Abstract
The cardiomyopathy associated 5 (CMYA5) gene was also called TRIM76, which was belonged to the tripartite motif super family of proteins (TRIM). It was a direct transcriptional target for MEF2A and it played an important role in myofibrillogenesis. In the present study, a 12056 bp cDNA sequence of the porcine CMYA5 gene was obtained by RT-PCR. The sequence encoded a large protein consisting of 4003 amino acids and the carboxyl terminus of the predicted CMYA5 protein comprised of a B-box coiled-coil, two fibronectin type III (FN3) repeats, and SPRY domains. The porcine CMYA5 gene was assigned to chromosome 2q21–24 by using the radiation hybrid (IMpRH) panel, and it was significantly linked to microsatellite Sw1602 with LOD scores of 6.74. Semi-quantitative RT–PCR revealed that the porcine CMYA5 gene was broadly expressed in all seven tissues(heart, liver, spleen, lung, kidney, skeletal muscle and adipose)harvested from different developmental stages(new born, five weeks and adult tongcheng pigs), with a high level in heart and skeletal muscle. One SNP (A7189C), leading to the amino acid alteration from the Ile residue to the Leu residue, was found and detected by BspTI PCR-restriction fragment length polymorphism. The association analysis revealed that the substitution of A7189C had significant associations with the percentage of ham (p < 0.05), water loss (p < 0.01) and intramuscular fat (p < 0.05). These results provide the evidence that the porcine CMYA5 gene can act as a potential candidate gene affecting pig meat quality.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The cardiomyopathy associated 5 (CMYA5) gene is also called TRIM76, Myospryn or genethonin 3, and it is drastically decreased in Duchenne muscular dystrophy (DMD) muscles in comparison with the normal muscle tissues [1]. CMYA5 encodes a novel 413-kDa protein and in the carboxyl terminus, it contains B-box coiled-coil (BBC), fibronectin type III (FN3) repeats, and SPRY domains in a configuration reminiscent of the tripartite motif protein family. CMYA5 and dysbindin, a major schizophrenia susceptibility factor, co-immunoprecipitate from muscle extracts and are extensively co-localized [2]. CMYA5 is also identified as a transcript down-regulated in MEF2A knock-out mice by microarray analysis [3]. The CMYA5 promoter contains four MEF2 cis-elements within 3.5 kb of the transcription start site [3]. CMYA5 localizes to the costamere at the periphery of the Z-disc complex and interacts with sarcomeric α-actinin-2 [3]. At the same time, CMYA5 is an anchoring protein for protein kinase A (PKA) (or AKAP) and serves as a substrate for PKA. CMYA5 co-localizes and interacts with RII alpha (a type II regulatory subunit of PKA) at the peripheral Z-disc/costameric region in striated muscle tissues [4]. The binding of desmin with CMYA5 is confirmed with glutathione S-transferase pull-down assays and co-immunoprecipitation experiments [5]. Further research has shown that CMYA5 co-localizes with desmin at the periphery of the nucleus using an antibody against the COOH terminus of CMYA5. Deletion analysis reveals that desmin binds to CMYA5 through the 24 amino acid-long carboxyl-terminal end of the SPRY domain [5]. Dystrophic muscle exhibits reduced PKA activity resulting, in part, from severely mislocalized CMYA5. Furthermore, CMYA5 and dystrophin also coimmunoprecipitate in native muscle extracts and interact directly in vitro [6].
Much of the available information about the CMYA5 gene was taken from studies on human and mice muscle disease. However, little has been known about the CMYA5 gene in the pig. The aim of this study was to clarify the molecular characterization, chromosomal localization, expression profile and associations of the porcine CMYA5 gene with pig meat quality.
Materials and methods
Isolation and cDNA sequence analysis of the porcine CMYA5 gene
The human CMYA5 gene mRNA (GenBank accession Number: NM_153610.3) was applied to compare with all sequences in the EST-others database using standard BLAST (http://www.ncbi.nlm.nih/gov/blast/), and the porcine ESTs which shared at least 85% identity with the corresponding human mRNA were selected to design gene specific primers (Table 1). The porcine CMYA5 gene mRNA sequence was obtained by reverse transcription PCR (RT-PCR). The PCR products were purified with the 3S Spin DNA Agarose Gel Purification system (Shenergy Biocolor, Shanghai, China) and cloned into the pMD18-T vector (Takara, Dalian, China), then sequenced by commercial service.
Chromosomal localization of porcine CMYA5 gene
The INRA/University of Minnesota porcine radiation hybrid panel (IMpRH) was used to assign CMYA5 gene to the porcine genome [7]. The primers of porcine-specific (Table 1) were designed to amplify porcine genomic DNA within the porcine IMpRH panels. The PCR reaction was performed in a mixtures (10 μl) contained 1× PCR buffer (Promega), 0.25 μM each primer, 1.5 mM MgCl2, 150 μM each dNTPs, 0.5 U Taq DNA polymerase (Promega, Madison, WI, USA) and 25 ng porcine genomic DNA. The PCR conditions were performed after an initial 5 min denaturation step at 95°C followed by 34 cycles of 94°C 30 s, 60°C for 30 s, 72°C for 20 s, and then 72°C for 5 min. PCR products were scored on a 1.5% agarose gel after ethidium bromide staining and the data were analyzed with the IMpRH mapping tools on http://www.toulouse.inra.fr/lgc/pig/RH [8].
Temporal and spatial expression patterns of porcine CMYA5 gene
Seven tissue samples for expression profile analysis were collected from different stages (new born, five weeks and adult) of Tongcheng (a typical indigenous Chinese breed) pigs’ heart, liver, spleen, lung, kidney, skeletal muscle and adipose tissues. The detailed method for cDNA preparation was described previously by Xu et al. [9]. The expression pattern of the porcine CMYA5 gene mRNA in different tissues from different stages was detected by semi-quantitative RT–PCR with the porcine RPL32 (pRPL32) gene as positive control. CMYA5 gene specific primers (CMYA5E-F and CMYA5E-R in Table 1) and pRPL32 specific primers (pRPL32-F and pRPL32-R in Table 1) amplified products of 266 bp and 284 bp, respectively, were used to detect the expression pattern.
SNP identification
Pooled genomic DNA from Landrace pigs, Large white pigs, Small Meishan pigs and Duroc pigs was amplified and sequenced directly for the identification of single nucleotide polymorphism (SNP) using gene specific primers (CMYA5SF and CMYA5SR in Table 1). The polymorphism site was analyzed by sequence comparisons using the DNAstar software (DNAstar Inc., Madison, WI, USA) and was further identified by the PCR-restriction fragment length polymorphism (PCR-RFLP) method.
Breeds used for testing the allele frequencies of the porcine CMYA5 gene
The allele frequency analysis included 202 unrelated animals from six breeds (Table 2): Qingping pigs (n = 26), Tongcheng pigs (n = 27), Small Meishan pigs (n = 36), Duroc pigs (n = 26), Landrace pigs (n = 40) and Large White pigs (n = 47) [10, 11]. A chi-squared test on the allele frequencies for the six pig breeds was performed using SAS V8.0.
Association analysis of the porcine CMYA5 gene with economic traits
Animals, traits and models used for analysis were described previously by Xu et al. [9] and Liu et al. [12]. Briefly, the linear model with the fixed effects is:
where Yijklmn is the ijklmn th traits observation value; μ is the mean; Gi is the effect of the ith genotypes; Bj is the effect of jth batch; Sk is the effect of jth sex; Cl is the effect of lth population; Fm(C) is family effects within breed and εijklmn is the random residual corresponding to the traits observation value with var (ε) =\( I\sigma_{e}^{2} \).
Results and discussion
Molecular cloning and sequence analyses of porcine CMYA5 gene
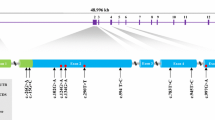
The 12,056 bp partial porcine CMYA5 cDNA (GenBank accession nos.FJ208850) was successfully obtained by RT-PCR procedures. The sequence contained an open reading frame of 12,009 bp, and encoded a protein of 4003 residues with an isoelectric point (pI) of 4.56 and calculated molecular mass of 442 kDa. It also contained a 47 bp of 3′-untranslated region (UTR). The analysis of CMYA5 amino acid sequences indicated that the protein contains BBC,FN3 and a SPRY domain in the C-terminal. Multiple alignment of the eight CMYA5 gene homology sequences indicated that these three domains are conservative among the species (Fig. 1). The three motifs comprise the TRIM region; they all have the ability to interact with the α-actinin independently [3]. TRIM proteins are involved in diverse cellular processes, including cell proliferation, differentiation, development, oncogenesis and apoptosis and, in some cases transcriptional regulation [13]. Mutations or rearrangements of the B-box family members cause abnormal cellular growth or aberrant cell function resulting in a variety of metabolic or growth regulatory dysfunctions [14].
The amino acid sequence-alignment of CMYA5 in different species. The protein sequences available from the GenBank were analyzed with ClustralX1.83, and their accession numbers are XP_00113794.7 (Pan troglodytes), FJ208850 (Sus scrofa), XP_001068814 (Rattus norvegicus), XP_536312.2 (Canis familiaris), XP_001079985.1 (Danio rerio), XP_424765.2 (Gallus gallus), XP_001503943 (Equus caballus). Different species shared the same domains: C terminus contains B-box coiled-coil (BBC), fibronectin type III (FN3) repeats, and domain in SPla and the RYanodine Receptor (SPRY)
Temporal and spatial expression patterns of porcine CMYA5 gene
RT-PCR was applied to detect the tissue distribution in different developmental stages (new born, five weeks and adult tongcheng pigs) of the porcine CMYA5 gene. The internal control, pRPL32, displayed a basically identical signal in each tissue. The CMYA5 gene was broadly expressed in all seven tissues, and significantly high expressed in the striate muscle tissues (Fig. 2). Our result was consistent with previous reports [2, 3]. Meanwhile, CMYA5 localized directly downstream of MEF2A at the costamere in striated muscle, so whether it plays a role in myofibrillogenesis needs further confirmation.
Chromosomal localization of porcine CMYA5 gene
Using the porcine RH panel, we assigned the porcine CMYA5 gene to porcine chromosome 2q21–24. It was significantly linked to microsatellite Sw1602 (distance 59 cR, LOD score value 6.74 and retention frequency 22%). The human CMYA5 gene has been mapped to 5q14.1 (http://www.ncbi.nlm.nih.gov/Louslink). Our assignment of the porcine CMYA5 gene to SSC2 is in agreement with previous comparative data [15, 16].
SNP identification and allele frequencies of the porcine CMYA5 gene in different breeds
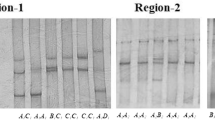
The amplification of one pair of primers (CMYA5SF and CMYA5SR in Table 1) on exon2 yielded to a 458 bp partial genomic DNA sequence. Sequencing and comparative analysis identified one SNP A383C that was detected by digestion with BspTI, resulting in a 458-bp PCR amplicon produced allele A (458) and allele C (383 and 75 bp) (Fig. 3).
One A383C SNP was detected in the Exon2. PCR products were digested with BspTI restriction enzyme to distinguish different alleles (458 bp for allele A, 383 and 75 bp for allele C). Two percent agarose gel showing the genotypes was indicated on the top of the column. M DL2000 (Jingmei BioTech Co. Ltd, China); PCR PCR products
Allele frequencies of the porcine CMYA5 gene SNP in 202 unrelated pigs indicated that all the breeds were polymorphic with the exception of the A383 C in Qingping pigs and Small Meishan pigs (Table 2).
Association analysis of the porcine CMYA5 gene with economic traits
The SNP (A383C) of the porcine CMYA5 gene was used for association analyses with the carcass and meat quality traits. According to the association analyses results, there were associations between the polymorphism (A383C) and the percentage of ham (p < 0.05), drop loss (p < 0.01) and intramuscular fat (IMF) (p < 0.05) (Table 3). The percentage of ham in pigs with the AA genotype was significantly higher than that of pigs with the CC genotype (p = 0.039), the drop loss of pigs with AC genotype was significantly higher than that of pigs with the CC genotype (p = 0.003), and IMF of pigs showed a similar trend between two different genotypes (p = 0.019).
We have assigned the porcine CMYA5 gene to porcine chromosome 2q21–24. Some interesting QTLs are located at the area, such as ham percentage QTL [17], IMF QTL [18], off-flavor score QTL [19], pH 24 h post mortem (Loin) [20] and muscle color score [21, 22]. Therefore, the association analyses are in accordance with the QTL distribution surrounding this gene.
In conclusion, the SNP identified in exon2 of the CMYA5 gene revealed an association between the genotypes and meat quality traits. It is suggested that the porcine CMYA5 gene could act as a candidate gene for meat quality traits, and thus may be useful as a genetic marker for pig breeding.
References
Tkatchenko AV, Piétu G, Cros N, Gannoun-Zaki L, Auffray C, Léger JJ, Dechesne CA (2001) Identification of altered gene expression in skeletal muscles from Duchenne muscular dystrophy patients. Neuromuscul Disord 11(3):269–277
Benson MA, Tinsley CL, Blake DJ (2004) Myospryn is a novel binding partner for dysbindin in muscle. J Biol Chem 279(11):10450–10458
Durham JT, Brand OM, Arnold M, Reynolds JG, Muthukumar L, Weiler H, Richardson JA, Naya FJ (2006) Myospryn is a direct transcriptional target for MEF2A that encodes a striated muscle, alpha-actinin-interacting, costamere-localized protein. J Biol Chem 281(10):6841–6849
Reynolds JG, McCalmon SA, Tomczyk T, Naya FJ (2007) Identification and mapping of protein kinase A binding sites in the costameric protein myospryn. Biochim Biophys Acta 1773(6):891–902
Kouloumenta A, Mavroidis M, Capetanaki Y (2007) Proper perinuclear localization of the TRIM-like protein myospryn requires its binding partner desmin. J Biol Chem 282(48):35211–35221
Reynolds JG, McCalmon SA, Donaghey JA, Naya FJ (2008) Deregulated protein kinase A signalling and myospryn expression in muscular dystrophy. J Biol Chem 283(13):8070–8074
Yerle M, Pinton P, Robic A, Alfonso A, Palvadeau Y, Delcros C, Hawken R, Alexander L, Beattie C, Schook L, Milan D, Gellin J (1998) Construction of a whole-genome radiation hybrid panel for high-resolution gene mapping in pigs. Cytogenet Cell Genet 82(3–4):182–188
Milan D, Hawken R, Cabau C, Leroux S, Genet C, Lahbib Y, Tosser G, Robic A, Hatey F, Alexander L, Beattie C, Schook L, Yerle M, Gellin J (2000) IMpRH server: an RH mapping server available on the web. Bioinformatics 16:558–559
Xu XL, Li K, Peng ZZ, Zhao SH, Yu M, Fan B, Zhu MJ, Xu SP, Du YQ, Liu B (2008) Molecular characterization, expression and association analysis with carcass traits of the porcine CMYA4 gene. J Anim Breed Genet 125:234–239
He X, Gao H, Liu C, Fan B, Liu B (2010) Cloning, chromosomal localization, expression profile and association analysis of the porcine WNT10B gene with backfat thickness. Mol Biol Rep. doi:10.1007/s11033-010-9978-4
Xu X, Qiu H, Du ZQ, Fan B, Rothschild MF, Yuan F, Liu B (2010) Porcine CSRP3: polymorphism and association analyses with meat quality traits and comparative analyses with CSRP1 and CSRP2. Mol Biol Rep 37(1):451–459
Liu K, Wang G, Zhao SH, Liu B, Huang JN, Bai X, Yu M (2009) Molecular characterization, chromosomal location, alternative splicing and polymorphism of porcine GFAT1 gene. Mol Biol Rep. 37:2711–2717
Nisole S, Stoye JP, Saïb A (2005) TRIM family proteins: retroviral restriction and antiviral defence. Nat Rev Microbiol 3(10):799–808
Torok M, Etkin LD (2001) Two B or not two B? Overview of the rapidly expanding B-box family of proteins. Differentiation 67(3):63–71
Goureau A, Yerle M, Schmitz A (1996) Human and porcine correspondence of chromosome segments using bidirectional chromosome painting. Genomics 36(2):252–262
Sun HF, Ernst CW, Yerle M, Pinton P, Rothschild MF, Chardon P, Rogel-Gaillard C, Tuggle CK (1999) Human chromosome 3 and pig chromosome 13 show complete synteny conservation but extensive gene-order differences. Cytogenet Cell Genet 85:273–278
Nezer C, Moreau L, Brouwers B, Coppieters W, Detilleux J, Hanset R, Karim L, Kvasz A, Leroy P, Georges M (1999) An imprinted QTL with major effect on muscle mass and fat deposition maps to the IGF2 locus in pigs. Nat Genet 21(2):155–156
Qu YC, Deng CY, Xiong YZ, Zheng R, Yu L, Su YH, Liu GL (2002) The construction of the genetic map and QTL locating analysis on chromosome 2 in swine. Yi Chuan Xue Bao 29(11):972–976
Kim JJ, Zhao H, Thomsen H, Rothschild MF, Dekkers JC (2005) Combined line-cross and half-sib QTL analysis of crosses between outbred lines. Genet Res 85(3):235–248
Lee SS, Chen Y, Moran C, Cepica S, Reiner G, Bartenschlager H, Moser G, Geldermann H (2003) Linkage and QTL mapping for Sus scrofa chromosome 2. J Anim Breed Genet 120(1):11–19
Malek M, Dekkers JC, Lee HK, Baas TJ, Prusa K, Huff-Lonergan E, Rothschild MF (2001) A molecular genome scan analysis to identify chromosomal regions influencing economic traits in the pig II. Meat and muscle composition. Mamm Genome 12(8):637–645
Rohrer GA, Thallman RM, Shackelford S, Wheeler T, Koohmaraie M (2005) A genome scan for loci affecting pork quality in a Duroc-Landrace F2 population. Anim Genet 37(1):17–27
Acknowledgements
We thank Dr Martine Yerle of INRA, France, for providing the RH panel. This research was supported by National Natural Science Foundation of China (30771536) and National High Science and Technology Foundation of China (2007AA10Z168).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Xu, X., Xu, X., Yin, Q. et al. The molecular characterization and associations of porcine cardiomyopathy asssociated 5 (CMYA5) gene with carcass trait and meat quality. Mol Biol Rep 38, 2085–2090 (2011). https://doi.org/10.1007/s11033-010-0334-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-010-0334-5