 18 and those without available chart data, including ICU admission or discharge time, are excluded from the study. Additionally, patients without clinical notes were also excluded. An encounter is classified as a case if a sepsis onset occurs at any point during the ICU stay. Controls, on the other hand, consist of patients without sepsis onset (although they may exhibit suspected infection or organ dysfunction independently). Moreover, controls must not possess any sepsis-related ICD-9 billing code to ensure they are not patients who recently developed sepsis before admission to the ICU. This study focuses on early sepsis detection, following the approach of Moor et al. [32], we exclude cases that manifest sepsis within the first 6 hours of ICU admission due to the limited availability of physiological indicators. To maintain a realistic class balance of approximately 10%, we apply this exclusion step after the case-control matching process (refer to the next paragraph). Consequently, after the data cleaning and filtering process, we ultimately analyzed a cohort consisting of 659 cases and 6268 controls. The statistical results of characteristics are shown in Table 1. As illustrated in Table 2, we considered 44 irregularly sampled laboratory and vital features and 4 demographics followed by the recommendation of Moor et al [32].
4.2.2 Case-control matching
Previous studies [32, 44] have indicated that an inadequate alignment of time series between sepsis cases and controls can result in a trivial classification task. For example, comparing a time window before sepsis onset to the final window (before discharge) of an ICU stay for controls makes the classification task significantly easier compared to comparing it to a reference time within a control’s stay that is more similar. To prevent the classification task from becoming trivial, we employ a case-control alignment in a matching procedure. Additionally, to accommodate the class imbalance, each case is assigned to 10 randomly selected unassigned controls, and the control onset is defined as the absolute time (in hours since admission) when the matched case fulfills the sepsis criteria.
4.3 Data preprocessing
4.3.1 Structured data
We addressed the irregularity of time series and missing values before inputting each instance into the model. Firstly, we applied the time bucket technique to handle the irregularity by aggregating the data into 1-hour time intervals. The raw data was partitioned into consecutive 1-hour buckets, and the measurement values were averaged within each bucket, resulting in a time series with 1-hour intervals. Subsequently, missing values were imputed using forward filling during the data imputation phase. If no preceding records were available, missing values were filled with the mean of the feature within the population. In case a feature had entirely missing data, it was filled with zero.
4.3.2 Unstructured data
We conducted additional preprocessing on the clinical notes. The clinical notes available in the MIMIC-III V1.4 NOTEEVETS table encompass various types, such as nursing notes, nursing_other notes, physician notes, radiology notes, respiratory notes, case management notes, consult notes, discharge summaries, ECG notes, echo notes, general notes, nutrition notes, pharmacy notes, rehab services notes, and social work notes. However, we excluded the discharge summary from our analysis as it provides limited value for sepsis prediction since it pertains to the end of the ICU stay. Furthermore, we specifically utilized clinical notes recorded prior to the onset of sepsis in patients. See method Section 3.2 for more details about clinical notes processing.
We perform three iterations of random splitting, allocating 75% of the samples for training and 15% each for validation and testing. For each random split, the time series were standardized by calculating the z-scores per channel based on the corresponding mean and standard deviation of the training set. Since the sepsis prevalence in the overall cohort is 10%, we employed the ’WeightedRandomSampler’ method, which is a sampling technique available in the PyTorch library, utilized to perform a weighted random sampling of data during the training process in order to achieve a balanced representation.
4.4 Baseline
To assess the improvement in model prediction performance following the ChatGPT summarization of medical texts, we selected several baseline models for comparison. First, we compared the performance of our model with other commonly used predictive scoring systems in clinical practice, including SIRS, SOFA, qSOFA, and modified early warning system (MEWS) [45], which are standardized scoring systems for sepsis prediction. According to a meta-analysis study [4], these four scoring systems demonstrate typical performance measures. The area under the curve (AUC) ranges from 0.50 to 0.78, the true positive rate (TPR) ranges from 0.56 to 0.80, and the false-positive rate (FPR) ranges from 0.16 to 0.50 when assessed 4 hours before the onset of sepsis.
Additionally, We selected five existing machine-learning approaches: 1) Support Vector Machine (SVM): Horng et al. [11] use the SVM model for early detection of sepsis. 2) Residual Network (ResNet). He et al. [46] introduced this highly effective deep learning framework, which effectively tackles the issue of vanishing/exploding gradients and achieves excellent performance across various classification tasks. 3) Deep SOFA-Sepsis Prediction Algorithm (DSPA): Asuroglu and Ogul [47] developed a hybrid deep learning model that combines CNN and random forest approaches to predict the SOFA scores of sepsis patients. 4) Time-phAsed: Li et al. [48] developed an XGBoost-based method that achieves excellent performance. 5) Multitask Gaussian Process Attention Time Convolutional Network (MGP-AttTCN): Rosnati et al. [49] employed this TCN-based deep learning model for early prediction of sepsis occurrence. 6) LSTM: Amrollahi et al. [9] utilized an LSTM model to integrate clinical notes representations and structured data for sepsis prediction.
4.5 Implementation details
We evaluate the performance of the proposed and baseline methods using 5-fold cross-validation. The patients and their labels are divided equally into five groups. Four of these groups are used to train the classifiers, while the remaining group is used for evaluation purposes. This process is repeated five times to ensure that all data are tested once. The models are implemented using the PyTorch framework and trained using an NVIDIA RTX 4090 GPU with 24GB memory.
The hyperparameters considered in this study include the mapping dimension of the first fully connected layer, the dimension of the hidden state, the number of LSTM layers, and the batch size. Dropout was employed as a regularization technique in order to mitigate overfitting. It randomly zeros out a portion of neuron activations during training in neural networks. The model was trained using the Adam optimizer with an initial learning rate of 0.005. Dropout and learning rate values were also treated as hyperparameters. A learning rate schedule based on plateau [50] is applied during the model training. The hyperparameter search is conducted using Bayesian optimization [51], which offers the advantage of efficiently exploring the search space by leveraging prior knowledge and adaptively selecting new points for evaluation. This approach leads to faster convergence and improved performance compared to grid search. The loss function employed in this study is cross-entropy.
For feature dimension, we select 44 irregularly sampled laboratory and vital features. Additionally, we incorporated several demographics, including gender, age, ethnicity, and type of first ICU unit. We applied the one-hot method to convert these static feature values into a vector format. Consequently, a structured data features vector of size 58 is obtained. By concatenating representations from the last hidden layer, we obtained a vector of size 768, which further expanded to a final feature vector of size 826 after incorporating structured data features. Subsequently, this vector was inputted into an LSTM-based model for sepsis prediction. The schematic diagram of the proposed model is illustrated in Fig. 1.
5 Results
We first conduct a human evaluation to verify the high quality of summaries generated by ChatGPT. Then, we perform experiments on our dataset to further test our model’s performance. Finally, we carried out an ablation study and analyzed the experimental results in detail.
5.1 Human evaluation
To ensure readability and verify the faithfulness of the content generated by ChatGPT, we invited the expertise of two professional ICU doctors to conduct a human evaluation of the generated content. We refrained from utilizing automated metrics for summarization fields due to previous studies [16, 21] demonstrating their inadequacy in evaluating prompt-based summaries. Human evaluation is regarded as the most efficient approach to assess the content generated by LLMs. Figures 4 and 5 present two examples of reports generated by ChatGPT that summarize clinical notes.
Figure 4 illustrates the generated report by ChatGPT and elucidates the abbreviations of medical terms found in the original clinical note. Furthermore, Fig. 5 illustrates that the generated report effectively eliminates a significant amount of extraneous information not pertinent to the diseases discussed in the original note. Importantly, the generated content remains highly faithful to the original text, accurately capturing the information within the clinical notes. To ensure generality, we conducted a random selection of 100 reports generated by ChatGPT, along with their corresponding original notes, for human evaluation by ICU doctors. Doctors all expressed satisfaction for the generated reports, affirming that the summary content provided by ChatGPT effectively encompassed the essential information found in the original clinical notes.
Furthermore, we examined how limiting the length of content in the prompt affects the quality of report generation (Fig. 6). Three distinct prompts were chosen, requiring summarization of provided clinical notes within word limits of 15, 30, or 45, alongside an unrestricted prompt used in our study: "Summarize the following clinical notes". Figure 6 demonstrates that limiting the generated report to 15 or 30 words resulted in poor quality and omitted disease-related content. Limiting the generated report to 45 words covers the original content but lacks an explanation for abbreviations, resulting in poor readability. Not limiting the length of the generated report allows correct interpretation of abbreviations (e.g., "F" as "female," "MICU" as "Medical Intensive Care Unit," and "HTN" as "hypertension"). This enhances readability and maintains faithfulness to the original clinical note.
5.2 Classification result
The primary performance metric is the AUC score, which evaluates the model’s ability to differentiate between sepsis and non-sepsis patients. Additionally, metrics such as specificity and accuracy were employed for further analysis of the model performance. Accuracy quantifies the model’s proficiency in correctly identifying patients, while specificity gauges its capability to accurately identify patients without sepsis.
ROC curve for 4 h prediction before sepsis onset. The ROCs represent the performance of the early prediction algorithm at 4 h prior to the onset of sepsis. “qSOFA”, “SIRS”, “MEWS”, and “SOFA” represent the TPR and FPR from these methods employed by physicians in prior studies at 0-4 h prior to the onset of sepsis
Firstly, we conducted a comparative analysis between our proposed method and baselines for sepsis prediction. The ROC curve of our method, specifically focusing on the early prediction of sepsis four hours in advance, was plotted against these baselines and clinical scoring systems. Analysis of the ROC curve demonstrates that our proposed method achieved the highest AUC value of 0.93 (95% CI, 0.92-0.93) for predicting sepsis onset 4 hours in advance (Fig. 7). In Fig. 7, the horizontal and vertical coordinates represent the false positive rate (FPR) and true positive rate (TPR), respectively. These coordinates specifically depict the proportion of negative samples erroneously classified as positive (FPR) and the proportion of positive samples accurately identified as positive (TPR). The larger the area between the horizontal and vertical coordinates on the AUC curve, the better the performance of the model in the classification task. So it is evident that our method outperformed the baselines and the accuracy rates typically reported by human-based scoring methods, such as SIRA, SOFA, qSOFA, and MEWS, as indicated in previous studies [4].
Table 3 displays the AUC score, accuracy, specificity, and sensitivity obtained by our method and baselines in this experiment for prediction windows of 4 h, 6 h, and 12 h. The results demonstrate that our method consistently achieves the highest AUC score across all prediction windows. However, as the prediction window increases in duration, the AUC score of our method decreases from 0.93 (95% CI, 0.92-0.93) for the 4 h prediction window to 0.92 (95% CI, 0.91-0.93) for the 12 h prediction window. Notably, our method exhibits a higher specificity of 0.89 (95% CI, 0.86-0.91) at the 4 h prediction window when compared to the LSTM’s specificity of 0.88 (95% CI, 0.86-0.89) and MGP-AttTCN’s specificity of 0.78 (95% CI, 0.75-0.81). This outcome aligns with the calculated AUC scores, which serve as measures of sensitivity and specificity. Additionally, our method demonstrates superior accuracy compared to all baselines for prediction windows of 4 h, 6 h, and 12 h. We also conducted comparisons with other state-of-the-art methods, and the results of these comparisons are presented in Table 4. Considering that a majority of these methods primarily target predicting sepsis six hours in advance, we assessed the performance of our approach against them specifically at the six-hour mark for fairness. Experimental findings demonstrate the consistent superiority of our approach over the compared methods in predicting sepsis six hours ahead.
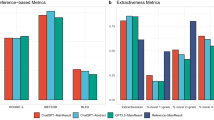
We also compared the summary reports generated by ChatGPT with reports generated by different summarization models. We selected the SOTA summarization models T0 [23] and BRIO [22] as comparison models, and the results of the experiment are shown in Fig. 8. To ensure a fair comparison, we opted for the versions of T0 and BRIO that were accessible during our research and employed an identical prompt. We independently employed the summary reports generated by these three models in the early prediction of sepsis. Experimental findings demonstrate that the summary reports generated by ChatGPT consistently outperform the others. In essence, the quality of ChatGPT-generated summary reports exceeds that of the comparative models. T0 and BRIO yielded similar outcomes, closely mirroring those achieved without the application of summarization models. This phenomenon may be explained by the fact that T0 and BRIO were originally designed for news summarization and may not be ideally suited for clinical notes summarization. This reinforces the robust capabilities of ChatGPT in the field of clinical notes summarization.
Furthermore, for enhanced validation of ClinicalBERT’s effectiveness, we employed the classic term frequency-inverse document frequency (TF-IDF) method as an alternative for contextualized embedding representations of ClinicalBERT. In TF-IDF embedding, the value of a word increases proportionally with its frequency within a document but is inversely adjusted by the number of documents containing that word. After excluding terms with unusually high or low frequency, we constructed a term-frequency matrix consisting of 1668 distinct medical terms. We conducted experiments on two NLP models using independent test sets to predict sepsis early within 4 h, 6 h, and 12 h prediction windows. The experimental results are presented in Fig. 9. The prediction outcomes achieved using ClinicalBERT for clinical note representation outperformed those obtained using TF-IDF across the 4 h, 6 h, and 12 h prediction windows. This superiority can be attributed to the limitations of TF-IDF, which represents all potential meanings of a word as a single vector and fails to disambiguate word senses based on contextual information and model negatives. In contrast, ClinicalBERT provides context-sensitive embeddings for each word in a given sentence, enhancing its predictive capabilities.
To assess the impact of processing text using ChatGPT on experimental results, we conducted an ablation study, and the results of this study are presented in Table 5. The findings presented in Table 5 reveal that the utilization of unstructured data alone leads to the lowest performance in each time window, which aligns with real-world clinical practices. In actual clinical operations, unstructured data serves as a supporting tool for doctors to assess a patient’s condition, while the primary focus lies on structured data, such as vital signs, for determining the patient’s condition. While the performance of using both clinical notes and structured data without employing ChatGPT surpasses that of using only structured data, our proposed method outperforms it in each scoring metric for the prediction windows of 4 h, 6 h, and 12 h. Specifically, when predicting sepsis four hours in advance, utilizing summary text with ChatGPT (Model IV 0.93, 95% CI, 0.92-0.93) resulted in a 2% improvement in AUC score compared to not employing ChatGPT (Model III 0.91, 95% CI, 0.90-0.93). This result provides evidence that the inclusion of noise in the clinical note, such as abbreviations and non-disease-related content, has a significant impact on the model’s predictions. Moreover, it confirms that ChatGPT successfully eliminates the noise present in the original clinical note and the generated summary report is faithful to the original clinical note.
Specifically, we also report the results obtained at 0 h prior to sepsis onset. Predictions within this time frame remain meaningful due to the potential delay between sepsis occurrence and the corresponding diagnosis. This delay can arise from the time required for laboratory tests to be conducted and results to be processed, which can span several hours [10]. Interestingly, our observations indicate that the use of structured data alone (Model I) achieves the best performance across all the evaluation metrics, while using unstructured data alone (Model II) yields the poorest performance. Even within the 0 h prediction window, our method (Model IV) exhibits an AUC score equivalent to that of using only structured data (Model I) and outperforms Model III in all evaluation measures. This finding suggests that as the onset of sepsis approaches, structured data increasingly reflect the patient’s health status, making unstructured data less influential, which aligns with the conclusion drawn by [8].
6 Discussion
Prior research and medical practice [55] have demonstrated the challenges associated with early sepsis detection, as sepsis patients are susceptible to rapid deterioration. Therefore, timely diagnosis is crucial in sepsis management. Accurate early prediction of sepsis can significantly improve the survival rate of septic patients and reduce hospital costs. Our LLMs-assisted deep learning algorithms have the potential to facilitate early sepsis detection, enabling clinicians to intervene and manage sepsis patients more effectively. Our study demonstrated that our method outperformed all baseline models and clinical score systems in predicting sepsis onset within 4 h, 6 h, and 12 h windows. Notably, the ablation study demonstrated that our method exhibited a performance improvement of 2% (0.93, 95% CI, 0.92-0.93) when predicting sepsis four hours in advance, surpassing the results obtained by utilizing only structured data (0.91, 95% CI, 0.91-0.91) and unstructured data without ChatGPT summary and structured data (0.91, 95% CI, 0.90-0.93). Furthermore, our method significantly outperformed traditional clinical scoring systems. This additional lead time in sepsis alert provides greater opportunities for physicians to commence treatment, thereby lowering mortality and cost.
We utilized ChatGPT to polish the noise contained in clinical notes and summarize the clinical notes. The resulting report was then used to generate patient-level representations using clinicalBERT, which were subsequently combined with structured data for sepsis early prediction. This approach yields improved results compared to not employing ChatGPT for processing clinical notes in order to achieve earlier sepsis prediction. To ensure the readability and faithfulness to the original notes of the content generated by LLMs like ChatGPT, we invited an expert ICU doctor to conduct a human evaluation of the reports generated by ChatGPT. The physician expressed satisfaction with the generated report, noting its high readability and faithfulness to the original clinical notes. Moreover, the generated report effectively captures the disease-related information from the original clinical note.
As expected, we achieved the best performance by utilizing only structured data to predict sepsis at the 0 h prediction window, which is consistent with prior study [8]. The findings indicate that as the onset of sepsis approaches, measurable symptoms, such as decreased blood pressure values, become evident in the structured variables. However, in this case, incorporating clinical notes yields only marginal improvements in prediction accuracy, given that the structured variables encompass the majority of sepsis symptoms. From a different perspective, our results in turn demonstrate the high quality of ChatGPT summaries. Given the inadequacy of existing automatic metrics in assessing the quality of content generated by LLMs [21], employing the generated content for downstream tasks and subsequently evaluating the quality of the generated content based on experimental results may offer an alternative approach for assessing the performance of LLMs in specific domains. This presents an interesting prospect.
However, this study has several limitations. Since our model is not end-to-end, additional resources and time are still consumed in the data preprocessing phase. Therefore, in future work, we will combine the whole data preprocessing stage with model training to construct a complete end-to-end prediction model to improve the timeliness of diagnosis. In light of time constraints, we have employed a traditional time series prediction model in this manuscript. To determine whether a BiLSTM-derived variant model can yield improved prediction results, further experiments are required. In future research, we will enhance the models employed in this manuscript and incorporate the unique attributes of medical data to enhance the early predictive accuracy of sepsis.
In conclusion, our approach has revealed the potential of LLMs in processing medical data and has demonstrated superior performance in predicting downstream tasks associated with sepsis compared to the baselines, which enables more time for sepsis intervention and management. Notably, our method can be easily applied to other tasks such as the prediction of morbidity and mortality of other diseases.
7 Conclusion
In this paper, we propose an LLMs-based deep learning algorithm for early sepsis prediction. We employ ChatGPT to denoise and summarize clinical notes, and then use the generated reports in combination with structured data to feed into an LSTM-based model for early sepsis prediction. Experimental results show that the proposed method outperforms traditional models and clinical scoring systems. Moreover, we conducted a comprehensive analysis of the impact of different prompts on the report generated by ChatGPT and performed a human evaluation of the generated reports. Experimental results show that in the domain of clinical note summarization, not limiting the length of ChatGPT generated report will achieve better results. Furthermore, ICU doctors perceive the generated summary reports as readable, faithful to the original notes, and accurately capture the information within the original clinical notes.
Data Availability
The datasets generated during and analyzed during the current study are available from the corresponding author upon reasonable request.
References
Singer M, Deutschman CS, Seymour CW, Shankar-Hari M, Annane D, Bauer M, Bellomo R, Bernard GR, Chiche J-D, Coopersmith CM et al (2016) The third international consensus definitions for sepsis and septic shock (sepsis-3). JAMA 315(8):801–810
Kumar A, Roberts D, Wood KE, Light B, Parrillo JE, Sharma S, Suppes R, Feinstein D, Zanotti S, Taiberg L et al (2006) Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit Care Med 34(6):1589–1596
Seymour CW, Gesten F, Prescott HC, Friedrich ME, Iwashyna TJ, Phillips GS, Lemeshow S, Osborn T, Terry KM, Levy MM (2017) Time to treatment and mortality during mandated emergency care for sepsis. N Engl J Med 376(23):2235–2244
Islam MM, Nasrin T, Walther BA, Wu C-C, Yang H-C, Li Y-C (2019) Prediction of sepsis patients using machine learning approach: a meta-analysis. Comput Methods Programs Biomed 170:1–9
Duan Y, Huo J, Chen M, Hou F, Yan G, Li S, Wang H (2023) Early prediction of sepsis using double fusion of deep features and handcrafted features. Applied Intelligence, 1–17
Reyna MA, Josef C, Seyedi S, Jeter R, Shashikumar SP, Westover MB, Sharma A, Nemati S, Clifford GD (2019) Early prediction of sepsis from clinical data: the physionet/computing in cardiology challenge 2019. In: 2019 computing in cardiology (CinC), p 1 . IEEE
Jensen K, Soguero-Ruiz C, Oyvind Mikalsen K, Lindsetmo R-O, Kouskoumvekaki I, Girolami M, Olav Skrovseth S, Augestad KM (2017) Analysis of free text in electronic health records for identification of cancer patient trajectories. Sci Rep 7(1):46226
Goh KH, Wang L, Yeow AYK, Poh H, Li K, Yeow JJL, Tan GYH (2021) Artificial intelligence in sepsis early prediction and diagnosis using unstructured data in healthcare. Nat Commun 12(1):711
Amrollahi F, Shashikumar SP, Razmi F, Nemati S (2020) Contextual embeddings from clinical notes improves prediction of sepsis. In: AMIA annual symposium proceedings, vol 2020, p 197. American medical informatics association
Qin F, Madan V, Ratan U, Karnin Z, Kapoor V, Bhatia P, Kass-Hout T (2021) Improving early sepsis prediction with multi modal learning. arXiv preprint arXiv:2107.11094
Horng S, Sontag DA, Halpern Y, Jernite Y, Shapiro NI, Nathanson LA (2017) Creating an automated trigger for sepsis clinical decision support at emergency department triage using machine learning. PLoS ONE 12(4):0174708
Apostolova E, Velez T (2018) Toward automated early sepsis alerting: identifying infection patients from nursing notes. arXiv preprint arXiv:1809.03995
Culliton P, Levinson M, Ehresman A, Wherry J, Steingrub JS, Gallant SI (2017) Predicting severe sepsis using text from the electronic health record. arXiv preprint arXiv:1711.11536
Alsentzer E, Murphy JR, Boag W, Weng W-H, Jin D, Naumann T, McDermott M (2019) Publicly available clinical bert embeddings. arXiv preprint arXiv:1904.03323
Yan MY, Gustad LT, Nytrø Ø (2022) Sepsis prediction, early detection, and identification using clinical text for machine learning: a systematic review. J Am Med Inform Assoc 29(3):559–575
Zhang T, Ladhak F, Durmus E, Liang P, McKeown K, Hashimoto TB (2023) Benchmarking large language models for news summarization. arXiv preprint arXiv:2301.13848
Brown T, Mann B, Ryder N, Subbiah M, Kaplan JD, Dhariwal P, Neelakantan A, Shyam P, Sastry G, Askell A et al (2020) Language models are few-shot learners. Adv Neural Inf Process Syst 33:1877–1901
Leiter C, Zhang R, Chen Y, Belouadi J, Larionov D, Fresen V, Eger S (2023) Chatgpt: A meta-analysis after 2.5 months. arXiv preprint arXiv:2302.13795
Wiering MA, Van Otterlo M (2012) Reinforcement learning. Adapt Learn Optim 12(3):729
Nie W, Wen X, Liu J, Chen J, Wu J, Jin G, Lu J, Liu A-A (2023) Knowledge-enhanced causal reinforcement learning model for interactive recommendation. IEEE Transactions on Multimedia
Goyal T, Li JJ, Durrett G (2022) News summarization and evaluation in the era of gpt-3. arXiv preprint arXiv:2209.12356
Liu Y, Liu P, Radev D, Neubig G (2022) Brio: Bringing order to abstractive summarization. arXiv preprint arXiv:2203.16804
Sanh V, Webson A, Raffel C, Bach SH, Sutawika L, Alyafeai Z, Chaffin A, Stiegler A, Scao TL, Raja A, et al. (2021) Multitask prompted training enables zero-shot task generalization. arXiv preprint arXiv:2110.08207
Polat G, Ugan RA, Cadirci E, Halici Z (2017) Sepsis and septic shock: current treatment strategies and new approaches. Eurasian J Med 49(1):53
Jaimes F, Garcés J, Cuervo J, Ramírez F, Ramírez J, Vargas A, Quintero C, Ochoa J, Tandioy F, Zapata L et al (2003) The systemic inflammatory response syndrome (sirs) to identify infected patients in the emergency room. Intensive Care Med 29:1368–1371
Levy MM (2003) Sccm/esicm/accp/ats/sis. 2001 sccm/esicm/accp/ats/sis. international sepsis definitions conference. Crit Care Med 31:1250–1256
Jones AE, Trzeciak S, Kline JA (2009) The sequential organ failure assessment score for predicting outcome in patients with severe sepsis and evidence of hypoperfusion at the time of emergency department presentation. Crit Care Med 37(5):1649
Calvert JS, Price DA, Chettipally UK, Barton CW, Feldman MD, Hoffman JL, Jay M, Das R (2016) A computational approach to early sepsis detection. Comput Biol Med 74:69–73
Desautels T, Calvert J, Hoffman J, Jay M, Kerem Y, Shieh L, Shimabukuro D, Chettipally U, Feldman MD, Barton C et al (2016) Prediction of sepsis in the intensive care unit with minimal electronic health record data: a machine learning approach. JMIR Med Inform 4(3):5909
Mao Q, Jay M, Hoffman JL, Calvert J, Barton C, Shimabukuro D, Shieh L, Chettipally U, Fletcher G, Kerem Y et al (2018) Multicentre validation of a sepsis prediction algorithm using only vital sign data in the emergency department, general ward and icu. BMJ Open 8(1):017833
Yang M, Wang X, Gao H, Li Y, Liu X, Li J, Liu C (2019) Early prediction of sepsis using multi-feature fusion based xgboost learning and bayesian optimization. In: The IEEE conference on computing in cardiology (CinC), vol 46, pp 1–4
Moor M, Horn M, Rieck B, Roqueiro D, Borgwardt K (2019) Early recognition of sepsis with gaussian process temporal convolutional networks and dynamic time warping. In: Machine learning for healthcare conference, pp 2–26 . PMLR
Shashikumar SP, Josef C, Sharma A, Nemati S (2019) Deepaise–an end-to-end development and deployment of a recurrent neural survival model for early prediction of sepsis. arXiv preprint arXiv:1908.04759
Liu R, Greenstein JL, Sarma SV, Winslow RL (2019) Natural language processing of clinical notes for improved early prediction of septic shock in the icu. In: 2019 41st annual international conference of the IEEE engineering in medicine and biology society (EMBC), pp 6103–6108 . IEEE
Armi L, Abbasi E, Zarepour-Ahmadabadi J (2021) Texture images classification using improved local quinary pattern and mixture of elm-based experts. Neural Computing and Applications, 1–24
Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, Kaiser Ł, Polosukhin I (2017) Attention is all you need. Advances in neural information processing systems. 30
Nie W, Chang R, Ren M, Su Y, Liu A (2021) I-gcn: incremental graph convolution network for conversation emotion detection. IEEE Trans Multimedia 24:4471–4481
Wen X, Nie W, Liu J, Su Y (2023) Mrft: Multiscale recurrent fusion transformer based prior knowledge for bit-depth enhancement. IEEE Trans Circ Syst Video Technol
Devlin J, Chang M-W, Lee K, Toutanova K (2018) Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv preprint arXiv:1810.04805
Mikolov T, Chen K, Corrado G, Dean J (2013) Efficient estimation of word representations in vector space. arXiv preprint arXiv:1301.3781
Pennington J, Socher R, Manning CD (2014) Glove: Global vectors for word representation. In: Proceedings of the 2014 conference on empirical methods in natural language processing (EMNLP), pp 1532–1543
Johnson AE, Pollard TJ, Shen L, L-wH Lehman, Feng M, Ghassemi M, Moody B, Szolovits P, Anthony Celi L, Mark RG (2016) Mimic-iii, a freely accessible critical care database. Scientific data 3(1):1–9
Seymour CW, Liu VX, Iwashyna TJ, Brunkhorst FM, Rea TD, Scherag A, Rubenfeld G, Kahn JM, Shankar-Hari M, Singer M et al (2016) Assessment of clinical criteria for sepsis: for the third international consensus definitions for sepsis and septic shock (sepsis-3). JAMA 315(8):762–774
Futoma J, Hariharan S, Heller K, Sendak M, Brajer N, Clement M, Bedoya A, O’brien C (2017) An improved multi-output gaussian process rnn with real-time validation for early sepsis detection. In: Machine learning for healthcare conference, pp 243–254 . PMLR
Subbe CP, Kruger M, Rutherford P, Gemmel L (2001) Validation of a modified early warning score in medical admissions. QJM 94(10):521–526
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 770–778
Aşuroğlu T, Oğul H (2021) A deep learning approach for sepsis monitoring via severity score estimation. Comput Methods Programs Biomed 198:105816
Li X, Xu X, Xie F, Xu X, Sun Y, Liu X, Jia X, Kang Y, Xie L, Wang F et al (2020) A time-phased machine learning model for real-time prediction of sepsis in critical care. Crit Care Med 48(10):884–888
Rosnati M, Fortuin V (2021) Mgp-atttcn: An interpretable machine learning model for the prediction of sepsis. PLoS ONE 16(5):0251248
Wang Z, Yan W, Oates T (2017) Time series classification from scratch with deep neural networks: A strong baseline. In: 2017 International joint conference on neural networks (IJCNN), pp 1578–1585 . IEEE
Frazier PI (2018) A tutorial on bayesian optimization. arXiv preprint arXiv:1807.02811
Hongyi Li G (1996) Maddala: Bootstrapping time series models. Economet Rev 15(2):115–158
Liu S, Fu B, Wang W, Liu M, Sun X (2022) Dynamic sepsis prediction for intensive care unit patients using xgboost-based model with novel time-dependent features. IEEE J Biomed Health Inform 26(8):4258–4269
Ding R, Rong F, Han X, Wang L (2023) Cross-center early sepsis recognition by medical knowledge guided collaborative learning for data-scarce hospitals. arXiv preprint arXiv:2302.05702
Rhodes A, Evans LE, Alhazzani W, Levy MM, Antonelli M, Ferrer R, Kumar A, Sevransky JE, Sprung CL, Nunnally ME et al (2017) Surviving sepsis campaign: international guidelines for management of sepsis and septic shock: 2016. Intensive Care Med 43:304–377
Acknowledgements
This work was supported by the Foundation of State Key Laboratory of Ultrasound in Medicine and Engineering (Grant No.2022KFKT004) and Tianjin Health Science and Technology Project (Grant NO TJWJ2023ZD006).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical standard
The authors state that this research complies with ethical standards. This research does not involve either human participants or animals.
Conflicts of interest
The authors have no competing interests to declare that are relevant to the content of this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, Q., Ma, H., Song, D. et al. Early prediction of sepsis using chatGPT-generated summaries and structured data. Multimed Tools Appl (2024). https://doi.org/10.1007/s11042-024-18378-7
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11042-024-18378-7