Abstract
The Pleurotus eryngii species-complex includes taxa of the northern hemisphere growing in association with plants of the family Apiaceae (umbellifers). In this study, 45 Pleurotus strains were isolated from five different host-plants: Ferula ovina, F. assa-foetida, Smyrniopsis aucheri, Kellusia odoratissima, and Cachrys ferulacea; all plant species, with the exception of C. ferulacea, are reported for the first time as hosts for this fungal group. Random amplified polymorphic DNA-PCR (RAPD) analysis and nucleotide sequence data from the internal transcribed spacer of the nuclear rRNA genes (ITS1, 5.8S and ITS2) were used for assessing genetic diversity and for determining phylogenetic relationships among the populations studied. Results permitted the grouping of the strains studied into three major clusters corresponding mainly to the nature of the host-plant: the first included isolates collected from Ferula spp. only, the second included isolates originating from C. ferulacea only but from various sampling locations, and the third included all K. odoratissima and S. aucheri associated strains plus a few isolates collected from F. ovina and C. ferulacea. The grouping of the Iranian material, in conjunction with the position in the resulting phylograms of other previously obtained P. eryngii complex sequences, revealed that the first cluster is related to the asiatic ‘P. nebrodensis’ (or to the asiatic Ferula spp. associated Pleuroti), the second forms a rather distinct lineage which is linked with reference strains originally classified as P. fossulatus, whereas the third cluster falls within the main part (or the “core”) of this complex, i.e., P. eryngii. Pleurotus populations growing on umbellifers in Iran seem either to have recently diverged through a sympatric speciation process based mainly on ecological factors (e.g., P. fossulatus), or they form part of a rather wide agglomerate associated with various host-plants where exchange of genetic material is still in progress (i.e., P. eryngii).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Iran is a country with vast mountainous regions, varied climatic and edaphic features, and a particularly rich plant biodiversity. Especially as regards the family Apiaceae (umbellifers), Iran constitutes a major center of diversification where a considerable level of taxa diversity and a high number of endemic species occur; notably, a total of 363 species and 114 genera of Apiaceae are recorded in this country, of which 114 species and 12 genera are endemic (Ajani et al. 2008; Pimenov and Leonov 2004). Among them, species of the genera Cachrys, Ferula, Smyrniopsis and Kelussia attract significant interest from both local scientists and village people towards their exploitation for various purposes, i.e., human food (e.g., K. odoratissima, commonly known as “mountain celery”), folk medicine applications, animal feed, production of essential oils for pharmaceutical and nutritive applications, etc. (Amiri 2007; Faridi et al. 2008; Ghorbani 2005; Khajeh et al. 2005). Of relevant importance is the consumption of Pleurotus mushrooms, which grow on umbellifers in Iran, either fresh as food readily available in local markets or in the form of dried powder used in traditional medicine.
The Pleurotus eryngii species-complex constitutes the only group of the genus Pleurotus whose members are associated with plants of the family Apiaceae by developing a facultatively biotrophic mode of growth (Hilber 1982; Joly et al. 1990; Zervakis and Balis 1996). Basidiomata of host-specific populations appear on the roots and stems of umbellifers (e.g., genera Eryngium, Ferula, Ferulago, Cachrys, Laserpitium, Diplotaenia, Elaeoselinum, Thapsia, etc.) singly or in groups from autumn until early summer. The distribution range of this species-complex extends through a rather well-defined zone in the Old World ranging from Morocco to the Netherlands in the west and extends to China eastwards, comprising regions of the Middle East and western Asia (e.g., Turkey, Israel, Armenia, Uzbekistan, Afghanistan) (Hilber 1982; Zervakis et al. 2001).
As regards Iran in particular, the existence of P. eryngii was first reported by Petrak (1939) [as P. fuscus (Batt.) Bres.] and Esfandiari (1948). Later, Heim (1960) recorded the presence of what he called ‘Pleurotus nebrodensis’ growing on root and stem residues of Diplotaenia cachrydifolia in the Fars mountains (Shiraz area, south Iran). Further information about the occurrence of P. eryngii species-complex in the Esfahan, Fars, Boyer-ahmad and Kohkiloyeh, and Tehran provinces was provided by Saber (1990, 1997), who reported that such fungal populations are abundant and widespread in central, southern and western regions. Recently, Abdollahzadeh et al. (2007) studied P. eryngii species-complex isolates from the Kurdistan region of northern Iran (at altitudes higher than 2,000 m asl in the areas of Sanandaj, Hane Gelan and Saral) and in association with the host plants Prangos sp., Pimpinella sp. and Ferula haussknechtii.
However, identification of Pleurotus isolates and establishment of taxonomic/phylogenetic relationships (together with assessment of intraspecific genetic variability/polymorphism) within this species-complex and among its various host-specific populations (‘ecotypes’) remain problematic since most morphological characteristics of basidiomata are influenced by environmental conditions, while initial misidentifications of the host-plant and/or of the associated fungal specimens have led to erroneous taxonomic conclusions (Hilber 1982; Zervakis and Balis 1996). Such problems were further aggravated by the introduction of new taxa (without the necessary support from robust taxonomic data originating from the use of additional criteria, e.g., ecomorphological characters, compatibility experiments, molecular analysis, etc.), and the complex processes governing speciation in this group (Zervakis et al. 2001).
The application of integrated approaches, usually employing one or more molecular techniques, have contributed to the elucidation of such ambiguous issues in Basidiomycota. Hence, for example, the use of random amplified polymorphic DNA (RAPD) permitted the assessment of population dynamics of Tricholoma spp. (Gryta et al. 2006), of the genetic variation in Hemileia vastatrix (Gouveia et al. 2005), and of the relationships among Lepista spp. (Stott et al. 2005). On the other hand, ribosomal DNA internal transcribed spacers (rDNA ITS-5.8S) sequencing provided valuable data for supporting species delimitations in the genera Amanita, Lactarius, Cystoderma and Cystodermella (Moreno et al. 2008; Nuytinck and Verbeken 2007; Saar et al. 2009), for assessing phylogeny within Cortinarius and Polyporus (Frøslev et al. 2007; Krüger et al. 2006), and for establishing long distance dispersal phenomena in Ganoderma and biogeography of Flammulina spp. (Ge et al. 2008; Moncalvo and Buchanan 2008).
This work aimed at determining the genetic polymorphism, the taxonomic identity and the phylogenetic relationships of Pleurotus isolates growing on several Apiaceae host-plants in poorly investigated areas of Iran by evaluating for the first time their host-plant associations and biogeographic distribution in conjunction with the data provided through the use of RAPD and ITS sequencing.
Materials and methods
Biological material
Basidiomata of 45 Pleurotus strains were collected directly from the roots and stems of their associated Apiaceae host-plants in 12 different locations of western Iran from April to May 2006 (Table 1). Sampling locations, their geographic coordinates and altitudes above sea level appear in Fig. 1.
Map of Iran with the main sampling region and individual sampling localities of the 45 Pleurotus eryngii complex strains marked in numbered circles: (1) Felard (host-plant: S. aucheri, longitude-latitude (°N–°E): 51.18–31.17, altitude (m): 2267); (2) Kohrig (C. ferulacea, 50.53–31.21, 2632); (3) Sardasht (C. ferulacea, 50.51–31.22, 2075); (4) Mangase (C. ferulacea, 50.49–31.31, 2297); (5) Gharun (C. ferulacea, 50.22–31.27, 3200); (6) Bazoft (K. odoratissima, 50.07–31.12, 2420); (7) Aligudarz (S. aucheri and F. assa-foetida, 49.41–32.24, 2034); (8) Fereidan (C. ferulacea, 50.06–32.56, 3585); (9) Sepidan (C. ferulacea, 51.47–30.17, 1620); (10) Dashtezarrin (C. ferulacea, 51.03–30.61, 2873); (11) Ahmadegharib (F. ovina, 51.19–30.43, 2860); (12) Pirmehran (F. ovina, 51.35–30.40, 1837)
For the establishment of dikaryons in pure culture, small pieces from the context of the isolated basidiomata were removed aseptically and transferred onto Petri dishes containing malt extract agar medium (MEA). After a few days of incubation at 27°C in the dark, hyphal tips were transferred to fresh medium. Pleurotus strains were maintained on MEA, and pure cultures were stored at −80°C in the fungal culture collection of the Department of Agronomy and Plant Breeding, Shahrekord University. All voucher specimens are deposited in the Herbarium of the Agricultural Research Centre, Shahrekord.
DNA extraction and RAPD-PCR analysis
Each fungal isolate was inoculated in Petri dishes with MEA medium and the resulting culture was incubated for two weeks at 27°C. Then, small pieces of mycelium were transferred into 250 ml flasks containing 100 ml of sterile malt extract and incubated at 23°C for 20 days (the cultures were shaken twice per day). When the fungus colonized the medium, mycelium was harvested through the use of a filter paper placed on the top of a funnel and then washed with sterile distilled water (Zervakis et al. 1994).
Isolation of total genomic DNA followed the protocol of Rogers and Bendich (1988) as modified by Zervakis et al. (2001). Mycelium (1.5–2.5 g) was frozen with liquid nitrogen and quickly homogenized into fine powder. The powder was mixed with 500–700 µl warm (60°C) DNA extraction buffer (50 mM Tris pH 8.0, 50 mM EDTA, 3% w/v SDS and 0.1 mg/ml proteinase K) into 2 ml microfuge tubes and extracted with phenol/chloroform/isoamyl alcohol (25:24:1). The DNA was precipitated with 0.1 vol of 3 M sodium acetate and 0.6 vol of isopropanol, washed with 70% ethanol and resuspended in water. The purity and quality of genomic DNA was determined spectrophotometrically and confirmed by use of 1.2% agarose gel electrophoresis. DNA stock solution was stored at −20°C until needed.
Initially, a subset of the isolates was used to perform a preliminary screening of 55 decamer oligonucleotide primers to identify those that provided reproducible marker profiles and to exclude those producing a very low proportion of polymorphic bands. Ten primers (OPG kit, Operon Technologies Inc., USA) were selected for further analysis of the 45 DNA samples: OPG 05, OPG 07, OPG 08, OPG 10, OPG 19, OPG 44, OPG 47, OPG 52, OPG 58, and OPG 68 (Table 2).
RAPD analysis was carried out by using the method of Williams et al. (1990). Amplification reactions were performed in a final volume of 25 μl containing 75 ng genomic DNA; the reaction solution consisted of 200 μМ each of dATP, dCTP, dGTP and dTTP, 15 ng oligonucleotide primer (Genset, France) and 1.2 units Taq DNA polymerase (Fermentas), 1.5 mМ MgCl2 and 1x PCR buffer. Amplification was performed in an Eppendorff Mastercycler gradient thermalcycler as follows: pre-denaturation stage at 94°C for 3 min followed by 45 cycles at 94 for 1 min, 37°C for 1 min, 72°C for 2 min, followed by an extension stage of 10 min at 72°C. Amplified fragments were resolved on a 1.2% agarose gel and stained by ethidium bromide. A 100 bp ladder DNA marker (Fermentas) was used as a size standard.
PCR, cloning and sequencing of the rDNA-ITS region
The entire region of nuclear ribosomal DNA which comprises both internal transcribed spacers ITS1 and ITS2 and the 5.8S subunit was PCR amplified in each of the isolates with primers ITS1 and ITS4 (White et al. 1990). Primer ITS1 (TCC GTA GGT GAA CCT GCG C) binds to the 3′ end of the small subunit (ssu) of the 18 S rDNA gene and primer ITS4 (TCC TCC GCT TAT TGA TAT GC) binds to the 5′ end of the large subunit (lsu) of the 28 S rDNA gene. A reaction mixture (50 μl) containing 10x reaction buffer, 2 mM MgCl2, 250 µM of each dNTP, 0.4 µM of each primer, 1.5 unit of Taq DNA polymerase (Fermentas International Inc., Canada) and 50 ng of purified total DNA was prepared. The amplification was performed using an Eppendorff Mastercycler gradient thermalcycler which was set to run at 94°C for 3 min for initial denaturation followed by 35 cycles of 94°C for 30 S, 50°C for 1 min, and 72°C for 1 min. A final extension of 7 min at 72°C was included. After confirming the amplification of single size fragment (∼680 bp) as revealed by the presence of a single band on 1% agarose gel, the agarose block containing DNA was excised from the gel with a razor blade. Purified DNA was recovered from the agarose using the DNA extraction kit (#K0513, Fermentas).
To obtain a clean sequencing, the ITS region was cloned. The PCR products of each of one of the 45 isolates were ligated into the pTZ57R/T vector and cloned using the InsT/A cloning kit (#K1214, Fermentas) following the manufacturer’s protocol. Recombinant plasmids were identified by colour selection after growth on Luria-Bertani plates containing X-Gal. The positive transformation of the bacterial clones was reconfirmed by using a small portion of a colony as the PCR template. Plasmid minipreps were performed using Genejet clean (Fermentas) following the manufacturer’s protocol. The plasmid DNA was used for PCR amplification of the ITS insert with both forward and reverse sequencing primers M13/pUC. Sequencing was performed by the Geneservice Ltd (Cambridge, UK) through the use of an ABI automated sequencer. Resulting chromatograms were assembled and edited with the DNAstar software (DNASTAR Inc., Madison, USA). The sequences have been deposited in GenBank/NCBI (accession numbers: FJ514549 to FJ514590, Table 1). Alignments were performed by using the CLUSTAL W (http://www.ebi.ac.uk/clustalw/). Gap regions with ambiguous alignment were excluded from the analyses.
Data scoring, statistical and phylogenetic analyses
For RAPD analysis, each amplification run included a negative control reaction without the addition of DNA, and each reaction was performed at least twice. Some variations in RAPD patterns were detected in the duplicate experiments. However, only distinct, clearly resolved and reproducibly amplified fragments were selected for RAPD analysis, most of which ranged in size from 0.25 to 3.5 kb (PCR products sized below 250 bp or above 3.5 kb yielded faint and irreproducible bands). Comparisons of RAPD profiles were made only between samples that were included in the same run, and which had been separated on the same agarose gel. There was no differential weighting for band intensity. The assumption was made that amplification products of the same size, which were present in the profiles generated by different isolates, represented products from equivalent loci. In total, 150 distinct and scorable bands were produced by the 45 Pleurotus isolates examined.
The presence or absence of DNA fragments generated by RAPD-PCR was assessed, and scored as present (1) or absent (0). This information was used to compile a binary data matrix. Genetic similarities were calculated among all possible pairs of strains using Jacard’s similarity index, and principal coordinate analysis (PCoA) was performed with NTSYS-PC 2.02 software (Rohlf 1998); PCoA starts with a matrix of similarities between the strains examined and depicts the data in three dimensions in such a way that the distances between points (strains) are as close as possible to the original state (Singh et al. 2006). Partitioning of molecular variance within and among populations was calculated by AMOVA (Excoffier et al. 1992) through the use of the ARLEQUIN software (Schneider et al. 2001).
The RAPD data matrix was also used for the calculation of distances between strains on the basis of mean genetic differences, and for the assessment of their phylogenetic relationships through the use of PAUP* (version 4.0b10) software developed for Macintosh PowerPC (Swofford 2002). All 150 characters were unordered and of equal weight. Cluster analysis was performed by the unweighted pair group method using arithmetic averages (UPGMA) with mean character difference as the distance measure; branch robustness of the derived cladograms was evaluated by PAUP* using two different methods: (a) bootstrapping (Felsenstein 1978), and (b) jackknife-resampling (Farris et al. 1996) with 1000 replicates. In the jackknife analysis, 50% of the characters were deleted in each replicate.
The PAUP* software was also used for ITS1, ITS2 and 5.8S rDNA sequence analysis, and maximum-parsimony was selected as the optimality criterion. The characters were unordered and weighted equally. Heuristic searches used 100 replicates of random addition sequence with tree-bisection-reconnection (TBR) branch-swapping. Other options in PAUP* were set as follows: starting trees obtained via stepwise addition, one tree held at each step during stepwise addition, MULPARS option in effect, steepest descent option not in effect, MAXTREES setting unlimited and branches having maximum length zero allowed to collapse to yield polytomies. Branch robustness was evaluated again by bootstrap and jackknife methods which used 100 replicates of heuristic searches with the same settings as above, except that in each replicate MAXTREES was set to 541 (i.e., number of all most-parsimonious trees derived from the respective heuristic searches).
In addition to the Iranian material, eight sequences obtained from NCBI were included in the analysis as “reference material” (taxon assignment was maintained as appearing in the NCBI information form): P. eryngii (strain code: A6690, origin: Austria, NCBI accession no.: AY450347), Pleurotus sp. (PHZAU18, DQ077883), P. eryngii (ATCC36047, ex-Czechoslovakia, AY368657), P. fossulatus (ATCC62885, India, AY368664), P. fossulatus (CCRC36238, AY265833), P. nebrodensis (S498, China, AY540331), P. nebrodensis (strain no. 4, AY720935), and P. nebrodensis (ACCC50869, host-plant: Ferula sinkiangensis, China, AY311408).
Results
Analyses of RAPD data
Ten primers were selected to assess the genetic diversity within a collection of 45 P. eryngii species-complex isolates (Table 2). A minimum of 9 (OPG8) and a maximum of 21 (OPG44) unambiguously amplified bands were generated furnishing a total of 150 bands ranging in size from 0.25 to 3.50 kb; 148 of these bands were polymorphic (98.67%). Only one band (size: 1350 bp) produced by primer OPG58, and one band (size: 510 bp) produced by OPG68 were monomorphic. All banding patterns were unique for each strain studied; in addition, a number of bands could be used as molecular markers for the identification of host-specific Pleurotus strains. Thus, all Pleurotus strains growing in association with F. ovina produced two common DNA bands, with approximate sizes 930 and 970 bp (primers OPG10 and OPG19 respectively); the latter was unique for this particular host, while the former was shared with strains growing on S. aucheri as well. Furthermore, all strains associated with C. ferulacea shared three DNA fragments with approximate sizes 1250, 980 and 850 bp (primers OPB7, OPG7 and OPG8 respectively); however, all of them appeared also in strains isolated from other host-plant species: the first in S. aucheri, the other two in K. odoratissima. All Pleurotus strains associated with K. odoratissima produced 16 common DNA bands in total, whereas isolates from S. aucheri shared 14 DNA fragments. In such cases, combinations of three or more DNA fragments could potentially discriminate among Pleurotus isolates originating from different host-plants.
Statistical (PAUP*) treatment of RAPD data produced a matrix of pairwise distances (not presented) between Pleurotus strains based on their mean character differences. It is interesting to note that the lowest distance values were obtained between strains isolated from the same host in the same location, i.e., the group of strains C10, C13 and C14 (distances: 0.013–0.040, C. ferulacea, Gharun), C44 and C45 (0.053, F. assa-foetida, Aligudarz), and C21 and C22 (0.100, F. ovina, Ahmadegharib). In contrast, the highest distances were observed mainly between strains isolated from different hosts and locations, e.g., C2 and C4 vs. C12 (0.500-0.520, F. ovina and C. ferulacea, Pirmehran and Gharun respectively), and C32 vs. C37 (0.507, K. odoratissima and C. ferulacea, Bazoft and Kohring respectively), or rarely from the same host but from distant geographic origins (e.g., C12 vs. C29, 0.513, C. ferulacea, Gharun and Fereidan).
RAPD data permitted the separation of Pleurotus strains into three large clusters: Clusters I, II and III (Fig. 3), which were supported by high bootstrap and jackknife resampling values (>96%/94%) in distance/UPGMA trees. The first (Cluster I) included eight strains (C1, C2, C18, C20, CP21, C22, C44 and C45) which were isolated from Ferula host plants only, i.e., six out of a total of ten from F. ovina, plus both strains originating from the same host-plant and locality (F. assa-foetida in Aligudarz). Positioning of the latter two (C44 and C45) was rather distinct and supported by high statistical value (100%), while strains C20, C21 and C22 associated with F. ovina (Ahmadegharib) were also closely related. Pairwise distances within this cluster ranged from 0.053 (C44 vs. C45) to 0.360 (C20 vs. C45). Cluster II included 14 strains isolated exclusively from C. ferulacea plants (C10–C15, C17, C24, C36–C40), with the only exception of strain C9 (unknown origin). In particular, strains C10 to C15 (all from C. ferulacea in Gharun), and the pairs of strains C36–C38 and C37–C39 (all from C. ferulacea and from adjoining geographic localities) demonstrated close affinity combined with relatively high bootstrap support. Pairwise distances within this cluster ranged from 0.013 (C13 vs. C14) to 0.307 (C9 vs. C36, and C12 vs. C24). The rest (23) of the strains investigated were positioned in Cluster III, i.e., all strains isolated from K. odoratissima and S. aucheri, plus a few strains from F. ovina and C. ferulacea. Within this cluster several strains were grouped closely together, i.e., C41, C42 and C43 (all from S. aucheri in Aligudarz), all but one isolates from K. odoratissima (in Bezoft), as well as strains C6 and C7 (from S. aucheri in Felard), and C28 and C30 (C. ferulacea in Fereidan). Pairwise distances within this cluster ranged from 0.127 (C41 vs. C42) to 0.393 (C27 vs. C5 and C25). “Inter-Cluster” comparisons of pairwise distances revealed considerably higher values, e.g., members of Clusters I vs. II (e.g., 0.520, C2 vs. C12), or II vs III (e.g., 0.513, C12 vs. C29, and 0.507, C32 vs. C37).
Results from application of PCoA separated strains under study into three major groups (Fig. 4) in accordance with the results of the clustering analysis presented above. The first group consisted of eight isolates (C1, C2, C18, C20, CP21, C22, C44 and C45) associated with Ferula ovina and F. assa-foetida plant-hosts, the second group was composed of 14 strains isolated from Cachrys ferulacea (C10–C15, C17, C24, C36–C40), with the only exception of strain C9 (unknown origin), and the remaining 23 isolates appeared in the third group. Eigenvalues and cumulative values of PCoA were obtained (data not presented); 24.28% of the total variation corresponded to the first principal component alone, while 80.84% of the total variation were obtained by the first 16 principal components.
Based on the average of 148 polymorphic loci, an analysis of molecular variance was performed taking into consideration either the ten geographic origins of the studied populations [Pirmehran, Felard, Gharun, Dashtezarrin, Ahmadegharib, Sepidan, Fereidan, Bazoft, Lordegan (incl. Mangese, Sardasht and Kohrig) and Aligudarz] or the five associated host-plants. AMOVA revealed that variance within localities accounted for 71.33% of total variance, whereas variance between localities accounted to only 28.67% of the total; the respective values among host-plants was 20.19% and within host-plants 79.81% (Table 3). Genetic diversity within P. eryngii complex strains, expressed as AMOVA mean square deviations, was positively correlated (r = 0.95; P < 0.001) with percentages of polymorphic markers detected per strain. The Bartlett’s test for population heteroscedasticity produced highly significant values indicating different levels of variability within different P. eryngii complex strains (Table 3).
ITS sequencing data analysis
Sizes of amplified products ranged from approximately 730 to 750 base pairs. Phylogenetic analyses were executed with 42 unambiguously aligned sequences of 709 base pairs (no sequences were obtained for strains C14, C21 and C43) plus the eight sequences obtained from the NCBI; 641 positions were constant, 48 variable characters were parsimony-uninformative and 20 variable characters were parsimony-informative.
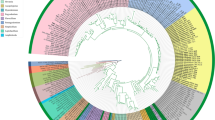
Eleven clades that were supported by 50% or higher bootstrap values (Fig. 5) were produced, whereas eight of these clades were also retained by jackknife-resampling with a statistical support higher than 50%. Four clades had bootstrap support values higher than 83% (and >78% with jackknife). Bootstrapping generally gave slightly higher statistical support than jackknifing, but both statistics were strongly correlated (R = 0.96; data not shown). Pleurotus strains were separated into three major clades (Clusters I, II and III, Fig. 5), or into three well-supported taxa that strongly (but not fully) correspond to the associated host-plants of the strains examined. The first (I) included six strains (C1, C2, C18, C22, C44 and C45) which were isolated from Ferula host plants only, i.e., four out of a total of nine from F. ovina, plus both strains originating from F. assa-foetida. Cluster II included 13 strains isolated exclusively from C. ferulacea plants, with the only exception of strain P9 of unknown origin. The rest (23) of the strains investigated were positioned in Cluster III, i.e., all strains isolated from K. odoratissima and S. aucheri, plus a few strains from F. ovina and C. ferulacea (as was the case in the RAPD grouping). All three clusters were supported by high bootstrap and jackknife resampling values (Fig. 5). For Cluster I in particular, C44 and C45 (from F. assa-foetida in Aligudarz) presented the closest affinity. Moreover, three previously studied strains (ACCC50869, Strain4 and S498), two of them originating from China and identified as P. nebrodensis, were grouped within this cluster (supported by bootstrap and jackknife values of 74% and 60% respectively). For Cluster II, identical sequences were obtained for strains C12, C13, C38 and C39 (all originating from C. ferulacea, and collected from the neighboring localities of Gharun and Shardasht). In addition, strains C15, C17 and C37, which were collected from the neighboring localities of Dashtezarrin and Kohrig, were grouped closely together. All Iranian strains of this cluster were linked together and were then affiliated with strains ATCC62885 (from India) and CCRC36238 (of unknown origin); the latter two previously identified as P. fossulatus appeared closely related. Last, in Cluster III, identical sequences were shared by strains C16, C19, C20 and C33, which originated from different hosts (C. ferulacea, K. odoratissima and F. ovina) but from nearby localities (Dashtezarrin, Bazoft and Ahmadegharib); and by strains C8, C26, C31 and C42, which were again isolated from various hosts (S. aucheri, C. ferulacea and K. odoratissima) and different origins. Eleven of the 23 Iranian strains of this cluster formed a sub-clade, while the following strains with sequences obtained from previous studies were also grouped within Cluster III: ATCC36047 (P. eryngii from former Czechoslovakia), PHZAU18 (deposited as Pleurotus sp.) and 6690 (P. eryngii from Austria). The whole clade was supported by high bootstrap and jackknife values (97% and 93% respectively), and this was also the case for the closely associated (99%) sister groups corresponding to Clusters II and III.
Partial 18S rRNA gene, the entire ITS1-5.8S rDNA-ITS2 and partial 28S rRNA gene sequences varied in length from 637 bp in strains C29 and C45 to 641 bp in strain C22. The highest divergence measured within one major cluster (I, II or III) or terminal clade was 1.59% (10 bp) for Cluster I (strain C2 vs. ACCC50869); the respective values among strains within Clusters II and III were 0.94% (6 bp, C11 vs. C37) and 1.41% (9 bp, PHZAU18 vs. C4 and C32). When comparisons were performed for pairs of sequences of the entire Pleurotus population examined, the highest values were obtained between members of Cluster I vs. those of Cluster III, e.g., 3.34% (21 bp, ACCC50 vs. PHZAU18), 3.03% (19 bp, ACCC50 vs. C27) or 2.35% (15 bp, C2 vs. C28) for Iranian material only. On the other hand, the lowest percentages of base differences in ‘inter-Cluster’ comparisons were noted between strains of Cluster II vs. those of Cluster III (e.g., 1.57%, 10 bp, C23 vs. C37; 1.41%, 9 bp, ATCC62885 vs. C32 and vs. C4), whereas relatively high values were calculated for comparisons between members of Cluster I vs. those of Cluster II (e.g., 2.39%, 15 bp, ACCC50 vs. ATCC62885 and vs. C40; 1.88%, 12 bp, C40 vs. C2 and vs. C44).
Discussion
Iran is a country which, despite its particularly rich plant biodiversity, remains largely under-investigated as regards the wealth of its plant-associated microbiota. Especially Pleurotus taxa, which grow as facultative biotrophs of Apiaceae species, present particular interest not only because of their particular growth habits and ecological preferences but also because of their economic importance to local communities. In the frame of the present study, 45 dikaryotic Pleurotus strains growing on umbellifers were isolated directly from five different host-plants in 12 locations of western Iran, and they were examined through the use of molecular techniques for determining their taxonomic identity and phylogenetic relationships, and for assessing the existing genetic polymorphism. Such novel results combined with ecological preferences and distribution data of the Iranian material could actually form a type of “connecting link” between pertinent information already available from European and East Asian populations of this species-complex (Kawai et al. 2008; Urbanelli et al. 2007; Zervakis and Balis 1996; Zervakis et al. 2001; Zhang et al. 2006).
The geographic distribution for the host-plants of the Iranian Pleurotus strains studied coincides with what for European countries is the distribution zone of the P. eryngii complex associated with C. ferulacea (this host-plant is the only common one among those examined in this work and in other previous relevant studies on this fungal group). In addition, K. odoratissima is endemic in Iran (found only in the western part), while S. aucheri, F. ovina and F. assa-foetida occur in central and western Asia only (Pimenov and Leonov 2004); these last four plant species are reported here for the first time as hosts for fungi of the P. eryngii complex. It should be noted that all 12 localities investigated for the occurrence of Pleurotus specimens growing in association in umbellifers formed part of mountainous regions in western Iran exceeding altitudes of 1620 m (Fig. 1). At these particular sites average annual values of temperatures ranged from 7 to 16°C, rainfalls from 384 to 900 mm, and relative humidities from 41 to 47%.
RAPD-PCR provides the means of assessing polymorphisms at a wide range of loci (Williams et al. 1990). Every strain examined in this study (through the use of 10 selected primers) showed a unique genotype, even within the individual clusters formed from the cladistic analysis of pertinent data (Figs. 2 and 3). Molecular variation among Pleurotus isolates yielded up to 98.67% polymorphic bands, which represents a particularly high degree of polymorphism. Similar polymorphism levels (99.4%) were observed in a study on Israeli P. eryngii complex strains assessed through the use of 12 primers, which amplified 164 scorable RAPD loci (Lewinsohn et al. 2001). Furthermore, high intraspecific genetic diversity values were noted in the tree endophyte Gnomonia setacea (Lappalainen and Yli-Mattila 1999), in the ant-symbiotic basidiomycete Leucoagaricus gongylophorus (Doherty et al. 2003), in the grass pathogen Claviceps purpurea (Jungehülsing and Tudzynski 1997), etc. Such cases were mainly attributed to the predominance of sexual reproduction, to adaptation to stressful and temporally heterogeneous environments and/or to the horizontal transmission system of propagules (airborne spores). On the other hand, adaptation of biotrophic and symbiotic fungi to different hosts causes accumulation of genetic differences within the same species due to isolation phenomena.
Statistical analysis of the RAPD data revealed that variation within host-specific populations was high as compared to variation between populations of the P. eryngii complex, which confirms previous results of a study with Italian strains of this complex (De Gioia et al. 2005). The relatively high genetic diversity detected within groups is probably due to an efficient gene flow among intercompatible strains within each group. Such levels of genetic variability are usually observed in populations which are of wild origin, reproduce sexually, have broad ecological niches and/or a wide geographical distribution (Babbel and Selander 1974; Hamrick and Godt 1990; James et al. 1999); populations of the P. eryngii complex share several of the above characteristics. In addition, what is more important, the congruence of RAPD data was tested against other sources of information (i.e., host-plant preferences and ITS sequence data), and they were demonstrated to adequately reflect relationships within the P. eryngii complex.
On the other hand, the internal transcribed spacer (ITS) region of the nuclear ribosomal repeat unit has proved to be a valuable tool for species delimitation and subgeneric phylogenetic inference in pertinent fungal studies (Kretzer et al. 1996; Taylor et al. 2000). The region is known to show certain variability even within species, and it might be also used to distinguish among strains of the same species (Chiu 2007; Healy et al. 2004). However, RAPD fingerprinting analysis seems superior in distinguishing closely related Pleurotus strains as was evidenced from this and previous studies (Ro et al. 2007).
ITS sequence divergence within host-associated isolates was lower than between host-associated populations. The highest intrataxon divergence measured was 1.59% (Cluster I) followed closely by 1.41% (Cluster III), which are similar to pertinent values noted for Lactarius deterrimus (1.35%; Nuytinck and Verbeken 2007) or Collybia s.str. (up to 1.22%; Hughes et al. 2001), higher than in Amanita porphyria (0.50%; Zhang et al. 2004) or in Pleurotus pulmonarius (0.58%; Vilgalys and Sun 1994), and significantly lower than in Pleurotus cystidiosus (up to 6.9%; Zervakis et al. 2004) or Suillus granulatus (6.04%; Kretzer et al. 1996). On the other hand, intertaxon sequence divergence values ranged from 1.41% to 3.34%, which are comparable to those noted between the very closely related Lactarius sanguifluus vs. L. vinosus (1.42 %) and L. deterrimus vs. L. fennoscandicus (1.37 %), and to the average interspecific distance (3.46%) within Lactarius sect. Deliciosi (Nuytinck and Verbeken 2007).
The use of RAPD-PCR and ITS sequence analyses permitted grouping of Iranian P. eryngii complex isolates into three principal clusters-lineages mainly in accordance with the separation of individual populations on host specialization (Figs. 3, 4 and 5). Two of the three large groups (Clusters I and II) were composed of strains isolated from Ferula spp. and C. ferulacea respectively, from various sampling locations; in addition, Pleurotus strains associated with the former host-plant produced distinctly higher mycelium growth rates (data not presented), possibly as a result of adaptation on this particular substrate type. In the derived phylograms, three strains originally classified as P. nebrodensis (two of them originating from China, and one of them collected from F. sinkiangensis) were included into a distinct subgroup of Cluster I, whereas two sequences of strains previously identified as P. fossulatus (one originating from India) also formed a distinct terminal clade within Cluster II. Therefore, the separation of Cluster I and II members from the rest of the Iranian specimens examined, and their classification as two distinct taxonomic entities at the species level, i.e., P. nebrodensis sensu lato (Cluster I) and P. fossulatus (Cluster II), seems well justified.
One of the 541 equally parsimonious trees found by parsimony analysis of the ITS data from the analysis of 42 Pleurotus eryngii complex strains produced with PAUP* (length: 77, CI: 0.935, RI: 0.965, HI: 0.065); the depicted tree coincides with the strict consensus tree. The numbers at branch points represent bootstrap/jackknife support (values appear only when exceeding 50%). Eight additional sequences were obtained from NCBI and included in the analysis as “reference material”: P. eryngii (strain code: A6690), Pleurotus sp. (PHZAU18), P. eryngii (ATCC36047), P. fossulatus (ATCC62885), P. fossulatus (CCRC36238), P. nebrodensis (S498), P. nebrodensis (strain no. 4), and P. nebrodensis (ACCC50869) (for more details see Materials and Methods)
Specimens from several geographic origins were grouped in Cluster III, including all K. odoratissima and S. aucheri associated strains plus the few remaining isolates collected from F. ovina and C. ferulacea. Three sequences obtained from previous studies were also grouped within Cluster III, two of them from P. eryngii var. eryngii from Europe (the third, PHZAU18, was deposited as Pleurotus sp.). Hence, Cluster III corresponds to P. eryngii, which is therefore expanded by engulfing Pleurotus isolates associated with additional previously unreported host-plants, such as K. odoratissima, S. aucheri and F. ovina. Especially as regards C. ferulacea, it has been until now exclusively linked with the appearance of P. nebrodensis in Europe and in western Asia (Hilber 1982; Joly et al. 1990; Zervakis et al. 2001). However, it seems that some of the Pleurotus mushrooms growing on the roots and stems of this plant in Iran share (at least partly) common gene pools with strains from other Apiaceae hosts as well. In the particular case of Cluster III, gene exchange is apparently under way among fungi associated with more than one host-plant (as our results demonstrated) irrespective of its nature and geographic origin. This hypothesis is confirmed by the findings of Abdollahzadeh et al. (2007), who demonstrated intercompatibility in the matings performed among P. eryngii strains isolated from three different host-plants (Prangos sp., Pimpinella sp. and Ferula haussknechtii) in northern Iran.
On the other hand, a more ‘typical situation’ for the P. eryngii complex, depicting strict host-specificity for distinct Pleurotus populations or taxa (‘ecotypes’), is the case for strains of Clusters I and II (P. nebrodensis s.l. and P. fossulatus), which seem to be reproductively isolated from members of the other groups examined due to their association with Ferula spp. and C. ferulacea plants respectively. Furthermore, P. nebrodensis and P. fossulatus are phenotypically similar taxa (morphological differentiation seems to follow genetic isolation, as usually happens with Pleurotus populations growing on umbellifers; Zervakis et al. 2001), albeit representing different phylogenetic lineages. This morphological and habit similarity may be a consequence of recent speciation events. In fungi, biological species do not necessarily correspond with phylogenetic species, at least when divergence time is short (Taylor et al. 2000), while with higher divergence times distinct phylogenetic groups are intersterile regardless of geography (Moncalvo and Buchanan 2008). Relevant cases of populations with low percentages of base differences in the ITS regions, which apparently have not had enough time to diverge genetically and develop distinct phenotypic characteristics, were observed in other closely related mushroom species as well, e.g., Collybia tuberosa, C. cookie and C. cirrhata (Hughes et al. 2001), and Lactarius spp. of the sect. Deliciosi (Nuytinck and Verbeken 2007).
Hence, the existence in Iran of three taxa of the Pleurotus eryngii complex is established:
-
(i)
P. nebrodensis sensu lato, which grows in association with Ferula assa-foetida and F. ovina only. Previous reports linked its occurrence in Iran with host-plants such as Diplotaenia cachrydifolia (Heim 1960), Ferula communis and Prangos sp. (Saber 1997; reported as P. eryngii var. nebrodensis). The morphological characteristics provided from the former author for the D. cachrydifolia specimen (with whitish-cream colored pilei) fits with the current notion of the species and with the pertinent features characterizing our isolates of Cluster I. On the other hand, for Saber’s (1997) specimens, no morphological description is provided, apart from the information that the F. communis associated strain was identified by Dr David Pegler. If these data are combined with the outcome of previous studies on P. nebrodensis (Kawai et al. 2008; Zervakis et al. 2001; Zhang et al. 2006), it can be deduced that the Asiatic populations of P. nebrodensis s. l. are associated with Ferula host-plants only (e.g., F. assa-foetida, F. communis, F. ovina, F. sinkiangensis), with the possible exception of Heim’s (1960) D. cachrydifolia. In contrast, European P. nebrodensis is related with C. ferulacea only (Zervakis et al. 2001).
-
(ii)
P. fossulatus, which grows in association with C. ferulacea only. Very limited information is presently available for this fungus with whitish to cream pilei, which grows in western and central Asia, i.e., Iran, Afghanistan, Pakistan, and India (Pegler 1977; Saber 1997); only Saber (1997) has reported an associated host-plant (Prangos sp., Prangos ferulacea is a synonym for C. ferulacea). Furthermore, Vilgalys and Sun (1994) included in their phylogenetic analysis two P. fossulatus strains originating from India (ATCC 62885 and 52666), which were reportedly intercompatible with P. eryngii strains; these two taxa formed distinct – albeit neighbouring – clusters in the resulting phylograms. Strain ATCC 62885 was also used in our study; it is clearly separated from what we here name P. eryngii (Cluster III), whereas it presented high affinity with CCRC 36238 (deposited at the NCBI as P. fossulatus too) and grouped into the same larger group (Cluster II) with Iranian isolates from C. ferulacea. Hence, P. fossulatus seems to form a distinct taxon (at the species level according to the phylogenetic species concept) associated in Iran with C. ferulacea only; local material forms a sister clade with P. fossulatus strains of other origins.
-
(iii)
P. eryngii, which grows in association with S. aucheri, K. odoratissima, C. ferulacea, and F. ovina. This taxon is characterized by the typical morphological features described elsewhere (Hilber 1982; Zervakis and Balis 1996); however, lighter coloured pilei (light brown to beige to buff) are present in many of the specimens examined as a result of the phenotypic plasticity due to local environmental conditions and habitat nature. Despite the fact that European P. eryngii strains produce basidiomata from autumn to early winter at low altitudes, i.e., from sea level to ca. 1,500 m (depending on the host plants, which are usually species of the genera Eryngium, Ferula, Eleoselinum, Thapsia, etc.) (Hilber 1982; Venturella et al. 2000; Venturella et al. 2002; Zervakis and Balis 1996), P. eryngii specimens from Iran are generally collected at high altitudes (often exceeding 2,000 m) in spring months only.
In conclusion, strong patterns of host-dependent subdivision in the genetic structure of the P. eryngii complex (P. nebrodensis s.l. and P. fossulatus cases) were evidenced, but occasional deviation from this situation seems to occur as well; it could be mainly attributed to ongoing gene-flow between certain populations in neighbouring areas irrespective of the host they are associated with (e.g., P. eryngii case). This is in contrast to what was observed in European populations of the P. eryngii complex, where molecular studies revealed strict host-dependent delimitation among different ‘ecotypes’ of the complex (Urbanelli et al. 2007; Zervakis et al. 2001). In our opinion, it is still early to draw conclusions about the systematics of the Asiatic populations of P. nebrodensis (or, of Pleurotus fungi growing in association with Ferula spp. in Asia) and of P. fossulatus, before results of a study including representative host-associated populations from the entire distribution range of the complex are evaluated.
References
Abdollahzadeh J, Asef MR, Mirmahmoodi T (2007) The Pleurotus eryngii species-complex in Kurdistan region of Iran. Pak J Biol Sci 10:3006–3009
Ajani Y, Ajani A, Cordes JM, Watson MF, Downie SR (2008) Phylogenetic analysis of nrDNA ITS sequences reveals relationships within five groups of Iranian Apiaceae subfamily Apioideae. Taxon 57:383–401
Amiri H (2007) Chemical composition and antibacterial activity of essential oil of Prangos ferulacea (L.) Lindl. J Medicinal Plants 6:36–41
Babbel GR, Selander RK (1974) Genetic variability in edaphically restricted and widespread plant species. Evolution 28:619–630
Chiu H-H (2007) Phylogenetic analysis of Antrodia species and Antrodia camphorata inferred from internal transcribed spacer region. Antonie Van Leeuwenhoek 91:267–276
De Gioia T, Sisto D, Rana GL, Figliulo G (2005) Genetic structure of the Pleurotus eryngii species complex. Mycol Res 109:71–80
Doherty KR, Zweifel EW, Elde NC, McKone MJ, Zweifel SG (2003) Random amplified polymorphic DNA markers reveal genetic variation in the symbiotic fungus of leaf-cutting ants. Mycologia 95:19–23
Esfandiari E (1948) Troisième liste des fungi ramassé en Iran. Entomol Phytopathol Appl 8:1–15
Excoffier L, Smouse P, Quattro J (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Faridi P, Ghasemi Y, Gholami A, Mehregan I, Mohagheghzadeh A (2008) Antimicrobial essential oil from Smyrniopsis aucheri. Chem Nat Compd 44:116–118
Farris JS, Albert VA, Källersjö M, Lipscomb D, Kluge AG (1996) Parsimony jackknifing out-performs neighbor-joining. Cladistics 12:99–124
Felsenstein J (1978) Number of evolutionary trees. Syst Zool 27:27–33
Frøslev TG, Jeppesen TS, Læssøe T, Kjøller R (2007) Molecular phylogenetics and delimitation of species in Cortinarius section Calochroi (Basidiomycota, Agaricales) in Europe. Mol Phylogenet Evol 44:217–227
Ge ZW, Yang ZL, Zhang P, Matheny PB, Hibbett DS (2008) Flammulina species from China inferred by morphological and molecular data. Fungal Divers 32:59–68
Ghorbani A (2005) Studies on pharmaceutical ethnobotany in the region of Turkmen Sahra, north of Iran (Part 1): general results. J Ethnopharmacol 102:58–68
Gouveia MMC, Ribeiro A, Várzea VMP, Rodrigues CJ Jr (2005) Genetic diversity in Hemileia vastatrix based on RAPD markers. Mycologia 97:396–404
Gryta H, Carriconde F, Charcosset J-Y, Jargeat P, Gardes M (2006) Population dynamics of the ectomycorrhizal fungal species Tricholoma populinum and Tricholoma scalpturatum associated with black poplar under differing environmental conditions. Environ Microbiol 8:773–786
Hamrick JL, Godt MJ (1990) Allozyme diversity in plant species. In: Brown AHD, Clegg MT, Kahler AL, Weir BS (eds) Plant Population Genetics - Breeding and Genetic Resources. Sinauer, Sunderland, MA, pp 43–63
Healy M, Reece K, Walton D, Huong J, Shah K, Kontoyiannis DP (2004) Identification to the species level and differentiation between strains of Aspergillus clinical isolates by automated repetitive-sequence based. PCR. Clin Microbiol 42:4016–4024
Heim R (1960) Le Pleurote des Ombellifères en Iran. Rev Mycol 25:242–247
Hilber O (1982) Die Gattung Pleurotus (Fr.) Kummer unter besonderer Berücksichtigung des Pleurotus eryngii-Formenkomplexes. Bibliotheca Mycologica 87. Cramer J, Vaduz
Hughes KW, Petersen RH, Johnson JE, Moncalvo J-M, Vilgalys R, Redhead SA, Thomas TA, McGhee LL (2001) Infragenic phylogeny of Collybia s. str. based on sequences of ribosomal ITS and LSU regions. Mycol Res 105:164–172
James TY, Porter D, Hamrick JL, Vilgalys R (1999) Evidence for limited intercontinental gene flow in the cosmopolitan mushroom. Schizophyllum commune. Evolution 53:1665–1667
Joly P, Cailleux R, Cerceau MT (1990) La sterilité male pathologique, élément de la co-adaptation entre populations de champignons et de plantes-hôtes: modèle des Pleurotus des Ombellifères. Bull Soc Bot Fr 137:71–85
Jungehülsing U, Tudzynski P (1997) Analysis of genetic diversity in Claviceps purpurea by RAPD markers. Mycol Res 101:1–6
Kawai G, Babasaki K, Neda H (2008) Taxonomic position of a Chinese Pleurotus “Bai-Ling-Gu”: it belongs to Pleurotus eryngii (DC.: Fr.) Quél. and evolved independently in China. Mycoscience 49:5–87
Khajeh M, Yamini Y, Bahramifar N, Sefidkon F, Reza Pirmoradei M (2005) Comparison of essential oils compositions of Ferula assa-foetida obtained by supercritical carbon dioxide extraction and hydrodistillation methods. Food Chem 91:639–644
Kretzer A, Li Y, Szaro T, Bruns TD (1996) Internal transcribed spacer sequences from 38 recognized species of Suillus sensu lato: phylogenetic and taxonomic implications. Mycologia 88:776–785
Krüger D, Petersen RH, Hughes KW (2006) Molecular phylogenies and mating study data in Polyporus with special emphasis on group “Melanopus” (Basidiomycota). Mycol Progress 5:185–206
Lappalainen JH, Yli-Mattila T (1999) Genetic diversity in Finland of the birch endophyte Gnomonia setacea as determined by RAPD-PCR markers. Mycol Res 103:28–332
Lewinsohn D, Nevo E, Wasser SP, Hadar Y, Beharav A (2001) Genetic diversity in populations of the Pleurotus eryngii complex in Israel. Mycol Res 105:941–951
Moncalvo J-M, Buchanan PK (2008) Molecular evidence for long distance dispersal across the Southern Hemisphere in the Ganoderma applanatum-australe species complex (Basidiomycota). Mycol Res 112:425–436
Moreno G, Platas P, Peláez F, Bernedo M, Vargas A, Daza A, Santamaría C, Camacho M, Romero de la Osa L, Manjón JL (2008) Molecular phylogenetic analysis shows that Amanita ponderosa and A. curtipes are distinct species. Mycol Progress 7:41–47
Nuytinck J, Verbeken A (2007) Species delimitation and phylogenetic relationships in Lactarius section Deliciosi in Europe. Mycol Res 111:1285–1297
Pegler DN (1977) Pleurotus (Agaricales) in India, Nepal and Pakistan. Kew Bull 31:501–510
Petrak F (1939) Fungi. In: Rechinger KH (ed), Ergebnisse einer Botanische Reise nach dem Iran, 1937. Annalen Naturhistorischen Mus Wien 40:414–521
Pimenov MG, Leonov MV (2004) The Asian Umbelliferae Biodiversity Database (ASIUM) with particular references to southwest Asian taxa. Turk J Bot 28:139–145
Ro H-S, Kim SS, Rye JS, Jeon C-O, Lee TS, Lee H-S (2007) Comparative studies on the diversity of the edible mushroom Pleurotus eryngii: ITS sequence analysis, RAPD fingerprinting, and physiological characteristics. Mycol Res 111:710–715
Rogers SO, Bendich AJ (1988) Extraction of DNA from plant tissues. In: Gevin SB, Schilperoort RA, Verma DP (eds) Plant Molecular Biology Manual. Kluwer Academic Publishers, New York, pp 1–11
Rohlf FJ (1998) NTSYS-PC. Numerical taxonomy and multivariate analysis system, Version 2.00. Exeter Software, Setauket, NY
Saar I, Põldmaa K, Kõljalg U (2009) The phylogeny and taxonomy of genera Cystoderma and Cystodermella (Agaricales) based on nuclear ITS and LSU sequences. Mycol Progress 8:59–73
Saber M (1990) Contribution to the knowledge of Agaricales, pleurotoid in habit in Iran. Iran J Plant Pathol 26:29–40
Saber M (1997) New records of Agaricales (pleurotoid in habit) for Iran. Iran J Plant Pathol 33:51–56
Schneider S, Roessli D, Excoffier L (2001) Arlequin: a software for population genetics data analysis, version 2.00. Genetics and Biometry Lab, Department of Anthropology, University of Geneva, Geneva
Singh BK, Munro S, Reid E, Ord B, Potts JM, Paterson E, Millard P (2006) Investigating microbial community structure in soils by physiological, biochemical and molecular fingerprinting methods. Eur J Soil Sci 57:72–82
Stott K, Desmerger C, Holford P (2005) Relationship among Lepista species determined by CAPS and RAPD. Mycol Res 109:205–211
Swofford DL (2002) PAUP*: phylogenetic analysis using parsimony (*and other methods), Version 4.0b10. Sinauer Associates, Sunderland, MA
Taylor JW, Jacobson DJ, Kroken S, Kasuga T, Geiser DM, Hibbett DS, Fisher MC (2000) Phylogenetic species recognition and species concepts in fungi. Fungal Genet Biol 31:21–32
Urbanelli S, Della Rosa V, Punelli F, Porretta D, Reverberi M, Fabbri AA, Fanelli C (2007) DNA-fingerprinting (AFLP and RFLP) for genotypic identification in species of the Pleurotus eryngii complex. Appl Microbiol Biotechnol 74:592–600
Venturella G, Zervakis G, La Rocca S (2000) Pleurotus eryngii var. elaeoselini var. nov. from Sicily. Mycotaxon 76:419–427
Venturella G, Zervakis G, Saitta A (2002) Pleurotus eryngii var. thapsiae var. nov. from Sicily. Mycotaxon 81:69–74
Vilgalys R, Sun BL (1994) Ancient and recent patterns of geographic speciation in the oyster mushroom Pleurotus revealed by phylogenetic analysis of ribosomal DNA sequences. Proc Natl Acad Sci USA 91:4599–4603
White TJ, Bruns T, Lee S, Taylor JW (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sininsky JJ, White TJ (eds) PCR Protocols: a guide to methods and applications. Academic Press, San Diego, pp 315–322
Williams JG, Kubelik AR, Livak KJ, Rafalski JA, Tinger SV (1990) DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res 18:6531–6535
Zervakis G, Balis C (1996) A pluralistic approach on the study of Pleurotus species, with emphasis on compatibility and physiology of the European morphotaxa. Mycol Res 100:717–731
Zervakis G, Sourdis J, Balis C (1994) Genetic variability and systematics of eleven Pleurotus species based on isozyme analysis. Mycol Res 98:329–341
Zervakis GI, Venturella G, Papadopoulou K (2001) Genetic polymorphism and taxonomic infrastructure of the Pleurotus eryngii species-complex as determind by RAPD analysis, isozyme profiles and ecomorphological characters. Microbiology UK 147:3183–3194
Zervakis GI, Moncalvo J-M, Vilgalys R (2004) Molecular phylogeny, biogeography and speciation of the mushroom species Pleurotus cystidiosus and allied taxa. Microbiology UK 150:715–726
Zhang JX, Huang CY, Ng TB, Wang HX (2006) Genetic polymorphism of ferula mushroom grown on Ferula sinkiangensis. Appl Microbiol Biotechnol 71:304–309
Zhang LF, Yang JB, Yang ZL (2004) Molecular phylogeny of eastern Asian species of Amanita (Agaricales, Basidiomycota): taxonomic and biogeographic implications. Fungal Divers 17:219–238
Acknowledgements
The authors are grateful to Shahrekord University for financial assistance. Special thanks are extended to Dr. Nabiolah Yarali for his help with the GIS software and the mapping of the samples studied.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ravash, R., Shiran, B., Alavi, AA. et al. Genetic variability and molecular phylogeny of Pleurotus eryngii species-complex isolates from Iran, and notes on the systematics of Asiatic populations. Mycol Progress 9, 181–194 (2010). https://doi.org/10.1007/s11557-009-0624-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11557-009-0624-2