Abstract
OsRac5 belongs to the rice Rho of plants family, and acts as the molecular switch in the signal pathway which is pivotally involved in the rice fertility control. One of its putative partners, OsMY1, was isolated by yeast two-hybrid screening from rice panicle cDNA library. Bioinformatics analysis shows that OsMY1 contains a coiled-coil domain which generally appeared in the partners of Rho GTPases. By yeast two-hybrid assay, it is confirmed that OsMY1 binds both the wild type (WT) and constitutively active (CA) OsRac5, but does not interact with dominantly negative OsRac5. In addition, the interactions between OsMY1 and WT-OsRac5 or CA-OsRac5 in vivo are demonstrated by bimolecular fluorescence complementation assay. Using PCR-mediated sequence deletion and point mutation of OsMY1, the interaction between OsMY1 and OsRac5 was identified to be mediated by the coiled-coil domain in OsMY1, and their binding was quantified by O-nitro-phenyl-β-d-galactopyranoside assay. Real-time PCR shows that OsMY1 and OsRac5 are coordinately expressed in rice leaves and panicles with similar expression patterns. Our results suggest that OsMY1 is an important target of OsRac5 and that these two genes are involved in the same biological processes in rice growth and development.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rho of plants (ROP) proteins, also known as RAC proteins, comprise one of the major regulatory proteins of polar growth of plant cells that has been verified as a molecular switch in the signalling pathway involved in various cellular processes, including cytoskeleton organization, vesicle movement, and Ca2+ gradient establishment during the polar growth of pollen tubes and root hairs [1, 2]. It is generally considered that actin cytoskeleton serves as an efficient molecular transportation track during tip growth. The actin cytoskeleton is regulated by actin-binding proteins, and the activities of these actin-binding proteins are regulated by some upstream signals, such as pH, [Ca2+] and ROP, to produce the proper actin configuration during the polar growth process [3]. In Arabidopsis, the AtROP1 gene activates two direct downstream targets, RIC3 and RIC4, involved in at least two downstream pathways that regulate the generation of the tip-focused [Ca2+]cyt gradients and the assembly of dynamic tip-localized actin microfilaments, respectively [4–6]. Therefore, pollen-tube oscillatory growth is assumed to function through the activation and/or deactivation of several actin-binding proteins [7, 8].
Myosin is the molecular motor of actin. Flowering plants generally possess two myosin classes: VIII and XI. Myosins of class XI have been implicated in plant growth, intracellular transport, and root-hair elongation [9]. Silencing the expression of myoXIa and myoXIb in 1-week-old moss resulted in growth inhibition, disrupted protonemal tip growth, and F-actin disorganization. When Arabidopsis myosin XI is knocked out, the cellular architecture and the orientation of prominent actin filament cables are altered from predominantly longitudinal to random positioning or are transversely oriented in the epidermal cells of the midveins of leaves [10]. A study of a photoperiod-sensitive rice using a dissociation (Ds) insertion mutant with a male sterile feature revealed that the defects in pollen development were due to the abnormal protein localization of a myosin XI [11]. Accordingly, dynamics of the actin cytoskeleton during plant polar growth is an extremely complex process, the underlying molecular mechanism of ROP, myosin and actin involved in the processes is still unclear.
The rice ROP gene OsRac5 (also named OsRacD) was isolated from young rice panicles by cDNA library screening [12]. This gene may participate in rice fertility control by regulating the polar growth of the pollen tube [13, 14]. To identify the function of OsRac5 as well as the related signalling pathways, yeast two-hybrid (Y2H) screening was performed to isolate the partners of OsRac5 in young panicles at the pistil and stamen formation stages. Some potential interaction protein-encoding genes, including of some known partners of ROP, such as OsRhoGDIs, OsRhoGAPs were obtained [15]. In the present study, we focused on OsMY1, one of these putative partners of OsRac5 from Y2H. The interaction between OsRac5 and OsMY1 was verified using Y2H assay and bimolecular fluorescence complementation (BiFC) assay. The mediated binding regions of OsMY1 were identified by sequence deletion, point mutation and binding assay, which based on the conserved domain analysis. The expression profiles of OsRac5 and OsMY1 during rice development were detected by qRT-PCR. The results show that OsMY1 may be an important target of OsRac5 in the signalling pathway, which associated with the actin cytoskeleton dynamics, and the two genes may participate in the same processes involved in rice growth and development.
Materials and methods
Plant material and growth conditions
Rice (Oryza sativa cv. Nipponbare) was planted in an experimental field of Henan Normal University (35°18′N, 113°52′E) on 5 May 2012. Rice leaves were collected at 15, 30, 60, and 90 days, and the young panicles at the pistil and stamen formation stage were also sampled.
Database search and sequence alignments
The gene sequence was submitted to the National Center for Biotechnology Information (NCBI; www. ncbi.nlm.nih.gov/BLAST) for identification and analysis. The conserved coiled-coil domain was identified using the online tools (http://www.ch.embnet.org/software/COILS_form.html). Sequence alignments were performed using DNAMAN software (Lynnon Biosoft).
RNA isolation and quantitative RT-PCR
Total RNA was isolated from rice leaves and panicles using TRIzol reagent (TaKaRa, Tokyo, Japan) according to the instructions. For real-time PCR analysis, first-strand cDNA was synthesized from DNaseI-treated total RNA using Prime Script reverse transcriptase (TaKaRa) according to the manufacturer’s instructions. Real-time PCR was performed on an optical 96-well plate with an ABI PRISM 7500 real-time PCR system (Applied Biosystems, Foster City, CA) with SYBR Green I (TaKaRa) and gene-specific primers, according to the manufacturer’s protocol. The rice house-keeping gene OsAct1 (accession no. X16280) was used as an internal control with primers 5′-TCTTCCAGCCTTCCTTCA-3′ and 5′-ATCCACGTCGCACTTCAT-3′. The primers for OsMY1 were 5′-TGGCTCCAGCACCCATAAACAACTA-3′ and 5′-ATGACAAAAATCCCAAAGGGCGATC-3′; the primers for OsRac5 were 5′-TTCGATGCTGCGATTAAGCTGGTTCTCC-3′ and 5′-TACAAACAAGCGACGATTGAAGCCAGAG-3′. Data were analyzed with 7500 System SDS software, version 1.4 (Applied Biosystems). The relative expression levels were determined using the 2−△△Ct method as described previously [16].
Yeast two-hybrid assays
Y2H experiments were performed using the HybriZAP2.1 system (Stratagene, Agilent Technologies, Santa Clara, CA). The yeast strain was Saccharomy cescerevisiae YRG-2, and the positive control plasmids were pBD-MUT and pAD-MUT. The series baits (pBD-OsRac5 [15], pBD-G15V-OsRac5 [17], pBD-T20N-OsRac5 [18], and pBD-D121N-OsRac5 [19]) were previously constructed by our laboratory. The prey plasmid pAD-OsMY1 was isolated as an putative partner of OsRac5 from rice panicle cDNA library by Y2H screening. YRG-2 yeast competent cells were co-transfected with pAD-OsMY1 and different pBD-bait combinations, then they were plated on SD/-Trp-Leu selection media. After 3–5 days, positive colonies were regrown on SD/-Trp-Leu-His selection media for 2–3 days. Significant growth on the selection media indicated a positive interaction, then the β-galactosidase enzyme activity was further analyzed by filter lift assay according to the instructions.
Bimolecular fluorescence complementation assay
The cDNA coding region of OsMY1 was fused to the C-terminus of YFP in pUC-SPYCE vector, and the cDNA coding region of OsRac5 or G15V-OsRac5 was fused to the N-terminus of YFP in pUC-SPYNE. All constructs were sequenced to confirm that no errors were incorporated and all fusions were in-frame with either C-terminus of YFP or N-terminus of YFP. The recombinant plasmid combinations OsMY1-YC and OsRac5-YN, OsMY1-YC and G15V-OsRac5-YN were transformed into Agrobacterium tumefaciens EHA105, respectively. The positive clones were cultured to logarithmic phase, centrifuged to collect bacteria, resuspended with sterile water which contain a final concentration of MgCl2(10 mM), MES(pH 5.6, 10 mM), acetosyringone (200 μM). Co-cultured with onion outer epidermis, which had been pre-cultured in 1/2MS medium for 30 min, then transferred the cultured the onion outer epidermis to a new 1/2MS medium for 1–2 days. The results were observed with converted fluorescence microscope (Nikon Ti-U) with excitation wavelength of 527 nm, and recorded by Nis-Elements AR software .
Sequence deletion and directed mutagenesis
The sequence encoding the coiled-coil domain of OsMY1 was amplified using the following primers: 5′-TTGAATTCGAAGAGGAGAGCATTGACCA A-3′ and 5′-AAGTCGACTCAGGCATCCCCGCCAATCACC-3′. Then the PCR products were inserted into the EcoRI and SalI sites in the pAD plasmid, fused in frame with the activation domain (AD), resulting in the recombinant vector pAD-OsMY1-Coil, which was verified by sequencing. The double-point mutation Q139P and A144P in the coiled-coil region of OsMY1 was introduced by PCR-based site-directed mutagenesis. The primers for 5′ terminal amplification were 5′-AGGGATGTTTAATACCACTAC-3′ and 5′-GCCGCCTCCGGGGCCTTTCCCATGGCTCGGTCT-3′. The primers for 3′ terminal amplification were 5′-GCACAGTTGAAGTGAACTTGC-3′ and 5′-AGACCGAGCCATGGCGAAAGGCCCCGGAGGCGGC-3′. The double-point mutant sequence was used to construct the recombinant vector pAD-OsMY1M, which was confirmed by sequencing.
Quantitative assay of the protein–protein interaction
YRG-2 competent cell was co-transformed using OsRac5 or G15V-OsRac5 as bait and OsMY1 or OsMY1-Coil as prey. The positive clones were screened using SD/-Trp-Leu-His, and at least three independent clones were cultured to OD600 0.5–0.8 in liquid medium, then collected by centrifugation. The yeast pellets were then suspended in Z-buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4) and repeatedly frozen in liquid nitrogen and thawed to 37 °C five times. After the Z-buffer (containing 0.27 % mercaptoethanol) and ONPG (O-nitro-phenyl-β-d-galactopyranoside) were added, the solution was incubated at 30 °C, and the reaction was terminated by adding 400 μL of 1 M Na2CO3 until the yellow color developed. The β-galactosidase quantities were calculated using the following equation: β-galactosidase units = 1,000 × OD420/(t × V × OD600), where t represents the reaction time and V represents the reaction volume.
Statistical analysis
In this study, all statistical analyses involved the use of Microsoft Excel software. Significant differences were determined using Student’s t test. Error bars represent standard error (SE).
Results
Sequence features of OsMY1
The NCBI database search revealed that the cDNA sequence of one positive clone from the Y2H assay contained a 1025-bp insert that shared 99 % nucleotide sequence identity and 99 % protein sequence identity with a myosin heavy chain gene (AY224532), accordingly we named it OsMY1 (accession no. DQ641916). The coiled-coil domain was identified in its protein sequence, but the conserved motor domain in the N-terminal of myosin could not be determined. It is very likely that OsMY1 is a truncated myosin that encodes the C-terminal sequence. Furthermore, the N-terminal amino acid sequence of OsMY1 could not be determined from our BLAST search against NCBI databases. We compared OsMY1 with 11 kinds of homologous proteins from our BLAST search with DNAMAN software, the results revealed a varying degrees of homology between OsMY1 and other proteins. OsMY1 had high degrees of similarity with the ROP1 interacting protein (77 %; XP_003569847) from Brachypodium distachyon and with ROP interactive partner 2 (75 %; NP_177964.2) from Arabidopsis thaliana (Fig. 1a). Although these results reveal that the OsMY1 amino acid sequence shares lower overall identity with these sequences, the well-conserved coiled-coil domain were apparent in all of these sequences, with conserved motifs of QWRKAA in the domain (Fig. 1b).
Phylogenetic analysis and sequence alignment of plant putative myosin proteins. a The deduced protein sequences of plant myosin were aligned with DNAMAN software, and the TreeView program was used to create a phylogenetic tree. The GenBank accession numbers of plant myosin genes used to build the phylogenetic tree are as follows: Brachypodium distachyon (XP_003569847), Zea mays (NP_001144325), Solanum demissum (AAT39303), Panax quinquefolius (ABX75363), Nicotiana tabacum (CAC84774), Oryza sativa japonica (CAD89536), Oryza sativa indica (EAY98684), Arabidopsis thaliana (NP_177964), Ricinus communis (XP_002525180), OsMY1 (DQ641916), Populus trichocarpa (XP_002300426), Medicago truncatula (XP_003623097). b Sequence alignment of the conserved amino acids in coiled-coil domain from plant putative myosin proteins, the black stars showed the Q139 and A144 in the coiled-coil domain of OsMY1
Analysis of the interaction between OsMY1 and OsRac5 in Y2H system
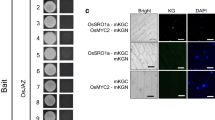
In an Y2H system, the interactions between OsMY1 and dominant-negative OsRac5 (DN-OsRac5, T20N-OsRac5 and D121N-OsRac5 in this study), constitutively active OsRac5 (CA-OsRac5, G15V-OsRac5 in this study), and wild-type OsRac5 (WT-OsRac5) were detected. The results show that OsMY1 can bind with both WT-OsRac5 and CA-OsRac5, but OsMY1 cannot interact with DN-OsRac5 (Fig. 2a, b). Therefore, the interaction between OsMY1 and OsRac5 may be constitutively independent depending on whether the latter is activated or not. When OsRac5 was locked in an inactivated state, it could not bind with OsMY1. Because myosin is the motor molecule of the actin filament, OsMY1 may be the necessary scaffold protein for the functional OsRac5 protein.
Analysis of the interaction between OsMY1 and OsRac5. a–c Yeast two-hybrid assay. d–f BiFC assay. a Yeasts co-transformed with prey and bait plasmids grown on SD/-Trp-Leu-His. 1 OsMY1 and OsRac5. 2 OsMY1 and G15V-OsRac5. 3 OsMY1 and T20N-OsRac5. 4 OsMY1 and D121N-OsRac5. 5 Positive control. b Filter lift assay of the plate replica. c Sketch of the Y2H assay. d Negative control. e OsMY1-YC and WT-OsRac5-YN. f OsMY1-YC and CA-OsRac5-YN
Verification of the interaction between OsMY1 and OsRac5 in planta
The interaction of OsMY1 and OsRac5 was further examined by BiFC assay. The OsMY1 was fused to the C-terminal half of the yellow fluorescence protein (YC), and the WT-OsRac5 and CA-OsRac5 were fused to the N-terminal half of the yellow fluorescence protein (YN), respectively. The YN-fused and YC-fused recombinant DNA constructs were introduced into onion (Allium cepa) epidermal layer by Agrobacterium-mediated method. As shown in Fig. 2, strong YFP fluorescence signals were observed with OsMY1-YC and OsRac5-YN combinations (Fig. 2e), OsMY1-YC and G15V-OsRac5-YN combinations (Fig. 2f) but were not observed when coexpressed with empty vector OsMY1-YC and YN or OsRac5-YN and YC (Fig. 2d). Taken together, these results show that OsMY1 is able to physically interact with WT-OsRac5 and CA-OsRac5 in plant cells. The fluorescence signals mainly distributed in cytoplasm suggest that OsMY1 has the potential to interact directly with WT-OsRac5 or CA-OsRac5 in the cytoplasm.
Identification of the binding domain mediated OsMY1 and OsRac5 interaction
The region mediating the interaction between OsMY1 and OsRac5 were investigated by sequence deletion and point mutation. The OsMY1 truncation that deleted the flank sequence of the coiled-coil domain (OsMY1-Coil) was generated by site-directed mutagenesis. The binding was then detected in an Y2H system. The results revealed that the coiled-coil domain of OsMY1 was responsible for the interaction with CA-OsRac5 or WT-OsRac5. Quantitative ONPG assay further indicated that the deletion mutation had almost no effect on binding, indicating that the interactions were mediated by the coiled-coil domain within OsMY1, independent of the adjacent sequence of the domain. In addition, there was almost no difference in binding capacities between OsMY1 and CA-OsRac5 versus OsMY1 and WT-OsRac5 (Fig. 3a).
Verification of the region mediated the binding between OsMY1 and OsRac5. a Quantitative analysis of the binding between OsMY1 and OsRac5. 1 Negative control. 2 Positive control. 3 WT-OsRac5 and OsMY1. 4 CA-OsRac5 and OsMYl. 5 WT-OsRac5 and OsMY1-Coil. 6 CA-OsRac5 and OsMY1-Coil. b Binding between OsMY1M and OsRac5 (2), G15V- OsRac5 (3), positive control (1). c Filter lift assay of the plate replica
To further verify that the coiled-coil domain is the region in which the interaction between OsMY1 and OsRac5 is mediated, the Q139P-A144P double point mutation (OsMY1M) was introduced into the conservative QWRKAA motif to abolish the function of the domain, because prolines are known helix breakers and can completely destroy the coiled-coil structure. The results indicated that OsMY1M cannot bind with WT-OsRac5 and CA-OsRac5 (Fig. 3b, c). Taken together, these results showed that the coiled-coil domain of OsMY1 is the key sequence that mediates binding with CA-OsRac5 or WT-OsRac5.
Expression profiles of OsMY1 and OsRac5
The gene transcripts of OsMY1 and OsRac5 in rice different development periods were determined by real-time PCR (Fig. 4). The expression of the two genes in rice leaves gradually increased along with the growth and development processes. However, the expression levels of both OsMY1 and OsRac5 in the leaves had declined at 90 days, the time at which rice has completed the transition from vegetative growth to reproductive growth. It is remarkably that the accumulations of the transcripts of these two genes were especially abundant in young rice panicles at the stamen and pistil formation stage, and it was more than twice compared with that in leaves at 60 days. These changes in expression of the two genes appear to have occurred co-ordinately, suggesting that there were important functional association between OsMY1 and OsRac5 during rice growth and development, especially may involve in rice floral organ development.
Discussion
In the present work, the rice OsMY1 gene was initially isolated as one of the putative partners of the small GTPase OsRac5 gene using an Y2H screening method. In a similar study, the interaction between Arabidopsis small GTPase AtRab and AtMYA2 was verified to be mediated by the tail sequence of AtMYA2, which had been identified belong to myosin XI in Arabidopsis [20]. According to the existing research results, the interacting proteins of small GTPases (e.g., ROP/RAC/RAB), including GEFs (regulators of the guanylate binding form) [21], GTPase activating protein (GAPs) [22], and Guanine nucleotide dissociation inhibitor (GDIs) [23], are involved in balancing small GTPases between the GTP and GDP binding states. Notably, coiled-coil domain containing proteins are also widely screened out as potential partners of small GTPases. Therefore, although different small GTPases are involved in various different physiological processes, signalling from small GTPases to downstream molecules in vivo may be similar, which implies that the signal relay mechanism of small GTPases may be conservative in evolution.
Coiled-coil domains are widely existed in the sequences of myosins. Some studies have shown that coiled-coil domains had the function of providing support for the functional complexs by forming steady structure [24], and the mutation of the domain disturbs the oligomerization of the myosin monomers, leading to disorder in the assembly of the actin-myosins in cells [25]. In this study, the function of the coiled-coil domain involved in protein–protein interactions between OsMY1 and OsRac5 was tested in an Y2H system by sequence deletion and point mutation, and was quantified by ONPG assays. The results indicate that the binding region of OsMY1 with OsRac5 is located only in the coiled-coil domain.
Recently, a novel Arabidopsis ROP/RAC effector, ICR1 (Interactor of Constitutive active ROP1), which composed of coiled-coil domain and no other recognizable catalytic or structural domains was reported [26, 27]. ICR1 is required for directional auxin transport and distribution, and thereby act as a regulator of polar pollen tube growth by recruiting active ROP to the plasma membrane [28, 29]. By BLASTp with OsMY1 sequences, we identified five potential rice XI type myosin related sequences XI B (AAW83512); XIE1 (AAQ87014); XIK (AAM44879); XIL (AAK98715) and two type VIII myosin VIII A1 (AAQ87012), VIII A2 (AAQ87013) from NCBI database, but the results can’t tell which subtype OsMY1 belongs to, based on the sequence alignments of their tail sequence. Owing to the huge difference in the plant myosin tail sequence, the classification of the existing myosin in plants mainly depend on their head conserved domain, hence cloning 5′ end sequence of OsMY1 is necessary for its classification and functional characterization.
ROP GTPases are a ubiquitous eukaryotic family of molecular switches within the RAS superfamily of monomeric GTPases. Specifically, plants possess a large family of genes encoding ROP, which have an important role in pollen tube growth and development. ROPs act as binary molecular switches that are turned on and off in response to a variety of extracellular stimuli. Only GTP-bound activated ROP can interact with a number of effectors to relay signals downstream, leading to diverse biological responses [24, 30]. When its bound GTP is hydrolysed to GDP, ROP returns to an inactive state. Under the normal cellular environment, although the concentration of GTP is higher than that of GDP, ROP normally exists in a GDP-bound state. Only when upstream signalling occurs does ROP convert to the activated GTP-binding form. In Arabidopsis, point mutations were introduced into the conserved sites of ROPs to mimic their GTP-bound CA or GDP-bound dominant negative (DN) state, which further revealed the functions of ROPs in the cell polarity establishment and growth [31]. In our previous studies, the WT-OsRac5 and three OsRac5 mutants [CA-OsRac5 (a G15V mutant) and DN-OsRac5 (T20N and D121N mutants)] were constructed, respectively, and verified by GTP hydrolysis activity [17–19]. In this study, the interaction between OsMY1 and WT-OsRac5, CA-OsRac5, or DN-OsRac5 were tested in an Y2H system. The results indicate that OsMY1 only binds with WT-OsRac5 or CA-OsRac5 and cannot interact with DN-OsRac5. Based on this evidence, we conclude that both WT-OsRac5 and CA-OsRac5 are targeted in the cytoplasm through the interaction with OsMY1. When OsRac5 was locked in the inactive GDP-bound state, there was no interaction between OsMY1 and DN-OsRac5. Therefore, binding with OsMY1 might be required for effective signal flow through OsRac5 to elicit downstream biological functions. Furthermore, it is found that OsMY1 was indeed co-localized with WT-OsRac5 or CA-OsRac5 in the cytoplasm by BiFC analysis. Based on the evidence and results, it is suggested that both WT-OsRac5 and CA-OsRac5 are targeted to cytoplasm through the interaction with OsMY1. When OsRac5 was locked in the molecular switch off state, which under the GDP-binding state, there was no interaction between OsMY1 and DN-OsRac5, suggesting that binding with OsMY1 was most likely the premise of the biological function of OsRac5.
Moreover, it’s worth noting that the expression patterns analysis revealed OsRac5 and OsMY1 were expressed co-ordinately, therefore it can be deduced that there is an important association between them during rice growth and development. Further investigations will be needed to ascertain the intrinsic biological significance of their expression correlation, and their functional relevance in the rice fertility control.
Conclusions
This study provides a new view of the molecular mechanisms involved in the ROP signalling in rice. This research on rice ROP protein OsRac5 and its putative partners OsMY1 showed that OsMY1 bound functional OsRac5 both in vitro and in vivo, and the protein–protein interaction was mediated by the high conservative colied-coil domain in the C-terminal of OsMY1. Moreover, the present results suggest that the expression of OsRac5 and OsMY1 is highly coordinated, implying that OsMY1 may function as a mediator protein of OsRac5 in signal pathway, which is involved in various growth and development processes in rice.
Abbreviations
- ROP:
-
Rho of plants
- Y2H:
-
Yeast two-hybrid
- CA:
-
Constitutively active
- DN:
-
Dominantly negative
- BiFC:
-
Bimolecular Fluorescence Complementation
- GEF:
-
Guanine nucleotide exchange factor
- GDI:
-
Guanine nucleotide dissociation inhibitor
- GAP:
-
GTPase-activating protein
- ONPG:
-
O-Nitro-phenyl-β-d-galactopyranoside
References
Nibau C, Wu HM, Cheung AY (2006) RAC/ROP GTPases: ‘hubs’ for signal integration and diversification in plants. Trends Plant Sci 11(6):309–315
Samaj J, Muller J, Beck M, Bohm N, Menzel D (2006) Vesicular trafficking, cytoskeleton and signalling in root hairs and pollen tubes. Trends Plant Sci 11(12):594–600
Papuga J, Hoffmann C, Dieterle M, Moes D, Moreau F, Tholl S, Steinmetz A, Thomas C (2010) Arabidopsis LIM proteins: a family of actin bundlers with distinct expression patterns and modes of regulation. Plant Cell 22(9):3034–3052
Hwang JU, Gu Y, Lee YJ, Yang Z (2005) Oscillatory ROP GTPase activation leads the oscillatory polarized growth of pollen tubes. Mol Biol Cell 16(11):5385–5399
Gu Y, Fu Y, Dowd P, Li SD, Vernoud V, Gilroy S, Yang ZB (2005) A Rho family GTPase controls actin dynamics and tip growth via two counteracting downstream pathways in pollen tubes. J Cell Biol 169(1):127–138
Fu Y, Wu G, Yang Z (2001) Rop GTPase-dependent dynamics of tip-localized F-actin controls tip growth in pollen tubes. J Cell Biol 152(5):1019–1032
Staiger CJ, Poulter NS, Henty JL, Franklin-Tong VE, Blanchoin L (2010) Regulation of actin dynamics by actin-binding proteins in pollen. J Exp Bot 61(7):1969–1986
Cheung AY, Wu HM (2008) Structural and signaling networks for the polar cell growth machinery in pollen tubes. Annu Rev Plant Biol 59:547–572
Prokhnevsky AI, Peremyslov VV, Dolja VV (2008) Overlapping functions of the four class XI myosins in Arabidopsis growth, root hair elongation, and organelle motility. Proc Natl Acad Sci USA 105(50):19744–19749
Ueda H, Yokota E, Kutsuna N, Shimada T, Tamura K, Shimmen T, Hasezawa S, Dolja VV, Hara-Nishimura I (2010) Myosin-dependent endoplasmic reticulummotility and F-actin organization in plant cells. Proc Natl Acad Sci USA 107(15):6894–6899
Jiang SY, Cai M, Ramachandran S (2007) ORYZA SATIVA MYOSIN XI B controls pollen development by photoperiod-sensitive protein localizations. Dev Biol 304(2):579–592
Mi ZY, Wang SS, Wu NH (2002) Isolation of OsRacD gene encoding a small GTP-binding protein from rice. Chin Sci Bull 47(20):1673–1679
Ye JR, Huang MJ, Wu NH (2003) Fertility analysis of the Arabidopsis transformed with antisense rice OsRacD gene. Prog Natl Sci 13(6):424–428
Ye JR, Huang MJ, Zhao SH, Wu NH (2004) The correlation analysis of the expression of OsRacD and rice photoperiod fertility transition. Prog Natl Sci 14(2):166–172 (in chinese)
Liang WH, Tang CR, Wu NH (2004) Cloning and characterization of a new actin gene from Oryza sativa L. Prog Natl Sci 14(10):867–874
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2 (−Delta Delta C (T)) method. Methods 25(4):402–408
Liang WH, Wu NH (2006) Effects of G15V point mutation on functions of rice OsRac5 protein. Chin J Biochem Mol Biol 22(4):338–342 (in chinese)
Liu XF, Liang WH (2010) Effects of T20N site directed mutation on GTPase activities of OsRacD from Oryza sativa. Chin Biotechnol 30(1):56–61 (in chinese)
Liang WH, Zhang LJ, Li L, Zhang F, Shi HH, Shang F, Li H, Li MM (2010) Effects of D121N point mutation on OsRac5 protein. In: 3rd international conference on future biomedical information engineering, vol III, pp 253–256
Hashimoto K, Igarashi H, Mano SJ, Takenaka C, Shiina T, Yamaguchi M, Demura T, Nishimura M, Shimmen T, Yokota E (2008) An isoform of Arabidopsis myosin XI interacts with small GTPases in its C-terminal tail region. J Exp Bot 59(13):3523–3531
Gu Y, Li S, Lord EM, Yang Z (2006) Members of a novel class of Arabidopsis Rho guanine nucleotide exchange factors control RhoGTPase-dependent polar growth. Plant Cell 18(2):366–381
Klahre U, Kost B (2006) Tobacco RhoGTPase ACTIVATING PROTEIN1 spatially restricts signaling of RAC/Rop to the apex of pollen tubes. Plant Cell 18(11):3033–3046
Kieffer F, Elmayan T, Rubier S, Simon-Plas F, Dagher MC, Blein JP (2000) Cloning of Rac and Rho-GDI from tobacco using an heterologous two-hybrid screen. Biochimie 82(12):1099–1105
Lavy M, Bloch D, Hazak O, Gutman I, Poraty L, Sorek N, Sternberg H, Yalovsky S (2007) A novel ROP/RAC effector links cell polarity, root-meristem maintenance, and vesicle trafficking. Curr Biol 17(11):947–952
Bloch D, Hazak O, Lavy M, Yalovsky S (2008) A novel ROP/RAC GTPase effector integrates plant cell form and pattern formation. Plant Signal Behav 3(1):41–43
Li S, Gu Y, Yan A, Lord E, Yang ZB (2008) RIP1 (ROP Interactive Partner 1)/ICR1 marks pollen germination sites and may act in the ROP1 pathway in the control of polarized pollen growth. Mol Plant 1(6):1021–1035
Hazak O, Bloch D, Poraty L, Sternberg H, Zhang J, Friml J, Yalovsky S (2010) A rho scaffold integrates the secretory system with feedback mechanisms in regulation of auxin distribution. PLoS Biol 8(1):e1000282. doi:10.1371/journal.pbio.1000282
Lupas A (1996) Coiled coils: new structures and new functions. Trends Biochem Sci 21(10):375–382
Lapierre LA, Kumar R, Hales CM, Navarre J, Bhartur SG, Burnette JO, Provance DW Jr, Mercer JA, Bähler M, Goldenring JR (2001) Myosin Vb is associated with plasma membrane recycling systems. Mol Biol Cell 12(6):1843–1857
Li H, Lin Y, Heath RM, Zhu MX, Yang ZB (1999) Control of pollen tube tip growth by a Rop GTPase-dependent pathway that leads to tip-localized calcium influx. Plant cell 11(9):1731–1742
Li H, Shen JJ, Zheng ZL, Lin Y, Yang ZB (2001) The Rop GTPase switch controls multiple developmental processes in Arabidopsis. Plant Physiol 126(2):670–684
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Nos. 31171182; U1204305), Innovation Scientists and Technicians Troop Construction Projects of Henan Province (No. 104100510012), Program for Innovative Research Team (in Science and Technology) in University of Henan Province (13IRTSTHN009) and Henan Natural Science Research project (No. 2010A180012, No. 132300410137). We thank MedSci for critical English editing of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Liang, WH., Wang, HH., Li, H. et al. Isolation and characterization of OsMY1, a putative partner of OsRac5 from Oryza sativa L.. Mol Biol Rep 41, 1829–1836 (2014). https://doi.org/10.1007/s11033-014-3032-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-014-3032-x