Abstract
This paper proposes a novel brain tumor segmentation algorithm that uses Active Contour Model and Fuzzy-C-Means optimization. In Active Contour model, the initial Contour selection is a challenging task for MRI brain tumor segmentation because the accuracy of active contour segmentation depends on initial contour. This method uses the two level morphological reconstruction processes such as Dilation and Erosion along with thresholding process for minimizing the non-tumor region. The segmented region thus obtained is not accurate that also contains non-tumor region. Also there is a chance of missing the tumor region along with background while performing two level morphological reconstructions. In order to overcome these issues, active contour model is used to segment the complete tumor part. The initial Contour for Active Contour model is detected by forming a circular region around the tumor region. The radius of the circular region is contracted or expanded based on the shape of the tumor. This proposed Radius Contraction and Expansion (RCE) technique is used to select the initial contour of active Contour model. Further Fuzzy-C-Means algorithm is used to optimize the edge pixels because the boundary of active contour model output also contains the non tumor pixels. The performance of the proposed segmentation algorithm was evaluated using the metrics such as specificity, sensitivity, dice score, Probabilistic Rand Index (PRI) and Hausdorff Distance (HD) on T1- weighted contrast enhanced image dataset. The experimental result shows that the proposed segmentation algorithm provides a good performance when compared to the state-of-the-art segmentation methods.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
The growth or proliferation of cells which destroys the normal cells, that forms a mass in brain is termed as brain tumor. Brain tumor [6] can be classified into two different types such as Benign (non-cancerous) and Malignant (cancerous). Benign is a primary type brain tumor that does not spread to other organs. Malignant is a secondary or metastatic type brain tumor that spread to other organs. The Intra Cranial Pressure (ICP) increases, when the growth of the tumor inside the brain gets increased. This increase in ICP can cause brain damage and leads to death. For a normal person, the new cells start growing when the old cell stop its growth and gets damaged. For a tumor person the growth of cells goes wrong ie., the new cell will grow when the old cells are not damaged. This leads to building of huge number of cells (extra cells) which forms a mass of tissue termed as tumor. A survey in the US states that in the year 2010, around 62,930 new cases found in adults and 4030 new cases found in children. It is necessary to diagnosis the tumor at the early stage, inorder to reduce the death caused due to brain tumor.
The tumors can be classified in four grades as Grade-I, Grade-II, Grade-III and Grade-IV. In Grade-I type the brain cells look normal and they grow slowly. In this grade the tumor is Benign type. In Grade-II, the tumor cells looks less like normal cells than the cells in Grade-I type. In Grade-III the tumor cells look different from normal cells, where the cells grow actively. In Grade-IV the tumor cells appear to be more abnormal and the growth of tumor is very high when compared to Grade-III type. Grade-II, Grade-III and Grade-IV are malignant type tumor. The tumors are named based on the part in which they originate. For example the tumor Glioma type originates in glial cells of brain.
Magnetic Resonance Imaging (MRI) [26] provides a high contrast between the tissues of tumor and non-tumor region. Detection of tumor [5] at the early stage is necessary to reduce the death rate caused by brain tumor. Diagnosing the tumor manually by the physician requires more time and also causes incorrect results in tumor segmentation. Therefore automatic brain tumor segmentation algorithm is essential for accurate and fast diagnosis of brain tumor. Most of brain tumor segmentation algorithm [13] can be categorized in four classes of segmentation which includes, (i) Edge detection (ii) Thresholding (iii) Region based segmentation (iv) Clustering approach.
Segmentation based on edges segments tumor region and non tumor region by detecting the edges where there is a high change in pixel intensity. Operator like Sobel operator, Canny operator, Prewitts operators and Laplacian operators are commonly used operators to detect the edges between tumor and non tumor region. Gradient based segmentation is an example of edge based segmentation, where the segmentation is done based on the intensity difference between the particular pixel and its neighbour pixel. Based on the intensity of the pixel present in the MRI image, the thresholding [15] approach partitions the images into different classes (two or more classes). Global thresholding and local thresholding algorithm are the two basic thresholding approaches that are used for brain tumor segmentation. Global thresholding is used in MRI images that has uniform and high contrast intensities between tumor and non-tumor region, since it uses a single threshold over the entire image. Otsu’s thresholding is an example of Global thresholding algorithm where it partitions the images into different classes of intensities.
The region based segmentation method, segments the image into two or more region based on certain rules. The method such as region growing, region splitting and merging are example for region based segmentation. Usually, a seed pixel is chosen in region growing method and the region begins to grow based on the difference between intensity of seed pixel region and the neighbouring region. Once the segmentation gets completed, the growing condition does not satisfy and thus the segmentation stops. Such region growing methods are effective where the Region of Interest (ROI) is small such as Brain tumor. In region splitting and growing approach, the image to be segmented is divided into smaller regions until the splitting condition fails. The merging process merges the splitted image based on the uniformity between the regions. The merging process also stops, if the merging condition is not satisfied. Clustering [2]–[12] methods groups the pixel that has large probability in the same category. This method does not require any training process and it clusters a pixel into different classes. This clustering methods are mainly used for optimized which includes K-means [35] and Fuzzy-C-Means [20]. A semi automatic system needs input from the physician mainly for three reasons, such as initializing the segmentation, feedback while segmenting and evaluation of segmented results. The proposed work is automatic segmentation method which does not need any additional input from the user.
The forthcoming sections of the paper are arranged as follows. Section 2 shows the related work, Section 3 shows the proposed Brain tumor segmentation algorithm, Section 4 shows the experimental results and analysis of the proposed segmentation algorithm and finally section 5 shows the conclusion of the paper.
2 Related works
Several research works on brain tumor segmentation which mainly includes the schemes such as K-means algorithm [35], Gaussian mixture model algorithm [22] and Fuzzy-C-Means algorithm [20] are going on,
The seed point selection is a major factor in K-means segmentation algorithm. The Gaussian Mixture Model (GMM) uses maximum likely-hood parameters and filtering approach for image segmentation. The FCM (Fuzzy C-Means) technique provides a high accurate result, if there is fuzziness in edge pixel intensities of the tumor. Apart from K-means, GMM and FCM techniques, the commonly used techniques are edge based segmentation, thresholding and region based methods. There are different types of region based methods [37] such as region growing, region splitting and merging, segmentation based on watershed, snake model, active contour model etc. In snake model, the initial contour or initial snake is initially placed around the Region of Interest (ROI). Due to external and internal energy, the snake moves and stops where there is minimum energy function.
Deep Neural Networks [8] [7] [11] can also be used for brain tumor segmentation where Mohamad Havaei et al. [16] proposed a convolutional Neural Network. This neural network uses both local features and global contextual features simultaneously. The last layer of neural network is a two phase training system with fully connected layer. Localized Active Contour Model (LACM) [19] balances the mean intensity between the tumor region. The LACM aims to reduce the movement of Contour in the undesired boundaries of the tumor. The LACM algorithm detects the tumor region with the help of Hierarchical Centroid Shape Descriptor (HCSD).
The Potential Field Segmentation (PFS) was proposed by Ivan Cabria et al. It also fuse the result of PFS to obtain the actual segmentation result. In [9] the potential field is calculated for every pixel and the pixel intensity is viewed as a mass. The potential field that is estimated for each pixel is compared with the adaptive potential threshold. If the potential field of a pixel is greater than the adaptive potential threshold, then the corresponding pixel will be considered as non- tumor pixel.
In [23] the brain tumor segmentation and its volumetric analyses were proposed using a Computer aided tool. In [19] optimal correlation is used to detect the initial contour. The initial contour is then used to detect the tumor region. Spectral clustering can be used to segment the tumor region which was proposed by Angulakshmi and Lakshmi Priya [3] . This method uses the super-pixels to identify the tumor region.
The tumor region can be clearly distinguished using the segmentation method proposed by Umit Ilhan et al. [18]. At the end of segmentation process, the tumor tissues will be clearly visible, so that a medical practitioner can easily identify the tumor region. So this method highly helps the medical practitioner. This segmentation method uses different process such as morphological operation, pixel subtraction, thresholding and filtering.
Specific fine tuning of deep learning was proposed by Guotai Wang et al. [38]. This method uses convolutional neural network along with interactive segmentation based deep learning. Further supervised image or unsuperivesed image fine tuning can be used. The unsupervised fine tuning does not require any user interaction, while supervised fine tuning requires additional user interaction. The additional user interaction can be done using interaction based uncertainty. The weighted loss function can be used to design the convolutional neural network. The 2D fetal MRI image segmentation for multiple organs can be achieved using this method. The 3D brain tumor segmentation can be done either by brain tumor core segmentation or whole brain tumor segmentation. The whole brain tumor based segmentation includes the edema region while the brain tumor core based segmentation exclude the edema.
In [34], the authors proposed a brain tumor segmentation algorithm based on ECNN (Enhanced Convolutional Neural Networks). In this method, the skull region present in the image is initially removed. The ECNN proposes an loss function optimization using BAT algorithm. This algorithm has the advantage of less weight with respect to over-fitting of convolutional neural network. In [17], Kai Hu et al. proposed a multi-cascaded convolutional neural networks and conditional random field (CRF). This method completes the segmentation process in two steps. In the first step, the intermediate results of the connected components are combined and this process leads to coarse segmentation of multi scale features by considering the local dependencies of labels. In the second step, fine segmentation is performed by eliminating some undesired outputs. This minimization is done by considering the spatial contextual information. This algorithm also uses image patches to train the segmentation models.
In [24], the authors proposed an end-to-end incremental Deep Neural Network based automatic brain tumor segmentation. This segmentation algorithm uses trial and error method which uses ensemble learning for efficient model design. Inorder to eliminate the problem of CNN training model, this method uses a new training strategy that considers only the most influencing hyper- parameters. This can be achieved by bounding and setting a roof to the parameters, which increases the speed of training process. In [25], the authors proposed a novel method for the classification of gait which uses an Extreme Learning Machine (ELM) algorithm. The ELM algorithm uses eight stages such as data collection, gait detection, trajectories smoothing, feature extraction, selection and classification. The authors compared the ELM results with the classification algorithms such as KNN, SVM and MLP.
In [27], the authors proposed biometric gait identification using multi-layered perception. They classified the gait into four classes such as crouch-2, crouch-3, crouch-4 and normal. The algorithm uses data collection of vision based gait and sensor based gait and the two gaits are fused to obtain the fused data, from which the principle features are extracted. The extracted features are trained using MLP algorithm. In [30], the authors have proposed vector fields of six joints in knee, hip and joints. In [28], the authors proposed a humanoid push recovery and classification. The features are extracted by using empirical mode decomposition. Four kinds of pushes are classified such as small, medium, moderately high and high using Deep Neural Network. Several classification algorithms are also used to classify the gait data [29, 31].
In [4], the authors proposed a brain tumor detection and feature extraction based on Berkeley Wavelet Transform (BWT) and support vector machine. This method, initially pre-processes the MRI image for image enhancement. The skull part present in the pre-processed image is removed and the tumor region is then segmented and features are extracted using mean, contrast, entropy and energy. The features are then classified using support vector machine to classify the normal tissue and abnormal tissue. In [39], the authors proposed infant brain image segmentation for multi modalities iso-intense using deep convolutional neural networks. In this, the hierarchical features are learned using deep learning models where low level features are transformed to high level features. The high level complex features are obtained from the row input images using trainable filters along with local neighbourhood pooling operation that are applied alternatively. The CNN first layer has 64 feature maps and each feature maps are linked to 3 input feature maps using the filter having a size of 5 × 5.
In [40], the author has proposed a brain tumor segmentation algorithm by integrating Fully Convolutional Neural Network (FCNN) and Conditional Random Fields (CRF). In this, they have trained the 2D images using deep learning based segmentation model. This learning algorithm has three major steps. In first step, the image patches are trained using fully convolutional neural network and in the second step,the image slices are trained using Conditional Random Field and Recurrent Neural Networks. In the third step, the fully Convolutional Neural Networks and Recurrent Neural Network are fine tuned. Here a voting based fusion strategy is used to combine the segmented brain tumors. In [36], the authors proposed an Automatic Brain Tumor segmentation method on Enhanced Darwinian Particle Swarm Optimization (EDPSO). The first two steps involved in this process are pre-processing and image enhancement. The enhanced image is segmented using EDPSO algorithm. The segmented image is trained and classified using ANFIS algorithm which is the combination of Artificial Neural Network and Fuzzy logic.
In [1], the authors proposed an ensemble brain tumor classification algorithm that uses Fuzzy anisotropic diffusion for brain tumor segmentation. This method initially processes the image for noise removals. Then the tumor region is then segmented from which the features are extracted. Two types of features are extracted which are first order histogram features and co-occurrence matrix based features. The segmentation process includes the steps such (i) Skull removal (ii) Brain region extraction (iii) Brain tumor extraction. The features extracted by feature extraction process are trained using ensemble based classification.
In [21], the authors proposed an MRI brain tumor segmentation algorithm using multi-modality aggregation network. The concept of Multi-Modality Aggregation Network (MMAN) is derived from the concept of deep learning. The MMAN algorithm can extract features like multi-scale features from brain tissues. It can also extract secondary information such as harness complementary information which is used to increase the speed and accuracy of segmentation. In [33], the authors proposed brain image segmentation from MRI images using optimal multi-level thresholding. In this, they have proposed a differential evolution algorithm that balances exploitation and exploration. The differential evolution was derived using a new adaptive approach and new mutation strategies. The optimal solution can be provided using the new adaptive approach by estimating the candidate solution quality.
2.1 Our previous work
In our previous work [32] we have proposed an automatic brain tumor segmentation using Greedy snake model and FCM optimization. In that we have used two levels of morphological reconstruction such as dilation and erosion. The erosion process is used as level 1 morphological reconstruction and dilation is used as level 2 morphological reconstructions. This two level morphological reconstruction process is used for background minimization. The segmentation of brain tumor is done using Greedy snake algorithm. The initial Contour of snake is estimated by applying thresholding and dilation to the background minimized image. The greedy snake algorithm is applied to the pre-processed image using the initial Contour obtained. The segmented brain tumor is not accurate and edges also has non-tumor edge pixel in the segmented Region of Interest obtained by the Greedy snake segmentation algorithm. The Greedy snake segmented output is optimized using Fuzzy-C-Means optimization algorithm. Finally, the accurate region is selected based on maximum perimeter from multiple regions to obtain the segmented output.
The proposed algorithm replaces Greedy snake algorithm by Active Contour model. Also the proposed paper proposes a novel initial Contour selection method known as Radius Contraction and Expansion (RCE) for active contour model. The next section shows the proposed brain tumor segmentation algorithm.
3 Proposed work
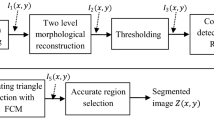
Figure 1 shows the block diagram of proposed brain tumor segmentation algorithm. The proposed segmentation algorithm has stages such as (i) Pre-processing (ii) Background Minimization (iii) Initial Contour Estimation using Radius Contraction and Expansion (RCE) (iv) Active Contour model Segmentation (v) Fuzzy C-means optimization and Region selection. The working of these blocks are discussed below.
3.1 Pre-processing
Before actual segmentation, the noise present in the MRI image must be removed for accurate segmentation result. While filtering, the important features present in the image must be preserved. Median filter is one of the commonly used filters that remove the noise present in the MRI image while it preserves the important features such as edge information. Let the original MRI image be represented as I1(x, y) and the pre-processed image (median filtered) be represented by
w is the 3 × 3 neighbourhood centered around the pixel position (x, y). The median filter replaces the pixel at (x, y) by the median value of the nearest neighbour intensities.
3.2 Background minimization
The non-tumor region can be eliminated by background minimization process, while it should preserves the tumor region. Two level morphological operations are used to minimize the background. The two level morphological operations include erosion as first level and dilation as second level.
-
(a)
Level 1 Morphological reconstruction
The mask for first level erosion is the median filtered image I1(x, y). For easy representation, the mask image is represented as U. Therefore the mask image U can be eroded from a marker image M can be represented as \( {r}_U^e(M) \), where \( {r}_U^e(M)={e}_U^{(n)}(M) \) with n so that \( {e}_U^{(n)}(M)={e}_U^{\left(n+1\right)}(M) \). Let G1(x, y) be the first level morphological dilated output which is represented as V for simplicity.
-
(b)
Level 2 Morphological reconstruction
Let V be the mask image for dilation process from the marker image M. Therefore the dilation process can be expressed as \( {r}_V^d(M) \). Where, \( {r}_V^d(M)={d}_V^{(n)}(M) \), with n so that \( {d}_V^{(n)}(M)={d}_V^{\left(n+1\right)}(M) \). The 3 × 3 structuring element for dilation and erosion is represented as, \( \left[\begin{array}{ccc}0& 1& 0\\ {}1& 1& 1\\ {}0& 1& 0\end{array}\right] \). The output of level 2 morphological reconstruction (erosion and dilation) be G2(x, y) which is used for initial Contour detection after thresholding process.
-
(c)
Thresholding
The output of level 2 morphological image is G2(x, y). This imageG2(x, y) is segmented using thresholding process given by the equation,
The threshold value is chosen as the maximum intensity present in the image. Therefore the intensity less than the maximum intensity are set as background. Let G3(x, y) be the output of thresholding process which is used for initial contour detection.
3.3 Initial contour detection
Let G3(x, y) be the background minimized image. The accuracy of active Contour model segmentation depends on the accuracy of initial Contour. So the proposed algorithm uses the following four steps for identification of accurate intial Contour. (a) Centroid estimation (b) Maximum radius calculation (c) Circular Contour formation (d) Radius contraction and expansion
-
(a)
Centroid estimation
Let (x1, y1), (x2, y2), ………. , (xn, yn) be the position of n number of edge pixels for the background minimized image G3(x, y). From the position of edges from the background minimized image G3(x, y), estimate the Centroid for the ROI (tumor). Let (xc, yc) be the Centroid estimated from the edge pixel position (x1, y1), (x2, y2), ………. , (xn, yn). Figure 2 (a) shows an example of Centroid estimation (Green color) from the edge pixel positions (Red color).
The Centroid can be calculated using the relation,
-
(b)
Maximum radius calculation
From the centroid (xc, yc) estimate the distance to the pixels (x1, y1), (x2, y2)………(xn, yn). Let r1, r2, …. . rn be the n number of distance between Centroid and edge pixels, where the distance between cetroid and edge pixel can be calculated using,
Where, i = 1, 2……n. Figure 2(b) shows the radius estimation for all edge pixels. From n number of radius r1, r2, …. . rn find the maximum radius rmax by using,
Figure 3a shows an image where Centroid is estimated (indicated by red) and Fig. 3b shows an image where maximum radius is estimated (indicated by green). Let (xmax, ymax) be the edge pixel that has maximum radius rmax as shown in Fig. 4.
-
(c)
Circular Contour formation
An example for Centroid estimation and Maximum radius calculation (a) Centroid indicated by red color (b) Green color is the point where the radius is maximum. In presence of two or more tumor regions, (i.e., if the background minimization returns two region), centroids are estimated for those two regions individually to segment the two tumor regions
With the Centroid (xc, yc) as centre and rmax as radius, draw a circle on a mask having a size same as MRI image I1(x, y) as shown in Fig. 4 (where brown color circle is the circular contour). Let the circular contour be represented by Ic(x, y). This circular Contour is Contracted and expanded to obtain the initial contour.
-
(d)
Radius Contraction and Expansion
The radius expansion is done after performing the radius contraction. The radius contraction brings the circular contour towards the Region of Interest. Similarly, the radius expansion moves the Contour that results in radius contraction outwards the ROI. The radius contraction is done on all position that lies on circular Contour. But the radius expansion is done on very closer positions of contracted pixels and the radius expansion is not performed on other contracted position. A contracted position is considered as closer pixel if the distance (di) between the background minimized edge pixel location (xi, yi) and contracted position (xi, yi) is less than the expansion threshold ∆1 (di < ∆1).
Let (x1, y1) (x2, y2)………(xn, yn) be the position of n number of pixel that lies on the edges of the background minimized image G3(x, y). Let (xc, yc) be the Centroid estimated by the Centroid estimation stage. The position corresponding to (x1, y1) (x2, y2)………(xn, yn) on circular contour be (X1, Y1) (X2, Y2)………(Xn, Yn) respectively. Where (X1, Y1) (X2, Y2)………(Xn, Yn)can be expressed as (Xi, Yi), i = 1, 2…. . n. The value of (X1, Y1) (X2, Y2)………(Xn, Yn) can be calculated using Eq. (7) to (14).
If xi > xc and yi ≥ yc
If xi ≥ xc and yi < yc
If xi < xc and yi ≤ yc
If xi ≤ xc and yi > yc
where
The positions (X1, Y1) (X2, Y2)………(Xn, Yn) or (Xi, Yi) lies on the circular contour as shown in Fig. 5 (a). The positions (X1, Y1) (X2, Y2)………(Xn, Yn) is contracted, such that the contracted location (ui, vi) or (u1, v2), (u1, v2), ……. (un, vn) lies to the midpoint of (Xi, Yi) and (xi, yi). The contracted new edge position (ui, vi) can be calculated as,
After contraction, the expansion is performed. The expansion is done based on the expansion threshold (∆1) The expansion is done if the distance between (xi, yi) and (ui, vi) is less than the threshold ∆1. After expansion the expanded position (ui, vi) gets modified using Eq. (17) to (24).
If xi > xc and yi ≥ yc and di < ∆1
If xi ≥ xc and yi < yc and di < ∆1
If xi < xc and yi ≤ yc and di < ∆1
If xi ≤ xc and yi > yc and di < ∆1
where ∆2 is the expansion factor (positive integers)
The position of edge pixels after radius contraction and expansion is (ui, vi) as shown in Fig. 5(b). Let the edge pixels be (ui, vi) = (u1, v1), (u2, v2)…………(un, vn).The pixels (u1, v1), (u2, v2)…………(un, vn) are linked to obtain the initial Contour of Active Contour model. The linking can be done by drawing a straight line between adjacent coordinates of edge pixels (ui, vi). Figure 6(a) shows an example for radius contraction and expansion process. Here green color indicates the edges of background minimized image G3(x, y), blue color indicates the circular contour, Cyan color indicates the contracted contour and Red color indicates the expanded contour. Figure 6(b) indicates the initial contour of active contour model detected after radius contraction and expansion (RCE). Let the contour obtained after radius contraction and expansion be represented by IR(x, y) Fig. 7 [14].
3.4 Active contour model
Let IR(x, y) be the initial contour for active contour model. The Active Contour model is a segmentation method that was proposed by Kass et al. The segmentation result in Active Contour model depends on the initial contour. In Active Contour model, the initial Contour IR(x, y) moves to the boundary of the tumor. The movement of Contour towards the boundary of tumor is driven by internal and external forces acting on the Contour. The movement of contour towards the tumor is done by external force, while the smooth deformation of contour at the boundary is done by internal forces. When the total energy of the contour is minimum the movement of the contour gets stopped. The total energy of the snake can be calculated using the expression,
Where v(s) is the parametric representation of curve given by,
Ein be the internal energy and Eex be the external energy of the snake. These internal and external energy are the two components of total energy Etot.
The internal energy of the snake has two components such as Contour energy (Econ) and spline curvature energy (Ecur). Therefore the internal energy can be expressed as,
The external energy of the Contour also has two components that include, image force (Eim) and external constraint force (Econstraint).
The snake will move and searches for boundary where Etot is minimum. The movement of snake stops when the total energy of the contour is minimum. Let I3(x, y) be the tumor region segmented by active contour model.
3.5 Optimization using fuzzy-C-means clustering
The output of Active Contour model is optimized using Fuzzy-C-means clustering algorithm. Let I3(x, y) be the segmented output of active Contour model. The boundaries pixel of active contour model is not accurate which may contain non-tumor region pixels. The Fuzzy-C-means algorithm aims to minimize the non-tumor region pixels which are segmented by active Contour model. Therefore the result of FCM contains the accurate (optimized) tumor region. Let the tumor segmented by active Contour model I3(x, y) contains the N number of pixels represented as,
In a Fuzzy-C-means clustering, the number of clusters is represented as u, where 2 ≤ u < N. Since the FCM optimization algorithm classify the image into two classes foreground and background, the number of cluster is u = 2. For any cluster j, the degree of membership for pixel set Pi be Mij. Let the cluster centre for jth cluster be Cj. The fuzzy C-means clustering is an iterative process which can be terminated based on the termination value ∆, where ∆ lies between 0 and 1. The membership function and cluster center are updated in each iteration. The Fuzziness index fand number of iteration T are real number greater than unity.
The FCM algorithm to optimize the output of active Contour model to obtain the accurate tumor region is as follows.
-
Step 1.
The membership matrix M must be initialized. Let the initial membership M = [Mij] be M(0).
-
Step 2.
The matrix for centre at any iteration T can be estimated as C(t) = [Cj]
-
Step 3.
$$ {C}_j=\frac{\sum_{j=1}^N{M}_{ij}^f{P}_i}{\sum_{i=1}^N{M}_{ij}^f} $$(32)
-
Step 4.
After identifying the cluster center Cj, update the membership M(T), M(T + 1) as,
-
Step 5.
$$ {M}_{ij}=\frac{1}{\sum_{T=1}^G{\left(\frac{d_{ij}}{d_{Tj}}\right)}^{\frac{2}{f-1}}} $$(33)
-
Step 6.
$$ {d}_{ij}=\sqrt{\sum_{i=1}^N\left({P}_i-{C}_j\right)} $$(34)
-
Step 7.
Repeat step 2 and 3, if the absolute difference between the membership values of any two successive iteration is greater than the terminating value ∆.
-
Step 8.
$$ \left\Vert {M}^{(T)}-{M}^{\left(T+1\right)}\right\Vert \ge \Delta $$(35)
-
Step 9.
Stop the iteration if the absolute difference ‖M(T) − M(T + 1)‖ is less than terminating value ∆.
Based on the distance between the cluster and pixel intensity Pi, the FCM clustering algorithm assigns membership to each pixel intensity Pi. If the distance between the pixel intensity Pi and cluster center is high, then the pixel intensity Pi have more membership to the opposite cluster (background). Let I4(x, y) be the output of Fuzzy-C-means clustering algorithm which has the optimized tumor region. As the result of clustering the non tumor pixels present in the boundary gets classified to the background cluster. Figure 8 (a) shows the input image to FCM algorithm, while fig. 8(b) shows the output of FCM optimization. For the proposed brain tumor segmentation algorithm, we have set the parameters such as fuzziness index f, the termination value ∆ and number of iteration as 2, 0.001 and 100 respectively.
3.6 Region selection
The image I4(x, y) contains multiple segmented regions inside a tumor as shown in Fig. 8 (c). These small regions inside the tumor are of small size and can be minimized. Let R1, R2, ……. . RL be the L number of regions present in the image I4(x, y) , which includes the actual tumor. Let the perimeter of the regions R1, R2, ……. . RL be Pe1, Pe2, ……. . PeL respectively.
The accurate region Rac can be selected from R1, R2, ……. . RL, where Rac is the region that has maximum perimeter. The accurate region forms the segmented image Z(x, y) as shown in Fig. 8(d). The next section shows the experimental results of the proposed brain tumor segmentation Fig. 9 [10].
4 Experimental results
The performance of the proposed segmentation algorithm was evaluated using T1-weighted contrast enhanced image dataset that contains the MRI images of 233 patients. These MRI images contain the tumor types such as Glioma, Meningioma and Pitutary. The dataset has 1426 Glioma, 708 Meningioma and 930 Pitutary type tumor images.The size of each MRI images are 512 × 512. The MRI images are filtered (pre-processed) using a 3 × 3 median filter. The pre-processed MRI image is subjected to actual segmentation. The two level morphological process such as dilation and erosion uses a 3 × 3 structuring element \( \left[\begin{array}{ccc}0& 1& 0\\ {}1& 1& 1\\ {}0& 1& 0\end{array}\right] \). The performance of proposed segmentation algorithm can be measured using the metrics such as True negative rate, True positive rate, Dice-score, Probabilistic rand index (PRI) and Hausdorff distance (HD). True negative rate (specificity) is the measure of negatives that are correctly identified. True positive rate (sensitivity) is the measure of positives that re correctly identified. The matching of proposed segmentation output and ground truth results can be estimated using Dice-score.
In Eq. (36), (37) and (38) the true positive (tp), true negative (tn), false positive (fp) and false negative (fn) are calculated by comparing, the segmented result with the ground truth result. True negative (tn) represents if the output is identified as tumor, while the ground truth is non-tumor. The the true positive (tp) represents if the output is identified as tumor, while the ground truth is also tumor. False negative (fn) represents, if the output is identified as non-tumor while the ground truth is tumor. False positive (fp) represents, if the output is identified as non-tumor, while the ground truth is also non-tumor.
Probabilistic Rand Index (PRI) estimate the fraction of pairs whose labelling such as tumor region and non-tumor region are consistent between the segmentation output and ground truth result. If PRI is 1, then it indicates that the segmentation result and ground truth results are identical. Similarly, if PRI is 0, then it indicates that there is no similarities between the segmentation result and ground truth result.
Where, B1 + B2 is the difference between the ground truth result and segmented pixel result, A1 + A2 is the similarly between the ground truth result and segmented pixel result.The performance of the proposed segmentation algorithm was compared with the stat-of-the-art segmentation algorithms such as Gaussian Mixture model [22], K-means algorithm [35], Fuzzy-C-means algorithm [20], New threshold approach [18], Interactive segmentation [38] and our previous work [32] using Greedy snake algorithm with FCM optimization.
For the experimental results and analysis, the expansion threshold ∆1 and expansion factor ∆2 are set as 10. The number of iteration in active contour model is 200 and for FCM clustering, the number of cluster is fixed as 2. Figure 10 shows the segmentation of Meningioma tumor, where Fig. 10 (l) shows the comparison of ground truth result (Red color) and proposed segmentation result (Green color). From the Fig. 10 (l), it is clear that proposed segmentation result closely matches with the ground truth result.
Experimental result for Meningioma type tumor segmentation (a) Input MRI image (b) Pre-processed image (c) Level 1 morphological output (d) Level 2 morphological output (e) Background minimized image (f) Maximum radius estimation (g) Radius contraction and expansion (h) Initial contour for active contour model (i) output of active contour model (j) FCM output (k) Region selection (l) segmented and ground truth output
Figure 11 shows the segmentation of Pituitary tumor, where Fig. 11 (k) shows the comparison of ground truth result (Red color) and proposed segmentation result (Green color). From the Fig. 10 (k), it is clear that proposed segmentation result closely matches with the ground truth result.
Experimental result for Pituitary type tumor segmentation (a) Input MRI image (b) Pre-processed image (c) Level 1 morphological output (d) Level 2 morphological output (e) Background minimized image (f) Maximum radius estimation (g) Radius contraction and expansion (h) Initial contour for active contour model (i) output of active contour model (j) FCM output (k) segmented and ground truth output
Figure 12 shows the segmentation of Glioma tumor, where Fig. 12 (i) shows the comparison of ground truth result (Red color) and proposed segmentation result (Green color). While comparing the output of Glioma tumor, Meningioma tumor and Pituitary tumor, the segmented output of Meningioma tumor and Pituitary tumor highly matches with the ground truth result.
Experimental result for Glioma type tumor segmentation (a) Input MRI image (b) Pre-processed image (c) Maximum radius estimation (d) Radius contraction and expansion (e) Initial contour for active contour model (f) output of active contour model (g) FCM output (h) Region selection (i) segmented and ground truth output
Table 1 shows the Dice-score comparison of proposed method with the traditional methods. The proposed work provides a Dice score of 0.825, 0.64 and 0.53 for the tumor Meningioma, Glioma and Pituitary respectively, which is higher than our previous work [32] that uses Greedy snake model and FCM optimization. Also the Dice score of the proposed method is higher than the traditional methods as depicted in Fig. 13.
Table 2 shows the Sensitivity Comparison of proposed method with the traditional methods. The Sensitivity of the proposed segmentation method is higher than the state-of-the art methods including our previous work. The sensitivity of the proposed method was found to be 0.71, 0.55 and 0.49 respectively for the tumor Meningioma, Glioma and Pituitary respectively. Figure 14 shows the graphical comparison of Sensitivity.
Table 3 shows the Specificity comparison of proposed work with the traditional method. The Specificity of the proposed method was found to be 0.975, 0.962 and 0.935 for the tumor types Meningioma, Glioma and Pituitary respectively. While comparing Table 1, Table 2 and Table 3 it is clear that the Dice-score, Sensitivity and Specificity of the proposed work is high than the traditional methods such as LACM (Localized active Contour model) [19], CNN (Convolutional Neural Network) [34], IDNN (Incremental Deep Neural Network) [24], FCNN+CRF [40],Multi-Modality [21], Optimal thresholding [33] and our previous work [32].
Figure 15 shows the Specificity comparison of proposed method with the state-of-the art methods. Table 4 shows the PRI comparison of our proposed work with our previous work. In both proposed and previous work, the FCM optimization increases the PRI value. The proposed method provides a PRI of 0.9106 and 0.9499 without and with FCM respectively.
Figure 16 shows some of the sample segmentation results of proposed algorithm. Here red color shows the ground truth results, while green color shows the segmentation results of proposed algorithm.
Figure 17 shows the graphical comparison of PRI for our proposed and previous work for the tumor types Meningioma, Glioma and Pituitary. The maximum surface distance can be estimated using Hausdorff distance. The Hausdorff distance specifies the maximum difference between the tumor size. Let the proposed segmentation output be represented as Pr and the ground truth result be represented as Gr, then the Hausdorff distance can be expressed as,
Fig 18 (a) shows the MRI input image where the RCE output, Initial contour, Active contour output, FCM output and the HD values are shown in Table 5 for different values of ∆1and ∆2. The expansion threshold ∆1 is varied as 10,5 and 7, while the expansion factor ∆2 is varied as 10,5 and 3. The HD is minimum for ∆1 = 10 or 5, while ∆2 = 10 . In Table 5 the HD is 3 for ∆1 = 10 or 5, with ∆2 = 10. This shows that, in proposed segmentation the accuracy of segmentation depends on the expansion threshold ∆1 and expansion factor ∆2.
Table 6 shows some of the input images and its segmented output and HD values for different types of tumor. For the Meningioma tumor the HD provides minimum value of 1.7321 and maximum value of 3.873. For the Pituitary tumor the HD provides minimum value of 3.7417 and maximum value of 4.899. For the Glioma tumor the HD provides minimum value of 3.873 and maximum value of 4.899.
The average Hausdorff distance for Meningioma, Glioma and Pituitary tumor was estimated as 2.92, 4.52 and 3.76 respectively. In all the tumor types the average HD values are less than 5 and it shows that the segmentation result highly matches with the ground truth results.
Table 7 shows the Confusion matrix for the proposed segmentation algorithm. The meningioma, Glioma and Pituitary has an accuracy of 0.97, 0.89 and 0.88 respectively. The proposed segmentation algorithm has a overall accuracy of 0.91. The next section shows the conclusion of the proposed segmentation algorithm.
5 Conclusion
The paper proposed a novel brain tumor segmentation algorithm on MRI images that uses Radius Contraction and Expansion approach (RCE). This method initially minimizes the background using two level morphological operations that includes dilation and erosion along with thresholding. The background minimized image is subjected to active Contour model segmentation. The initial Contour of Active Contour model is estimated by process of Radius estimated and Radius Contraction and Expansion. This proposed initial Contour detection method increases the segmentation accuracy in active Contour model. The boundary pixels of active Contour output also contain the pixels that belong to non-tumor region. Therefore Fuzzy-C-means algorithm is used to optimize the output of Active Contour model. The performance of the proposed Active Contour model was evaluated using the metrics such as Sensitivity, Specificity, Dice-score, PRI and Hausdorff distance. The experimental results of the proposed algorithm were compared with the traditional brain tumor segmentation algorithm. The sensitivity, Specificity and Dice-score are found to be higher than the traditional brain tumor segmentation methods. In the proposed system, the meningioma tumor provides a high Dice-score, Sensitivity and Specificity when compared to Glioma and Pituitary tumor. The Dice-score, Sensitivity and Specificity of the Meningioma tumor was found to be 0.825,0.71,0.975 respectively. The PRI of proposed work is higher than the previous work. Also the average HD values are less than 5 in all the tumor types. This shows that the proposed segmentation result highly matches with the ground truth results. Thus the experimental results reveal that the proposed brain tumor segmentation algorithm shows a better performance that the State-of-the art brain tumor segmentation methods.
References
Ain Q, Jaffar MA, Choi TS (2014) Fuzzy anisotropic diffusion based segmentation and texture based ensemble classification of brain tumor. Appl Soft Comput 21:330–340. https://doi.org/10.1016/j.asoc.2014.03.019
Andrew YN, Jordan, M, Yier, W 2001 et al., “ On spectral clustering: analysis and an algorithm”, Adv Neur In 2, 849–856
Angulakshmi, M., Lakshmi priya, G.G 2017 et al., “Automated Brain Tumor Segmentation Techniques—A Review”, Int J Imaging Syst Technol. 27, 66–77
Bahadure NB, Ray AK, Thethi HP (2017) Image analysis for MRI based brain tumor detection and feature extraction using biologically inspired BWT and SVM. Int. J. Biomed. Imaging 2017:1–12. https://doi.org/10.1155/2017/9749108
Bauer, S Et al. 2011, “Fully automatic segmentation of brain tu- mor images using support vector machine classification in combination with hierarchical conditional random field regularization” , In: MICCAI, Vol. 6893, pp. 354–361
Bauer S et al (2013) A survey of mri-based medical image analysis for brain tumor studies. Phys Med Biol 58:97–129
Bengio, Y , Courville, A 2013, et al ,“Representation learning: a review and new perspectives” Pattern Anal Mach Intell IEEE Trans 35, 1798–1828
Bengio, Y et al. 2012, “Practical recommendations for gradient-based training of deep ar- chitectures in: neural networks”, Tricks of the Trade Springer, pp. 437–478
Cabria, I., Gondra, I 2017 et al., “MRI segmentation fusion for brain tumor detection”. Information Fusion 36, 1–9
Cheng, J.u.n. (2017) Brain Tumor Dataset. https://figshare.com/articles/brain_tumor_dataset/1512427, 5, https://figshare.com/articles/brain_tumor_dataset/1512427
Ciresan, D, Giusti, A , Gambardella 2012 et al., “ Deep neural net- works segment neuronal membranes in electron microscopy images”, Ad- vances in Neural Information Processing Systems, pp. 2843–2851
Clark, M , Hall, L , Goldgof, D , Velthuizen, RP 1998 et al, “Automatic tumor segmentation using knowledge-based clustering”, IEEE Trans Med Imag 17, 187–201
Dass R, Priyanka, Devi S (2012) Image segmentation techniques. International Journal of Electronics & Communication Technology (IJECT) 3(1):2230–7109
Elyasi A et al (2011) Active contours in Brain tumor segmentation. Journal of American Science 7(7)
G. Evelin Sujji, YVS. Lakshmi and G. Wiselin Jiji 2013, “MRI Brain Image Segmentation based on Thresholding”, International Journal of Advanced Computer Research, vol. 3, no. 1, issue 8, pp. 2249–7277
Havaei M, Davy A, Warde-Farley D, Biard A, Courville A, Bengio Y, Pal C, Jodoin PM, Larochelle H (2017) Brain tumor segmentation with deep neural networks. Med Image Anal 35:18–31
Hu K, Gan Q, Zhang Y, Deng S, Xiao F, Huang W, Cao C, Gao X (2019) Brain tumor segmentation using multi-cascaded convolutional neural networks and conditional random field. IEEE Access 7:92615–92629. https://doi.org/10.1109/ACCESS.2019.2927433
Umit Ilhan, Ahmet Ilhan 2017 et al “brain tumor segmentation based on a new threshold approach” international conference on theory and application of soft computing, Prog. Comput. Sci. 120, 580–587
E. Ilunga-Mbuyamba, JG. Avina-Cervantes 2017 et al, “Localized active contour model with background intensity compensation applied on automatic MR brain tumor segmentation”, Neurocomputing 220, 84–97
Jeetashree, A, PradiptaKumar, N, Niva 2016 et al., “Modified possibilistic fuzzy C-means algorithms for segmentation of magnetic resonance image”, Appl Soft Comput. 41, 104–119
Li J, Yu ZL, Gu Z, Li Y (2019) MMAN: multi-modality aggregation network for brain segmentation from MR images. Neurocomputing. 358:10–19. https://doi.org/10.1016/j.neucom.2019.05.025
Liang, Z. , Wei, W., Jason, JC 2012 et al, “Brain tumor segmentation based on GMM and active contour method with a model-aware edge map”, Proceedings of MICCAI BRATS 24–2
S. Morales, A. Bernabeu-Sanz, F. Lopez-Mir 2017 et al., “BRAIM: a computer-aided diagnosis system for neurodegenerative diseases and brain lesion monitoring from volumetric analyses”, Comput Methods Prog Biomed 145, 167, 179.
Naceur MB, Saouli R, Akil M, Kachouri R (2018) Fully automatic brain tumor segmentation using end-to-end incremental deep neural networks in MRI images. Comput. Methods Prog. Biomed 166:39–49. https://doi.org/10.1016/j.cmpb.2018.09.007
P. Patil, KS. Kumar, N. Gaud and VB. Semwal 2019, “Clinical Human Gait Classification: Extreme Learning Machine Approach," 1st international conference on advances in science, engineering and robotics technology (ICASERT), Dhaka 2019, pp. 1–6.
Selvaraj D et al (2013) MRI brain image segmentation techniques - a review. Indian Journal of Computer Science and Engineering (IJCSE) 4(5):0976–5166
Semwal VB, Raj M, Nandi GC (2015) Biometric gait identification based on a multilayer perceptron. Robot Auton Syst 65:65–75. https://doi.org/10.1016/j.robot.2014.11.010
Semwal VB, Mondal K, Nandi GC (2017) Robust and accurate feature selection for humanoid push recovery and classification: deep learning approach. Neural Comput. & Applic. 28:565–574. https://doi.org/10.1007/s00521-015-2089-3
Semwal, VB., Singha, J., Sharma, P. (2017) et al. An optimized feature selection technique based on incremental feature analysis for bio-metric gait data classification. Multimed Tools Appl 76, 24457–24475 https://doi.org/10.1007/s11042-016-4110-y
Semwal VB, Kumar C, Mishra PK, Nandi GC (2018) Design of Vector Field for different subphases of gait and regeneration of gait pattern. in IEEE Trans. Autom. Sci. Eng. 15(1):104–110
Semwal VB, Gaud N, Nandi GC (2019) Human gait state prediction using cellular automata and classification using ELM. In: Tanveer M, Pachori R (eds) machine intelligence and signal analysis. Advances in intelligent systems and computing, vol 748. Springer, Singapore
Sheela CJJ, Suganthi G (2019) Automatic Brain Tumor Segmentation from MRI using Greedy Snake Model and Fuzzy C-means Optimization. Journal of King Saud University -Computer and Information Sciences
Tarkhaneh O, Shen H (2019) An adaptive differential evolution algorithm to optimal multi-level Thresholding for MRI brain image segmentation. Expert Syst Appl 138:112820. https://doi.org/10.1016/j.eswa.2019.07.037
Thaha MM, Kumar KPM, Murugan BS, Dhanasekeran S, Vijayakarthick P, Selvi AS (2019) Brain tumor segmentation using convolutional neural networks in MRI images. J Med Syst 43:294. https://doi.org/10.1007/s10916-019-1416-0
Tuhin, UP, Samir, KB 2012 et al., “Segmentation of Brain Tumor from Brain MRI Images. Reintroducing K–Means with advanced Dual Localization Method”, Int J Eng Res Appl. 2 (3), 226–231
Vijay V, Kavitha AR, Rebecca SR (2016) Automated brain tumor segmentation and detection in MRI using enhanced Darwinian particle swarm optimization(EDPSO). Prog. Comput. Sci. 92:475–480. https://doi.org/10.1016/j.procs.2016.07.370
Viji KA, JayaKumari J (2013) Modified texture based region growing segmentation of MR brain images. In: Information & Communication Technologies (ICT), IEEE Conference on, pp. 691–695. IEEE
Guotai Wang, Wenqi Li, Maria A. Zuluaga 2018 et al., “Interactive medical image segmentation using deep learning with image-specific fine-tuning”, IEEE Transactions on Medical Imaging, Interactive Medical Image Segmentation Using Deep Learning With Image-Specific Fine Tuning.
Zhang W, Li R, Deng H, Wang L, Lin W, Ji S, Shen D (2015) Deep convolutional neural networks for multi-modality isointense infant brain image segmentation. NeuroImage 108:214–224. https://doi.org/10.1016/j.neuroimage.2014.12.061
Zhao X, Wu Y, Song G, Li Z, Zhang Y, Fan Y (2018) A deep learning model integrating FCNNs and CRFs for brain tumor segmentation. Med Image Anal 43:98–111. https://doi.org/10.1016/j.media.2017.10.002
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Sheela, C.J.J., Suganthi, G. Brain tumor segmentation with radius contraction and expansion based initial contour detection for active contour model. Multimed Tools Appl 79, 23793–23819 (2020). https://doi.org/10.1007/s11042-020-09006-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-020-09006-1