Abstract
The aims of this study are to assess the utility of the internal transcribed spacer (ITS) region, and partial translation elongation factor (EF1α) and RNA polymerase II (RPB2) genes, for differentiation of Bailinggu, P. eryngii, and P. nebrodensis; to reconstruct phylogenetic relationships between the three species; and to confirm the taxonomic status of Bailinggu based on ribosomal and protein-coding genes. Pairwise genetic distances between Bailinggu, P. eryngii, and related Pleurotus strains were calculated by using the p-distance model, and molecular phylogeny of these isolates was estimated based on ITS, RPB2, and EF1α using maximum parsimony and Bayesian methods. Differences in ITS, RPB2, and EF1α sequences show that Bailinggu, P. eryngii, and P. nebrodensis are distinct at the species level. Phylogenetic analyses reveal that P. eryngii is closer to P. nebrodensis than to Bailinggu. Sequence analyses of ribosomal and protein-coding genes confirm that P. eryngii var. tuoliensis is identical to Bailinggu. P. eryngii var. tuoliensis should be raised to species level or a new name should be introduced for Bailinggu after a thorough investigation into Pleurotus isolates from Ferula in Xinjiang Province. This study helps to resolve uncertainty regarding Bailinggu, P. eryngii and P. nebrodensis, improving the resource management of these strains. ITS, EF1α, and RPB2 sequences can be used to distinguish Bailinggu, P. eryngii and P. nebrodensis as three different species, and P. eryngii var. tuoliensis should be the scientific name for Bailinggu at present.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pleurotus is one of the most diverse groups of cultivated mushrooms. In this group, “Xingbaogu” and “Bailinggu” are two popular edible mushrooms of high commercial value that are widely cultivated in China. Xingbaogu was introduced from Europe about 20 years ago, and it is commonly accepted that P. eryngii (DC.) Quél. is the scientific name for this popular edible mushroom. Bailinggu, a famous edible mushroom, was known as “Awei Mo” in Xinjiang Province, where they were first cultivated and widely exported. Bailinggu was a trade name for the cultivated Awei Mo proposed by Mao in 1997 [1]. Although the name has become widely accepted, the taxonomic status of Bailinggu remains uncertain. P. eryngii var. nebrodensis, P. eryngii var. tuoliensis, and P. nebrodensis have all been used as scientific names for Bailinggu [1–8]. After its successful cultivation, Bailinggu was morphologically identified as P. nebrodensis (Inzenga) Quél. by Mao [1]. P. eryngii var. nebrodensis has also been used for Bailinggu by some researchers because of taxonomic disagreements regarding P. nebrodensis [9, 10]. P. eryngii var. tuoliensis, also known as Awei Mo, was discovered on Ferula sp. in Tuoli and Mulei counties of Xinjiang Province by Mou et al. [11], and was first cultivated in that region. Awei Mo is a general designation of Pleurotus spp. on Ferula, and two different taxa, P. eryngii and P. eryngii var. tuoliensis were involved [11, 12]. Mao [2] accepted that Bailinggu was actually the cultivated strain of P. eryngii var. tuoliensis, and concluded that P. eryngii var. tuoliensis should be a synonym of P. nebrodensis, as did Jia and Qin [7]. P. nebrodensis has become dominant in the last few decades and the name has appeared in many publications and commercial catalogs by Chinese researchers, which have proliferated since the 1990s. More recently, P. eryngii var. tuoliensis C. J. Mou was resurrected as the scientific name for Bailinggu at the variety level by Kawai et al. based on analysis of internal transcribed spacer (ITS) sequences [5].
The relationship between Bailinggu, P. eryngii and P. nebrodensis remains controversial. Prior to this study, some attempts have been made to clarify the relationship between them. Based on PCR–RFLP analysis of partial 28S rDNA and monokaryon–monokaryon mating tests, Bao et al. [13, 14] concluded that P. eryngii and “P. nebrodensis” (Bailinggu) from China were independent and incompatible species. Zhang et al. [12] investigated the genetic polymorphism of Ferula Mushroom growing on Ferula sinkiangensis using ITS sequences and IGS1-RFLP analyses, and their results showed that P. eryngii and Bailinggu (as “P. nebrodensis”) are two different Pleurotus species growing on Ferula sinkiangensis. Kawai et al. [5] indicated that Bailinggu and P. nebrodensis belong to the same species, P. eryngii, because hybridization occurred between them.
However, the aforementioned studies were mainly based on a single ribosomal gene, and the results did not resolve the uncertainty regarding Bailinggu, P. eryngii, and P. nebrodensis. There is not as yet a unified scientific name for Bailinggu, and many strains labeled “P. eryngii” or “P. nebrodensis” in culture collection centers are actually representatives of Bailinggu. These have caused much confusion for phylogenetic, genetic, and breeding programs, which hindered strategies for scientific research, breeding, and commercial activities of Bailinggu. In contrast, the utility of ITS sequences for species identification in Pleurotus has been questioned, and we hesitate to agree with the treatment of Bailinggu and P. nebrodensis as different varieties of the same species P. eryngii. It is important to clarify the taxonomic status of Bailinggu and the relationship between Bailinggu, P. eryngii, and P. nebrodensis based on multiple molecular markers.
ITS sequences have been widely used to reconstruct phylogenetic relationships and for species discrimination in some Agaricales groups at or below the species level. According to previous studies [15, 16], ITS sequences cannot be used to unequivocally distinguish varieties in the genus Pleurotus, whereas a more recent study revealed that ITS might be a desirable DNA barcode for cultivated oyster mushroom [17]. The two protein-coding genes RPB2 and EF1α have been used in combination with other genomic regions to infer phylogenetic relationships at the species level for some mushrooms [18], and their utility for discrimination and phylogenetic reconstruction among members of Pleurotus at the species or variety level has also been explored [19]. Furthermore, RPB2 yielded better resolution than ITS for species discrimination in fungal groups [20]. Nevertheless, RPB2 and EF1α have not been used to distinguish Bailinggu from P. eryngii and P. nebrodensis in previous studies.
In the present study, ITS, RPB2, and EF1α sequences of Bailinggu, P. eryngii, P. nebrodensis, and related species were analyzed: (1) to assess the utility of the three genes for differentiation of Bailinggu, P. eryngii, and P. nebrodensis; (2) to clarify relationships between the three species; and (3) to confirm the scientific name of Bailinggu based on ribosomal and protein-coding genes. This study would help to improve the resource management of Bailinggu, P. eryngii, and P. nebrodensis, and provide a theoretical basis for phylogenetic, genetic, and breeding research.
Materials and methods
Sampling, GenBank data
Three wild Awei Mo strains (“Awei”, “Pnh529” and “HM777041”), and cultivated Bailinggu and P. eryngii strains were tested in this study (Table 1). The wild strains were isolated from Awei Mo on Ferula in Xinjiang Province, and the rest cultivated Bailinggu and P. eryngii strains were obtained from Soil and Fertilizer Institute, Sichuan Academy of Agricultural Sciences (SAAS Table 1). Isolates of Bailinggu and P. eryngii were grown on potato dextrose agar medium in a Petri dish at 25 °C for 7–10 days. These strains were stored at 4 °C.
Accession numbers of the tested sequences with codes “GU” and “GQ” presented in Table 1 were from Rodriguez Estrada et al. [19].
DNA extraction, PCR and sequencing
Genomic DNA was extracted from mycelia using a Biospin Fungus Genomic DNA Extraction Kit according to the manufacturer’s instructions. ITS sequence was amplified using the primers ITS4 and ITS5 [21]. The primers used for RPB2 were fRPB2 5F (5′-GAYGAYMG WGATCAYTTYGG-3′) and bRPB2 7.1R (5′-CCCATRGCYTGYTTMCCCATDGC-3′), and b6.9F (5′-TGGAC NCAYTGY GARATYCAYCC-3′) and b11R1 (5′-TGGATYTTG TCRTC CACCAT-3′) [22, 23]. Primers b6.9F and b11R1 were used to amplify a region between domains 3–11; and fRPB2 5F and bRPB2 7.1R were used for amplification of domains 5–11. Primers EF116OR (5′-CCGAT CTTGTA GACGT CCTG-3′) and EF595F (5′-CGTGACTTCAT CAAGAAC ATG-3′) were used to amplify a portion of the EF1α gene [24, 25]. Amplification was performed in 30-µl volumes containing 1 µl template DNA, 12 µl distilled water, 1 µl of each primer and 15 µl PCR mix (DreamTaq™ Green PCR Master Mix [2×], Fermentas). Amplifications for the three genes were carried out under the same conditions: 94 °C/5 min; 35 cycles of 94 °C/1 min, 55 °C/1 min, 72 °C/90 s; and a final extension step of 72 °C/10 min. Primers for sequencing are the same as amplification. Sequences generated in the present study are deposited in GenBank.
Phylogenetic and statistical analyses
The sequences used in phylogenetic analysis were aligned in Muscle 3.6 [26] and ClustalX [27], and manually modified in BioEdit 7.0.9.0 where necessary [28]. Sites judged to be too ambiguous in alignment, as well as spliceosomal introns in RPB2, were excluded from phylogenetic analysis.
The ITS and the combined EF1α and RPB2 dataset (Table 1) were analyzed respectively using maximum parsimony (MP). MP analyses were conducted in PAUP* version 4.0b10 [29]. All characters were treated as unordered and of equal weight. Gaps were treated as missing data. Bootstrap values were calculated from 1000 replicates.
Bayesian analyses were also performed on ITS and the combined dataset using MrBayes 3.1.2 [30]. Six Markov chains were run for two runs from random starting trees for one million generations and sampled every 10 generations. Every time the diagnostics were calculated, 25 % of the samples from the beginning of the chain were discarded. A majority rule consensus tree of all remaining trees was calculated.
Nucleotide differences between Pleurotus species were calculated. Genetic distances were inferred by computing pairwise distances based on the p-distance (nucleotide substitution) model in Mega 4 [31].
Results
ITS sequence analysis of Bailinggu, P. eryngii, P. nebrodensis, and Pleurotus spp.
ITS sequences obtained in this study consist of 592 bp, comprising 232 bp for ITS1, 145 bp for 5.8S, and 215 bp for ITS2. The genetic distances among Bailinggu, P. eryngii, P. nebrodensis, and the other Pleurotus spp. (Fig. 1) were inferred based on the p-distance (nucleotide substitution) model. The lowest divergence (0.005) among these species was between P. populinus (AY 450346) and P. ostreatus (AY 450345, epitype of P. ostreatus), and the highest (0.178) was observed between P. eryngii and P. djamor (GU 722277). Among the 18 taxa tested, the divergence value between Bailinggu and P. eryngii (0.016) is higher than those between Bailinggu and either P. populinus or P. ostreatus (0.013 and 0.009 respectively), but identical to that between P. populinus and P. pulmonarius. The divergence between Bailinggu and P. nebrodensis is 0.009, which is identical to the distance between Bailinggu and P. ostreatus. The distance between P. eryngii and P. nebrodensis (0.011) is identical to the divergence between P. ostreatus and P. pulmonarius but higher than that between P. ostreatus and Bailinggu.
Among these tested Pleurotus species, 11 nucleotides could be used to distinguish the least divergent species P. populinus (AY 450346) and P. ostreatus (AY 450345); 12 for P. nebrodensis and P. ostreatus; 13 for Bailinggu and P. ostreatus; and 14 for P. eryngii and P. ostreatus. Regarding P. nebrodensis, Bailinggu, and P. eryngii, the ITS similarities of Bailinggu and P. eryngii are 97–98 %, and 14–16 nucleotides were found to be different between these isolates (Table 2). A total of 12 different bases were observed in the ITS sequences of P. eryngii and P. nebrodensis, and 14 base substitutions could distinguish Bailinggu from the European P. nebrodensis. Additionally, six base substitutions were also found in partial ITS1 sequences of the epitype of P. nebrodensis and Bailinggu.
ITS sequences recovered from wild Awei Mo strains were almost identical to those of the cultivated Bailinggu strains, differing in no more than four bases. However, up to 16 and 14 different nucleotides were found between P. eryngii and wild Awei Mo, and between P. nebrodensis and wild Awei Mo, respectively. As mentioned previously, two taxa (P. eryngii and P. eryngii var. tuoliensis) are involved in Awei Mo. Therefore, the wild Awei Mo tested in the present study should be P. eryngii var. tuoliensis, and Bailinggu could be the cultivated strains of P. eryngii var. tuoliensis.
Sequence analysis of EF1α and RPB2 genes
Two portions of RPB2 sequences were amplified using the two pairs of RPB2 primers, and they were edited and trimmed following Rodriguez Estrada et al. [19]. Of the RPB2 sequences, 1253 sites remained, and the two portions were joined by a string of 369 Ns estimated from alignments with the RPB2 sequence of P. ostreatus (AY786062). In total, 33 sites were able to distinguish Bailinggu and P. eryngii from each other; 26 base substitutions allowed discrimination between P. nebrodensis and Bailinggu; and a total of 20 sites differentiated P. nebrodensis from P. nebrodensis (Table 3).
Amplification of the EF1α gene yielded a segment of 538 bp. EF1α showed less variation than RPB2. Eleven base substitutions (95, 155, 172, 181, 188, 241, 271, 334, 346, 480, 483) discriminated Bailinggu from P. eryngii. Eight base substitutions at positions 116, 155, 172, 181, 334, 346, 394, 483 discriminated P. nebrodensis from Bailinggu, and seven sites (95, 116, 188, 241, 271, 394, 480) could distinguish P. nebrodensis from P. eryngii.
Only two different sites were observed between RPB2 sequences of wild Awei Mo strains and the cultivated Bailinggu strains; and four different bases for EF1α sequences. The results based on protein-coding genes are in accord with those of ITS sequence analysis.
Phylogenetic analysis inferred from ITS, and the combined partial sequences of EF1α and RPB2 genes
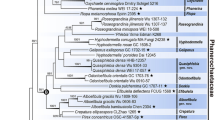
The parsimony analysis of ITS sequences was conducted using 48 taxa, with Hohenbuehelia grisea (Peck) Singer and H. mastrucata (Fr.) Singer designated as outgroups. The aligned dataset contained 694 nucleotide sites, among which 355 characters were constant, 250 were parsimony-informative, and 89 variable characters were parsimony-uninformative. Parsimony analysis resulted in 15 most parsimonious trees, and one of them is shown in Fig. 2.
The combined dataset of RPB2 and EF1α consisted of 2152 nucleotide sites, among which 1751 characters were constant, 199 were parsimony-informative, and 202 variable characters were parsimony-uninformative. Parsimony analysis resulted in eight most parsimonious trees, and one of the most parsimonious trees is shown in Fig. 3. Bayesian analyses produced topologies almost identical to those from parsimony analyses.
In the ITS tree (Fig. 2), all the Pleurotus species we tested formed a monophyletic group with a bootstrap value of 100 %, divided into two different clades. Clade I was composed of P. cystidiosus var. abalonus, P. cystidiosus, P. australis, P. dryinus, P. levis, P. djamor, P. calyptratus, P. opuntiae, P. cornucopiae, and P. citrinopileatus. Bailinggu, P. eryngii, and P. nebrodensis clustered in Clade II with P. abieticola, P. pulmonarius, P. ostreatus, P. sapidus, P. ostreatus var. florida, P. populinus, and P. albidus. Isolates of Bailinggu, P. eryngii, and P. nebrodensis clustered together in the same clade with low support (bootstrap value 65 %), and the latter two groups (P. eryngii, and P. nebrodensis) were nested in an internal clade with a bootstrap value of 81 %. Isolates of cultivated Bailinggu, the wild Awei Mo strains collected from Xinjiang, “GQ456052, P. nebrodensis” and three Iranian “P. nebrodensis” (FJ514550, FJ514589, FJ514568) were clustered in the same clade with strong support (92 %). P. eryngii var. eryngii, P. eryngii var. ferula, and var. elaeoselini were grouped as a monophyletic group (bootstrap value 94 %), while the different varieties could not be separated by their ITS sequences. Additionally, P. nebrodensis from Italy was nested in another clade with a support of 87 %, and it was placed as the sister species to the P. eryngii complex.
Topologies of the P. eryngii complex, P. nebrodensis, and Bailinggu in the trees inferred from the combined EF1α and RPB2 are similar to those from ITS sequences. The three groups formed a monophyletic clade with a support of 95 %, while isolates of Bailinggu, P. eryngii, and P. nebrodensis were nested in different subclades with strong support. The phylogenetic trees placed P. nebrodensis as the closest relative of the P. eryngii species complex. Furthermore, the analysis showed that the representatives of P. eryngii var. eryngii and P. eryngii var. ferula could be separated based on the combined partial sequences of EF1α and RPB2, as previously reported by Rodriguez-Estrada et al. [19].
The results showed that Bailinggu, P. eryngii, and P. nebrodensis are three distinct species rather than varieties within the same species P. eryngii, and that P. nebrodensis is the species most closely related to the P. eryngii complex.
Discussion
Because the application of an incorrect scientific name for Bailinggu may hinder strategies for breeding and create complications for publications, patents and products, the application of a correct scientific name for commercial and research-oriented Bailinggu strains is important. Although doubts concerning the relationship between Bailinggu, P. eryngii, and P. nebrodensis, and the identity of Chinese Bailinggu, have been expressed by some researchers [5, 13, 14], none of them have proved that Bailinggu is a distinct species separate from P. eryngii; or that P. nebrodensis is closer to P. eryngii. There is no doubt that more breeding and scientific research materials of Bailinggu could be uncovered based on the analyses of ITS, RPB2, and EF1α sequences. This study would improve the uncertainty around Bailinggu, P. eryngii, and P. nebrodensis, promoting scientific research and breeding programs for these mushrooms.
Although Bailinggu is the cultivated strain of P. eryngii var. tuoliensis, and that molecular analyses suggest that it is a distinct species from P. nebrodensis and P. eryngii, we hesitate to raise P. eryngii var. tuoliensis to the species leve or to introduce a new name for the well-known Bailinggu, because the holotype of P. eryngii var. tuoliensis is lost, and neither additional materials from the type locality nor from adjacent areas are available for morphological or molecular phylogenetic comparison. For the time being, we propose that the name P. eryngii var. tuoliensis be used for Bailinggu.
P. eryngii var. tuoliensis has been recorded on Ferula sinkiangensis and F. ferulaeoides (Steud.) Korov. in Xinjiang Province, and some researchers traced the cultivation history of Bailinggu and concluded that it is the cultivated strain of P. eryngii var. tuoliensis. Zhang et al. [12] found that there are only two different Pleurotus species, P. eryngii and Bailinggu (as “P. nebrodensis”), on Ferula sinkiangensis. In the present phylogenetic analysis based on ITS sequences, a voucher specimen HM77041 labeled as P. eryngii var. tuoliensis was collected on Ferula in Xinjiang Province, and its morphological characters correspond to the original descriptions of P. eryngii var. tuoliensis except for variation in stipe length. Additionally, ITS, RPB2, and EF1α sequences of this voucher specimen and other two wild Awei Mo strains are almost identical with the cultivated Bailinggu. It seems obvious that Bailinggu should be the cultivated P. eryngii var. tuoliensis. Three Iranian “P. nebrodensis” were also nested in the Bailinggu clade, which were collected on Ferula [32]. In conjunction with previous studies [5, 12, 33], it can be deduced that the distribution of wild Bailinggu is not restricted to China, but that it is also found in other Asiatic areas on Ferula host-plants. Unlike Bailinggu, which is restricted to Ferula, the European P. nebrodensis is associated only with Cachrys ferulacea, and they were placed in a distinct “nebrodensis” clade in the analysis (Fig. 3). P. eryngii has lower host specificity than P. nebrodensis or P. eryngii var. tuoliensis, and various host-plants of P. eryngii have been reported in previous studies [32, 34]. In contrast, the morphological traits of Bailinggu are also different from P. eryngii. Bailinggu always has a white cap, while P. eryngii has a pale brown cap; and the basidiospores of Bailinggu are larger than those of P. eryngii. Differences in habitat, morphological characters, and sequences of ribosomal and protein-coding genes distinguish Bailinggu from P. nebrodensis and P. eryngii.
The relationship among P. eryngii var. tuoliensis, P. nebrodensis, and P. eryngii has been controversial. The present results uncover that they are three different species, and P. nebrodensis rather than P. eryngii var. tuoliensis shows the closest relationship to the P. eryngii species complex. Despite previous studies based on morphological evidence and RAPD analysis supporting the separation of P. nebrodensis from the P. eryngii complex and its classification within a distinct taxonomic entity at the species level [32, 33, 35], Kawai et al. [5] indicated that P. eryngii var. tuoliensis, P. eryngii var. eryngii, and P. eryngii var. nebrodensis should be regarded as different varieties of the same species P. eryngii because propagation occurred among them. In their study, Kawai et al. [5] over-emphasized the biological species concept, but reproductive barriers may not be absolute, and two different species may be compatible when the divergence time between them is short [36]. Additionally, mating rates observed in the laboratory could be greater than those that occur in nature, because gene flow can be reduced because of host specificity and allochrony. The molecular evidence furnished by this work and that of Rodriguez Estrada et al. [19] have further confirmed that P. nebrodensis should be considered a different species rather than a variety. In our phylogenetic analysis, P. eryngii, P. nebrodensis, and P. eryngii var. tuoliensis form a monophyletic group, showing a close relationship between them. This result suggests that the morphological or habit similarity may be a consequence of recent speciation events, and relatively minor genetic differences were found between these species.
In conclusion, ITS, RPB2 and EF1α sequences can be used to discriminate between Bailinggu, Xingbaogu (P. eryngii), and P. nebrodensis. P. eryngii is more closely related to P. nebrodensis than to Bailinggu. Although the name P. nebrodensis has been widely applied to the Chinese species of Bailinggu in recent decades, it is a misapplied name. P. eryngii var. tuoliensis should be recognized as the scientific name for Bailinggu for the time being. P. eryngii var. tuoliensis represents a distinct species, but thorough investigation into Pleurotus isolates from Ferula in Xinjiang Province are needed before raising the taxonomic status of P. eryngii var. tuoliensis or introducing a new name for Bailinggu. Additional specimens of P. eryngii var. tuoliensis collected in the type locality or its adjacent areas need to be examined.
References
Mao XL (2000) Agaricales. In: Mao XL (ed) The macrofungi in China. Henan Science and Technology Press, Zhengzhou, pp 64–66
Mao X (2005) Promoting a new development for precious mushroom Pleurotus nebrodensis (in Chinese). China (Guang Shui) Symposium on Standardization Production for Edible Mushroom & Products Fair for Rare Mushroom (Pleurotus nebrodensis), Hubei, China, January 17–18, pp 25–27 (in Chinese)
Zhang J, Huang C, Li C (2005) The cultivars of P. nebrodensis in China. In: Tan Q, Zhang J, Chen M, Cao H, Buswell JA (eds) Mushroom biology and mushroom products, vol 12. Shanghai Xinhua Printing Co., Ltd, Acta Edulis Fungi, Shanghai, pp 350–353
Huang N (1998) Colored illustrations of macrofungi (mushrooms) of China. China Agricultural Press, Beijing, p 96 (in Chinese)
Kawai G, Babasaki K, Neda H (2008) Taxonomic position of a Chinese Pleurotus ‘‘Bailinggu’’: it belongs to Pleurotus eryngii (DC.: Fr.) Quél. and evolved independently in China. Mycoscience 49:75–87
Zhao M, Huang C, Chen Q, Wu X, Qu J, Zhang J (2013) Genetic variability and population structure of the mushroom Pleurotus eryngii var. tuoliensis. PLoS ONE 8:e83253
Jia SM, Qin M (2006) Domestication and cultivation of Pleurotus nebrodensis in China. Edible Fungi China 25:3–7 (in Chinese)
Xu ML (2010) Sexual distant-crossbreeding between P. eryngii and P. nebrodensis. Master’s Thesis, Fujian Agriculture and Forestry University
Li GX, Shao SG, Li YJ (2004) Effects of different hormones on growth and yield of Pleurotus eryngii var. nebrodensis. Edible Fungi of China 23:37–38
Xu JY (2010) The preliminary study on the structure of A mating type loci in Pleurotus eryngii var. nebrodensis and Pleurotus eryngii var. ferulae. Master’s Thesis, Huazhong Agricultural University
Mou C, Cao Y, Ma J (1987) A new variety of Pleurotus eryngii and its cultural characters. Acta Mycol Sin 6:153–156 (in Chinese)
Zhang JX, Huang CY, Ng TB, Wang HX (2006) Genetic polymorphism of ferula mushroomgrown on Ferula sinkiangensis. Appl Microbiol Biotechnol 71:304–309
Bao D, Kinugasa S, Kitamoto Y (2004) The biological species of oyster mushrooms (Pleurotus spp.) from Asia based on mating compatibility tests. J Wood Sci 50:162–168
Bao D, Ishihara H, Mori N, Kitamoto Y (2004) Phylogenetic analysis of oyster mushrooms (Pleurotus spp.) based on restriction fragment length polymorphisms of the 5′ portion of 26S rDNA. J Wood Sci 50:169–176
Ro HS, Kim SS, Ryu JS, Jeon CO, Lee TS, Lee HS (2007) Comparative studies on the diversity of the edible mushroom Pleurotus eryngii: ITS sequence analysis, RAPD fingerprinting, and physiological characteristics. Mycol Res 111:710–715
Rodriguez Estrada AE (2008) Molecular phylogeny and increases of yield and the antioxidants selenium and ergothioneine in basidiomata of Pleurotus eryngii. Ph.D. Dissertation, The Pennsylvania State University
Avin FA, Bhassu S, Tan YS, Shahbazi P, Vikineswary S (2014) Molecular divergence and species delimitation of the cultivated oyster mushrooms: integration of IGS1 and ITS. Sci World J. doi:10.1155/2014/793414
Froslev TG, Matheny PB, Hibbett DS (2005) Lower level relationships in the mushroom genus Cortinarius (Basidiomycota, Agaricales): a comparison of RPB1, RPB2, and ITS phylogenies. Mol Phylogenet Evol 37:602–618
Rodriguez Estrada AE, del Mar Jimenez-Gasco M, Royse DJ (2010) Pleurotus eryngii species complex: sequence analysis and phylogeny based on partial EF1α and RPB2 genes. Fungal Biol 114:421–428
Schoch CL, Seifert K, Huhndorf S et al (2012) Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proc Natl Acad Sci USA 109:241–6246
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenies. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols, a guide to methods and applications. Academic Press, San Diego
Liu YJ, Whelen S, Hall D (1999) Phylogenetic relationships among Ascomycetes: evidence from an RNA polymerase II subunit. Mol Biol Evol 16:1799–1808
Matheny PB (2005) Improving phylogenetic inference of mushrooms with RPB1 and RPB2 nucleotide sequences (Inocybe; Agaricales). Mol Phylogenet Evol 35:1–20
Marongiu P, Maddau L, Frisullo S, Marras F (2005) A multigene approach for the taxonomic determination of Pleurotus eryngii isolates. In: Tan Q, Zhang J, Chen M, Cao H, Buswell JA (eds) Mushroom Biology and Mushroom Products, vol 12. Shanghai Xinhua Printing Co., Ltd, Acta Edulis Fungi, Shanghai, pp 89–91
Wendland J, Kothe E (1997) Isolation of tef1 encoding translation elongation factor EF-1α from the homobasidiomycete Schizophyllum commune. Mycol Res 101:798–802
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Hall TA (1999) Bioedit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Swofford DL (2003) PAUP*: phylogenetic analysis using parsimony (*and other methods) version 4.0b10. Sinauer, Sunderland
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Ravash R, Shiran B, Alavi A-A, Bayat F, Rajaee S, Zervakis GI (2010) Genetic variability and molecular phylogeny of Pleurotus eryngii species-complex isolates from Iran, and notes on the systematics of Asiatic populations. Mycol Prog 9:181–194
Zervakis G, Venturella G, Papadopoulou K (2001) Genetic polymorphism and taxonomic infrastructure of the Pleurotus eryngii species complex as determined by RAPD analysis, isozyme profiles and ecomorphological characters. Microbiology 147:3183–3194
Zervakis G, Balis C (1996) A pluralistic approach on the study of Pleurotus species, with emphasis on compatibility and physiology of the European morphotaxa. Mycol Res 100:717–731
Venturella G (2000) Typification of Pleurotus nebrodensis. Mycotaxon 75:229–231
Taylor JW, Jacobson DJ, Kroken S, Kasuga T, Geiser DM, Hibbett DS, Fisher MC (2000) Phylogenetic species recognition and species concepts in fungi. Fungal Genet Biol 31:21–32
Acknowledgments
Dr. Egon Horak is acknowledged for valuable suggestions to improve the manuscript. The research was financed by the Applied Basic Research, Science and Technology Department of Sichuan Province (Project No. 2013JY0114), National Public Welfare (Agriculture) Science and Technology Project (201503137), Sichuan Provincial Infrastructure of Microbial Resources (15010302) and Sichuan Provincial Innovation Ability Promotion Engineering (Project No. 2014LWJJ-005).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
He, XL., Wu, B., Li, Q. et al. Phylogenetic relationship of two popular edible Pleurotus in China, Bailinggu (P. eryngii var. tuoliensis) and Xingbaogu (P. eryngii), determined by ITS, RPB2 and EF1α sequences. Mol Biol Rep 43, 573–582 (2016). https://doi.org/10.1007/s11033-016-3982-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-016-3982-2