Abstract
Breast cancer has become an important factor affecting human health. Diagnosis based on pathological images is considered the gold standard in the clinic. In this paper, an automatic breast cancer detection method based on hybrid features is proposed for pathological images. To obtain better segmentation results under conditions of crowded and chromatin-sparse nuclei, a 3-output convolutional neural network (CNN) is employed to segment the nuclei. Due to the weak correlation between the hematoxylin (H) and eosin (E) channels, texture features are separately extracted for the two channels, which provides more representative results. From multiple perspectives, the morphological features, spatial structural features and texture features are extracted and fused. Using a support vector machine (SVM) classifier with improved generalization, the pathological image is classified as benign or malignant on the basis of the relief method for feature selection. For the University of California, Santa Barbara database (UCSB), the classification accuracy of the method is 96.7%, and the area under the curve (AUC) is 0.983. The experimental results show that the proposed method yields superior classification performance compared with existing techniques.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Cancer has increasingly become a chronic disease that affects human health. According to the statistics of World Cancer Research Fund International [1], breast cancer is the most common cancer in women worldwide, with nearly 1.7 million new cases diagnosed in 2012 (second most common cancer overall). Breast cancer represents approximately 12% of all new cancer cases and 25% of all cancers in women. It is the fifth most common cause of death from cancer in women. In addition, the incidence rate is increasing, with a decrease in the age of onset. Clinically, compared with mammography, magnetic resonance imaging (MRI) and other imaging techniques, pathological imaging is the most important criterion for the final diagnosis of breast cancer. The accurate classification of pathological images provides an important basis for doctors to formulate optimal treatment plans.
In general, a classification method is used to train a classifier on the structural, morphological, and texture features of the extracted region of interest (ROI) or the entire image to detect a malignant tumour. The features of the nuclei can be used to distinguish between benign and malignant tumours. To extract these features, the nuclei should first be segmented. Segmentation methods can be divided into supervised and unsupervised methods. State-of-the-art nuclei segmentation techniques include watershed segmentation [2], the threshold method [3], active contours [4,5,6] and region growing [7]. Ali et al. [8] proposed a novel synergistic boundary and region-based active contour model that incorporates shape priors in a level-set formulation with automated initialization based on watershed segmentation. Better performance compared with the traditional active contour method was reported, with a segmentation accuracy of more than 90%. To handle diffuse intensities along object boundaries, Beevi et al. [9] provided a nuclei segmentation method by combining a localized active contour model (LACM) with Krill Herd algorithm (KHA)-based optimal thresholding. This method gave a segmentation sensitivity of 94.36%, an accuracy of 93.54%, and a F1-score of 93.79%. However, such methods cannot be generalized across a wide spectrum of tissue morphologies due to inter- and intra-nuclear colour variations in crowded and chromatin-sparse nuclei. Supervised segmentation methods include learning-based feature methods and deep learning-based methods. Learning-based feature segmentation typically uses features such as colour histograms, colour textures, and geometric features to train a classifier [10,11,12,13]. Deep learning-based segmentation does not require the extraction of complex artificial features. Hatipoglu et al. [14] proposed a segmentation method based on a deep learning model with 2-output, using cellular and extracellular patches of various sizes. The experimental results showed that convolutional neural networks (CNNs) and partly stacked auto encoders (SAEs) had better cell-segmentation performance than traditional methods, but some touching and overlapping remained. Neeraj Kumar et al. [15] proposed a three-class CNN-based method for nuclei segmentation with an accuracy of 92%. This method showed an improvement in the segmentation of crowded and chromatin-sparse nuclei. The CNN model employed a large size of convolution kernels, which increased the calculations.
Improved segmentation has provided a good foundation for increasing classification accuracy. Anuranjeeta et al. [16] proposed a method that combined the morphological features of the nuclei and a Rotation Forest classifier to obtain an accuracy of 85.7%. However, the morphological features were too simple to explain the nuclear arrangement and chromatin characteristics. Doyle et al. [17] used an SVM classifier to distinguish between cancer or non-cancer images based on the combination of the texture features of grayscale images and the architectural features of the nuclei. The system achieved an accuracy of 95.8% in distinguishing cancer from non-cancer and 93.3% in distinguishing high from low grades of cancer. However, the highest accuracy was achieved when only Gabor filter features were used instead of all feature sets. With the improvement of computing power, attention has increasingly focused on the classification of deep learning [18, 19]. To reduce the dependence on feature engineering and improve the automatic learning ability of features, Spanhol et al. [20] employed AlexNet to classify benign or malignant tumours from breast cancer pathological images. Their classification results were 6% more accurate than those obtained by traditional machine learning classification algorithms [21]. Bayramoglu et al. [22] proposed a CNN-based method to classify breast cancer histopathology images. It reported that the classification performance did not degrade when mixing images with different magnifications in the training stage. However, the complexity of this system was increased, thus requiring longer training time. Cavalin et al. [23] conducted transfer learning through a modified Alexnet model. Combining full-connection layer features, the classification accuracy is about 85% to 90%.
In this paper, an automatic breast cancer detection method based on hybrid features is proposed for pathological image. Nuclei may be crowded and chromatin-sparse. To get more accurate nuclei segmentation result, a 3-output CNN model and a post processing scheme are designed. The benign and malignant nuclei are different from shape, texture and spatial distribution. To comprehensively describe the difference, morphological features, spatial features, and texture features are extracted and fused. According to the Lambert-Beer law, the colour in the H&E-stained image is a superposition of two independent vectors. The correlation between the H and E channels is weak. Based on this observation, texture features are extracted for these two channels, respectively.
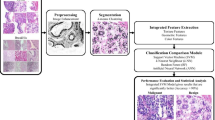
The flow chart of the proposed method is shown in Fig. 1. First, the nuclei in the pathological image are segmented by combining a 3-output CNN and a post-processing scheme. Then, the morphological features and spatial structural features are extracted based on the segmented nuclei. These features are fused with texture features of the H E images. Finally, the relief method is employed for feature selection, and an SVM classifier is adopted.
2 Database
The pathological image database from the Center for Bio-image Informatics [24], University of California, Santa Barbara (UCSB), includes 26 malignant cell images and 32 benign cell images. As the segmentation results and classification results are both provided, this database is selected to verify the proposed method. All images are stored in 24-bit TIFF format with a resolution of 896×786 and a magnification of 40×. As the images in this database are prepared by the same pathology laboratory, there is no spectral variation in the imaging illuminant [25]. Only a size of 200×200 ground truth (GT) nuclei segmentation are labelled for each image, as shown in Fig. 2. Touching and overlapping nuclei are not further segmented.
3 Method
3.1 Nuclei segmentation based on deep learning
Deep learning-based segmentation can automatically discover features from images, which is convenient for automatic detection and produces relatively good results. Instead of a CNN model with 2 classes including foreground and background, a 3-class CNN model with nuclei, nuclear boundary and background is employed [15]. In the 2-output CNN model, unnecessary touching may occur between the nuclei. In the 3-output CNN model, the nuclear boundary is extracted and then used in re-determination to alleviate segmentation errors. The process of segmentation based on the 3-output CNN is shown in Fig. 3.
3.1.1 Structure of the CNN model
The designed CNN model consists of 3 convolutional layers, 3 pooling layers, 2 fully connected layers and 1 output layer. The structure is shown in Table 1.
Convolutional layer
This layer is used to learn features from images. The advantages of the CNN model are local perception and weight sharing, which are fully reflected in the convolutional layer. The size of the scan window is the same as that of the convolution kernel, and only partial images or feature maps are scanned at one time. A feature map shares a convolution kernel.
A convolution kernel is convolved with a number of feature maps from the previous layer. Then, the corresponding elements and a bias are added. Finally, the weighted sum are conveyed to a nonlinear activation function to obtain a new feature map [26]. The activation function can be a rectified linear unit (ReLU) function or sigmoid function. The process of feature extraction is given by the following:
In (1), \( {x}_j^l \) denotes the jth feature map of the lth layer, and \( {w}_{ij}^{(l)} \) shows the weights between the jth feature map of the lth layer and the ith feature map of the (l − 1)th layer. \( {b}_j^{(l)} \) represents the offset of the jth feature map in the lth layer, and ml − 1 indicates all feature maps in the (l − 1)th layer.
The convolution kernel size of the CNN model is 3×3 pixels, which is beneficial for extracting the finer features from the image and reducing the number of parameters and the amount of calculation.
Pooling layer
This layer is used to compress the feature map. Feature compression is performed to extract the main features, which reduces the size of the feature map and simplifies the network. In general, the feature map of the convolutional layer is downsampled by taking the regional maximum or average value.
Fully connected layer
All features are connected and prepared for classification. The convolution can be transformed into a global convolution with the size of the previous convolution kernel if the previous layer is the convolutional layer, otherwise, the convolution is converted to a convolution with a 1×1 convolution kernel.
Output layer
The softmax classifier is used to obtain the classes. The softmax classifier is an algorithm that classifies samples into multiple classes. Suppose there are N input images \( {\left\{{x}_i\right\}}_{i=1}^N \), each image is labelled with {yi ∈ {1, 2, 3, … , k}, k ≥ 2}, for a total of k classes. For each input xi, there will be a probability for each class.
In (2), \( \frac{1}{\sum_{j=1}^k{e}^{\theta_j^T{x}_i}} \) represents the normalization of the probability distribution, that is, the sum of all the probabilities is 1. θ represents the parameters of the softmax classifier.
3.1.2 Post processing
The probability growing method is used to convert the three probability maps obtained by the CNN model into fine nuclei [15]. The probability maps are shown in Fig. 4. First, the nuclear probability map is binarized at the threshold Thn to obtain a reduced nucleus. Second, the morphological dilation operation is used to amplify the nucleus by 1 pixel. Then average probability of the nuclear region is obtained by multiplying the boundary probability map by the nuclear region. Probability growing is not complete until the average probability of the nucleus reaches the average probability of the boundary probability map by iteration. The images after post processing are shown in Fig. 4.
3.2 Feature extraction
In general, the benign nucleus is basically elliptical or round-like, with a consistent shape and size, smooth edges, abundant cytoplasm, fine chromatin and a uniform distribution. Malignant nuclei have irregular shapes, with spiky rather than smooth edges. The volume is generally 1–4 times larger than that of a normal nucleus. The number of nuclei is greater, and the staining is deeper and more uneven [27].
Based on the above information, a feature vector v is created for each image I that includes morphological features, spatial features, and texture features. Because of the weak correlation between the H and E channels, the image is first decomposed to H channel and E channel according to Lambert-Beer law. The texture features are extracted for these two channels separately.
3.2.1 Morphological features
Ten morphological features are calculated from image I, including the number of nuclei, the proportion of nuclei to the entire area, and the average, standard deviation, maximum, and minimum of nuclei areas and perimeters. Since the overlap and touch are not serious, the separation of connected nuclei is ignored. The number of all connected components in the segmentation result is taken as the number of nuclei. In calculating the perimeter and area, only the discrete nuclei are employed.
3.2.2 Spatial features
Benign and malignant images have different spatial distributions of nuclei. The Voronoi diagram, Delaunay diagram and minimum spanning tree are employed to extract the spatial features.
Voronoi diagram
Given a set of two or more but a finite number of distinct points in the Euclidean plane, all locations in that space are associated with the closest member of the point set with respect to the Euclidean distance, which is called the Voronoi diagram [28]. The Voronoi diagram is shown in Fig. 5.
Since the Euclidean distance is used to divide these points, the distances between overlapping and touching nuclei are so small that they can be ignored for a moment. Centroids of connected regions are used to plot the Voronoi diagram. Then, 12 spatial features are calculated from image I, including the average, standard deviation, maximum and minimum of areas, side lengths and perimeters of each Euclidean polygon region. The Voronoi diagram assigns infinity points to the outermost centroid, resulting in an infinite area of the Voronoi polygon formed by the outermost points. Thus, the calculation of the outermost regions is neglected.
Delaunay diagram
The Delaunay triangulation of a discrete point set P in a general position corresponds to the dual graph of the Voronoi diagram for P. The planar Voronoi graph has a straight line between two nodes if the corresponding cells share an edge. All lines constitute the Delaunay graph. Three points di, dj, dk ∈ D are vertices of the same face of the Delaunay graph of D if and only if the circle through di, dj, dk contains no point of D in its interior. Two points di, dj ∈ D form an edge of the Delaunay graph of D if and only if there is a closed circle C that contains di and dj on its boundary and does not contain any other point of D [29]. The Delaunay diagram is shown in Fig. 6.
The average, standard deviation, maximum and minimum of areas, side lengths and perimeters of each triangle as well as the number of areas and side lengths are calculated to obtain a total of 14 features.
Minimum spanning tree
The minimum spanning tree is one of the applications of the Delaunay diagram. For a set T of n points in the plane, the Euclidean Minimum Spanning Tree is the graph with the minimum summed edge length that connects all points in T and has only the points of T as vertices [29]. The minimum spanning tree is generated by taking the distance between nodes as the weight of the edge. Multiple minimum spanning trees constitute the minimum spanning forest. A diagram of a minimum spanning forest is shown in Fig. 7.
Benign nuclei often have cluster distributions. For example, benign ductal tumour cells are confined within the duct, and the basement membrane is intact. However, malignant cancer cells break through the epithelial basement membrane and widely invade surrounding tissues [27]. The minimum spanning tree is used to simulate this distribution. Based on the Delaunay graph, the side length threshold is set as Thl, the side length that exceeds Thl is deleted, and the number of minimum spanning trees in the minimum spanning forest is calculated. At the same time, the average, standard deviation, maximum, and minimum of the side lengths in the minimum spanning forest are calculated, and there are 5 features in total.
3.2.3 Texture features
The benign nuclear chromatin is fine, with a uniform distribution and many interstitial fibrous tissues. The malignant nucleus is vacuolated, with obvious nucleoli and less interstitial fibrous tissue. Speeded Up Robust Features (SURF), Grey-level co-occurrence matrix (GLCM), and Local binary patterns (LBP) features are extracted to represent the texture changes in the nucleus and fibrous tissues. In terms of colour space, the H and E channels are employed as the basis for extracting texture features. According to the Lambert-Beer law, the colour in the H&E-stained image is a superposition of two independent vectors, namely the absorption spectra of H and E in the optical density domain. The smaller cross information in the H&E model means that the channels in the H&E model are less dependent on each other than the channels in other colour spaces [25].
Colour decomposition
According to the Lambert-Beer law, the reverse operation by which a colour represented in RGB colour is converted to the H&E model is called colour decomposition [30]. The relation between light transmission and the amount (A) of stain with dye absorption factor C in stained samples is given by the following:
In (3), Iiis the intensity of light detected after passing the specimen, I0, i is the intensity of light entering the specimen, and subscript i indicates the detection channel.
The optical density (OD) for each channel is defined as follows:
Since the optical density (OD) of each channel is linear with the concentration of absorbing material, it can be used to separate the contributions of multiple stains in a specimen.
If R is a 3×1 vector of the amounts of the three stains at a particular pixel and the optical density matrix (OD) is Q, then the OD vector detected at that pixel is y = RQ. After transformation, the vector is R = Q−1y, and then the colour deconvolution matrix can be easily defined as follows:
Utilizing the colour deconvolution matrix, the RGB image (shown in Fig. 8a) is converted into the H channel image (shown in Fig. 8b) and E channel image (shown in Fig. 8c).
SURF
SURF is a robust local feature point detection and description algorithm [31] that includes the construction of a Hessian matrix and scale space, the location of feature points, assignment of the main direction of feature points and the generation of feature descriptors. The Hessian matrix is the core of the SURF algorithm.
Given a point x = (x, y) in an image I, the Hessian matrix \( \mathcal{H}\left(\mathrm{x},\upsigma \right) \) in x at scale σ is defined as follows:
In (6), Lxx(x, σ) is the convolution of the Gaussian second-order derivative \( \frac{\partial^2}{\partial {x}^2}g\left(\sigma \right) \) with the image I in point x; Lxy(x, σ) and Lyy(x, σ) are derived in a similar manner.
In the main direction, a square region round the feature point is split up regularly into smaller 4×4 square sub-regions. For each sub-region, we compute Haar wavelet responses at 5×5 regularly spaced sample points. Each sub-region has a four-dimensional descriptor vector V, V = (∑dx, ∑dy, ∑ | dx| , ∑ | dy| ), which represents the sum of the responses in the horizontal and vertical directions and the sum of the absolute values of the responses in the horizontal and vertical directions, resulting in 64 features. The 64 features of multiple interest points of each image are summed so that each image has SURF features with the same dimension.
GLCM
GLCM [32] is often used to represent image texture features. A GLCM is a matrix in which the number of rows and columns is equal to the number of grey levels in the image. The matrix element P(i, j| Δx, Δy) is the relative frequency separated by a pixel distance (Δx, Δy). Then four features are obtained, including contrast, energy, correlation and homogeneity [33]. The contrast reflects the sharpness and texture of the image. The energy is the sum of the squares of the elements in the grey-level co-occurrence matrix and is also a measure of the stability of the grayscale variations of the image texture. The correlation is used to measure the similarity of the grey level of the image in the row or column direction. Homogeneity transfers the tightness of the element distribution to the diagonal of the GLCM [34]. The directions include 0∘, 45∘, 90∘ and 135∘. There are 16 features for each image.
LBP
LBP consists of computing the distribution of binary patterns in the circular neighbourhood of each pixel. The neighbourhood is characterized by a radius R and a number of neighbours P. The principle is to threshold neighbouring pixels compared to the central pixel. The value 1 is assigned to each of the P neighbours if the current pixel intensity is superior or equal to the central pixel intensity. Otherwise, the value 0 is assigned. Thus, for each pixel, a binary pattern is obtained from the neighbourhood [21]. The number of neighbours P is set as 8, and 59 features are obtained. All features and numbers are shown in Table 2.
3.3 Feature selection and classification
The number of features is large, and there are some redundant and irrelevant features. It is necessary to select useful and important features for classification. The relief-based feature selection method [35], which is a kind of feature weighting algorithm, is chosen. Different features are assigned weights based on the relevance of each feature and class, and features with weights less than a certain threshold are removed.
A SVM classifier is used to classify benign and malignant tumours, which satisfies the requirements of small sample, fast computation, generalization and robustness. Moreover, a few support vectors can determine the final result. It can not only grasps key samples and eliminates a large number of redundant samples, but also reduces the computational complexity.
4 Experiment and result
4.1 Experiment settings
A 10-fold cross-validation algorithm is used to obtain credible results. For each fold, the training set consists of approximately 90% of the images, resulting in the set of 29 benign images and 23 malignant images, the remaining images are used as the testing set.
Training strategy
As only a segmentation size of 200×200 is labelled for each image, training and quantitative measurement for segmentation is only computed for the 200×200 ROI for each image. For each pixel, a 25×25 patch is taken. A total of 200,000 patches with 3 classes are obtained from the training images. CNN is trained using Torch [36] on a graphics processing unit (GPU). The initial learning rate for the network is 0.01. The CNN model is trained for 50 epochs with a batch size of 128, and the time of each epoch is approximately 1 min 30 s.
For classification, the entire image with a size of 896×768 is first segmented using the 3-CNN model. Afterwards, the features are extracted and used to train the SVM classifier in MATLAB 2016a on a central processing unit (CPU). Feature extraction and selection require about 20 min. The time of classifier training is about 0.5 s.
4.2 Segmentation result
The proposed segmentation method is compared with threshold processing [37], the marker controlled watershed algorithm [38], the fuzzy C-means clustering method (FCM) [39] and CNN with 2 classes (2-CNN) [14]. The segmentation performance is quantitatively evaluated based on accuracy, sensitivity, specificity and precision and the F1-score. All measure metrics are derived from the confusion matrix. The confusion matrix containing information about the actual and predicted classification results is defined in Table 3. True positive (TP) indicates that an originally positive class is assigned to a positive class. False positive (FP) represents that an originally negative class is assigned to a positive class. True negative (TN) means that an originally negative class is assigned to a negative class. False negative (FN) indicates that an originally positive class is assigned to a negative class. The foreground and background are considered positive and negative, respectively.
Accuracy represents the proportion of correct classification in all samples.
Sensitivity indicates the proportion of positive categories that are correctly classified, which is also known as recall.
Specificity shows the proportion of negative categories that are correctly classified.
Precision represents the proportion of positive classes in all samples that are classified as positive.
The F1-score is the harmonic mean of accuracy and sensitivity. It reflects the robustness of the classification.
A quantitative comparison of the different segmentation methods is shown in Table 4. As shown in Table 4, the accuracy and F1-score of the proposed method are 0.9082 and 0.8164, obviously higher than those of the unsupervised methods OTSU, Watershed and FCM. Compared with 2-CNN, the accuracy and F1-score are increased by 0.98% and 0.28%, respectively. Some segmentation examples for different methods are shown in Fig. 9, which illustrates that the proposed method can produce better nuclei segmentation than the 2-CNN model in terms of overlapping nuclei.
4.3 Classification result
After feature extraction, the best 50 features are selected using the relief algorithm to train the SVM classifier. The classifier performance is evaluated by accuracy, sensitivity, specificity, F1-score, Matthews correlation coefficient (MCC) and AUC. Malignant and benign are considered positive and negative, respectively.
MCC is used to measure the binary classifier. Its value ranges between −1 and + 1, where −1, +1, and 0 correspond to worst, best and at random prediction [16].
AUC is the area covered by the receiver operating characteristic (ROC) curve. The x-coordinate of the ROC curve is the false-positive rate (FPR), and the y-coordinate is the true-positive rate (TPR). A larger value of TPR indicates that more positive samples are correctly detected, and a larger value of FPR signifies that more negative samples are misdiagnosed. Thus, AUC reflects the classifier performance.
Five existing methods are employed for comparison, including hand-crafted features [17, 18, 31], CNN trained from scratch [38] and transfer learning [40]. The comparison results are shown in Table 5. The accuracy and AUC of the proposed method are 0.967 and 0.983, which are higher than the five existing methods. The specificity of the proposed method is 1, indicating that all benign pathological images of breast cancer are correctly identified. The MCC of the proposed method is 0.932, showing excellent classification performance. Moreover, as shown in Fig. 10, the ROC curve is close to the upper left corner, and AUC is close to 1.
The reason for the higher performance may be as follows. For CNN-based method, just high level features are extracted blindly from the entire image. However, medical heuristic characteristics is important for medical image classification. Moreover, the parameter setting of CNN model is also a big challenge. The proposed method uses medical heuristic characteristics in feature extraction. The extracted features including morphology, spatial distribution and texture, which is comprehensive for discriminating benign and malignant pathological images.
5 Discussion
In classification, only malignant breast pathological images were wrongly classified as benign pathological images, as shown in Fig. 11a and b). The feature vectors for these two images were analysed. The areas and perimeters of nuclei, GLCM features and features extracted from the minimum spanning tree are shown in Figs. 12 and 13. Specifically, these features of the misclassified images are obviously more similar to those of benign images, and thus it is easy for these images to be classified as benign images. The features of the image in Fig. 11a are shown in Fig. 12. The shape and stability of the grayscale variations of the image texture are clearly similar to those of the ordinary benign image (shown in Fig. 11c). The boundaries between benign cells and surrounding tissues are clear, with no metastasis, and thus the integrity of the extracellular matrix structure is maintained. Adhesions and connection-related components between malignant cells are mutated or absent, and cells lose their association with the intercellular and extracellular matrices. Therefore, the texture in malignant images is more blurred with a lower value of energy. The features of the image in Fig. 11b are shown in Fig. 13. Apart from similar morphological features, the nuclei in the misclassified image are less widely distributed, thus more similar to those in an ordinary benign image than the nuclei in an ordinary malignant image (shown in Fig. 11d). For nuclei in an ordinary benign image are generally distributed in some regions, and few nuclei invade into other tissues. Therefore, these images may be less malignant.
Classification based on handcrafted features requires effective professional knowledge. It is specific for each problem and has limited applicability in other domains. CNN-based approach does not take exact difference between images into consideration, giving it the ability to self-learn. However, due to the complexity of model-tuning, the training time is longer. To achieve better results, large dataset and parameter tuning are needed. Compare with natural image, medical image is with small dataset and has some heuristic characteristics. In breast cancer classification, the nuclei number size, nuclei distribution and the texture of stroma can be observed. The proposed method utilized these medical characteristics to classify benign or malignant tumours and a better performance is obtained.
6 Conclusion
In this paper, a classification method based on 3-output CNN segmentation and hybrid feature extraction is proposed. In the process of segmentation, the probability growing method is used to fuse the boundary probability map and the nuclear probability map to achieve the effect of fine segmentation. For benign and malignant classification, hybrid features representing the differences in morphological, spatial and texture are extracted. In addition, the classification of breast pathological images with limited annotations is very effective. It is not necessary to obtain a perfect segmentation effect by segmenting the overlapping nuclei, and the benign and malignant images can also be distinguished, which not only reduces the complexity of image processing but also reduces the workload of pathologists annotating images.
Change history
09 April 2019
The article Breast cancer classification in pathological images based on hybrid features, written by Cuiru Yu, Houjin Chen, Yanfeng Li, Yahui Peng, Jupeng Li and Fan Yang, was originally published electronically on the publisher’s internet portal (SpringerLink) on March 16, 2019 with open access.
References
Ferlay J, Soerjomataram I, Ervik M et al (2015) Breast cancer statistics. World Cancer Research Fund International. https://www.wcrf.org/int/cancer-facts-figures/data-specific-cancers/breast-cancer-statistics. Accessed 16 January 2015
Veta M, van Diest PJ, Kornegoor R et al (2013) Automatic nuclei segmentation in h&e stained breast cancer histopathology images. PLoS One 8(7):e70221
Petushi S, Garcia FU, Haber MM et al (2006) Large-scale computations on histology images reveal grade-differentiating parameters for breast cancer. BMC Med Imaging 6(1):1–11
Vink JP, Van Leeuwen MB, Van Deurzen CHM et al (2013) Efficient nucleus detector in histopathology images. J Microsc 249(2):124–135
Sabeena BK, Nair MS, Bindu GR (2014) Automatic segmentation and classification of mitotic cell nuclei in histopathology images based on Active Contour Model. International Conference on Contemporary Computing and Informatics on IEEE, pp 740–744
Qi X, Xing F, Foran DJ et al (2012) Robust segmentation of overlapping cells in histopathology specimens using parallel seed detection and repulsive level set. IEEE Trans Biomed Eng 59(3):754
Zucker SW (1976) Region growing: childhood and adolescence. Computer Graphics & Image Processing 5(3):382–399
Ali S, Madabhushi A (2012) An integrated region-, boundary-, shape-based active contour for multiple object overlap resolution in histological imagery. IEEE Trans Med Imaging 31(7):1448–1460
Sabeena BK, Nair MS, Bindu GR (2016) Automatic segmentation of cell nuclei using krill herd optimization based multi-thresholding and localized active contour model. Biocybernetics & Biomedical Engineering 36(4):584–596
Kong H, Gurcan M, Belkacem-Boussaid K (2011) Partitioning histopathological images: an integrated framework for supervised color-texture segmentation and cell splitting. IEEE Trans Med Imaging 30(9):1661
Plissiti ME, Nikou C (2012) Overlapping cell nuclei segmentation using a spatially adaptive active physical model. IEEE Transactions on Image Processing A Publication of the IEEE Signal Processing Society 21(11):4568
Chang H, Han J, Borowsky A et al (2013) Invariant delineation of nuclear architecture in glioblastoma multiforme for clinical and molecular association. IEEE Trans Med Imaging 32(4):670–682
Zhang M, Wu T, Bennett K (2015) Small blob identification in medical images using regional features from optimum scale. IEEE Trans Biomed Eng 62(4):1051–1062
Hatipoglu N, Bilgin G (2017) Cell segmentation in histopathological images with deep learning algorithms by utilizing spatial relationships. Med Biol Eng Comput 55(10):1–20
Kumar N, Verma R, Sharma S et al (2017) A dataset and a technique for generalized nuclear segmentation for computational pathology. IEEE Trans Med Imaging 36(7):1550–1560
Anuranjeeta A, Shukla KK, Tiwari A et al (2017) Classification of histopathological images of breast cancerous and non cancerous cells based on morphological features. Biomedical & Pharmacology Journal 10(1):353–366
Doyle S, Agner S, Madabhushi A et al (2008) Automated grading of breast cancer histopathology using spectral clustering with textural and architectural image features. IEEE International Symposium on Biomedical Imaging: From Nano To Macro 29:496–499
Hu Z, Tang J, Wang Z et al (2018) Deep learning for image-based cancer detection and diagnosis - A survey. Pattern Recogn 83(2018):134–149
Wu G, Lin Z, Han J, et al (2018) Unsupervised Deep Hashing via Binary Latent Factor Models for Large-scale Cross-modal Retrieval. Proceedings of the Twenty-Seventh International Joint Conference on Artificial Intelligence, pp 2854–2860
Spanhol FA, Oliveira LS, Petitjean C et al (2016) Breast cancer histopathological image classification using Convolutional Neural Networks. International Joint Conference on Neural Networks, Vancouver
Spanhol F, Oliveira L, Petitjean C et al (2016) A Dataset for Breast Cancer Histopathological Image Classification. IEEE Trans Biomed Eng 63(7):1455–1462
Bayramoglu N , Kannala J, Heikkilä J (2017) Deep learning for magnification independent breast cancer histopathology image classification. International Conference on Pattern Recognition IEEE, pp 2440–2445
Spanhol F, Cavalin P, Oliveira L S, et al (2017) Deep Features for Breast Cancer Histopathological Image Classification. 2017 IEEE International Conference on Systems, Man and Cybernetics (SMC)
Gelasca ED, Byun J, Obara B, et al (2008) Evaluation and benchmark for biological image segmentation. IEEE International Conference on Image Processing, pp 1816–1819
Li X, Plataniotis KN (2015) Color model comparative analysis for breast cancer diagnosis using H and E stained images. SPIE Medical Imaging. International Society for Optics and Photonics 9420:94200L–94200L-6
He X, Han Z, Wei B (2000) Breast cancer histopathological image auto-classification using deep learning. Computer Engineering and Applications:1–000
Xiangsheng Z, Hong B, Chengquan Z (2014) Pathological diagnosis and differential diagnosis of breast. People’s Medical Publishing House, China
Okabe A, Boots B, Sugihara K et al (2001) Spatial Tessellations: Concepts and Applications of Voronoi Diagrams. John Wiley & Sons, Inc 3(4):357–363
De Berg M, Van Kreveld M, Overmars M et al (2000) Computational geometry: algorithms and applications. Math Gaz 19(3):333–334
Ruifrok AC, Johnston DA (2001) Quantification of histochemical staining by color deconvolution. Anal Quant Cytol Histol 23(4):291–299
Bay H, Ess A, Tuytelaars T et al (2008) Speeded-up robust features (surf). Computer Vision & Image Understanding 110(3):346–359
Zulpe NS, Pawar VP (2012) Glcm textural features for brain tumor classification. International Journal of Computer Science Issues 9(3):354–359
Haraclick RM (1973) Texture Features for Image Classification. IEEE Trans Syst Man Cybern 3(6):610–621
Tahir MA, Bouridane A, Kurugollu F (2005) An fpga based coprocessor for glcm and haralick texture features and their application in prostate cancer classification. Analog Integr Circ Sig Process 43(2):205–215
Liu X (2014) Mass classification in mammogram with semi-supervised relief based feature selection. International Conference on Graphic and Image Processing, International Society for Optics and Photonics 9069:361–368
Collobert R, Bengio S, Marithoz J (2002) Torch: A Modular Machine Learning Software Library. Idiap, Switzerland
Otsu NA (1979) Threshold Selection Method from Gray-Level Histograms. IEEE Trans Syst Man Cybern 9(1):62–66
Veta M, Huisman A, Viergever M A, et al (2011) Marker-controlled watershed segmentation of nuclei in H&E stained breast cancer biopsy images. IEEE International Symposium on Biomedical Imaging: From Nano To Macro, pp 618–621
Lei T, Jia X, Zhang Y et al (2018) Significantly fast and robust fuzzy c-means clustering algorithm based on morphological reconstruction and membership filtering. IEEE Trans Fuzzy Syst PP(99):1
Argandacarreras I, Kaynig V, Rueden C et al (2017) Trainable weka segmentation: a machine learning tool for microscopy pixel classification. Bioinformatics 33(15):2424
Song Y, Li Q, Huang H, et al (2016) Histopathology Image Categorization with Discriminative Dimension Reduction of Fisher Vectors. Computer Vision - ECCV 2016 Workshops, Springer International Publishing, pp 306–317
Acknowledgements
This work was supported in part by the National Natural Science Foundation of China under no. 61872030 and no. 61571036.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original version of this article was revised due to retrospective open access.
Rights and permissions
About this article
Cite this article
Yu, C., Chen, H., Li, Y. et al. Breast cancer classification in pathological images based on hybrid features. Multimed Tools Appl 78, 21325–21345 (2019). https://doi.org/10.1007/s11042-019-7468-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-019-7468-9