Abstract
Gene Rlr etb , derived from the potato species Solanum etuberosum, confers resistance to potato leafroll virus (PLRV). Mapping of this gene would aid in developing marker-assisted selection protocols to facilitate its introgression into cultivated potato. One RFLP marker and 45 cleaved amplified polymorphic markers (CAPs) markers were used to screen an etuberosum-derived BC3 family segregating for PLRV resistance conferred by Rlr etb . Nine markers from linkage group 4 of the tomato map displayed linkage with Rlr etb , however, eight additional markers from linkage group 4 that should have been syntenic with Rlr etb were not. Instead they segregated with 12 markers previously mapped to linkage group 9 of the tomato map, indicative that chromosomes 4 and 9 of S. etuberosum have translocated regions relative to the potato and tomato genomes. These chromosomal translocations have placed Rlr etb beyond the end of the published map of linkage group 4 of tomato with the closest marker, C2_At1g42990, mapping 13.6 cM from Rlr etb .
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Solanum etuberosum Lindl., a wild, non-tuber-bearing relative of cultivated potato (S. tuberosum L. subsp. tuberosum), is highly resistant to several potato pathogens, including potato leafroll virus (PLRV), potato virus Y (PVY), potato virus X (PVX), and green peach aphid, Myzus persicae (Valkonen et al. 1992a, b; USDA, ARS, National Genetic Resources Program 2003). It is a member of sect. Etuberosum which also comprises the non-tuber-bearing species S. palustre (formerly classified as S. brevidens) and S. fernandezianum. These three species are taxonomically and sexually isolated from sect. Petota to which cultivated potato belongs (Spooner and Hijmans 2001), and have been characterized as having an E-genome distinct from the A-genome of S. tuberosum (Ramanna and Hermsen 1981; Matsubayashi 1991; Perez et al. 1999).
Barriers to hybridization of S. etuberosum with cultivated potato were overcome through the use of somatic hybridization (Novy and Helgeson 1994a; Thieme et al. 1999). High levels of resistance to PVY were reported in somatic hybrids of S. etuberosum and their sexual progeny (Novy and Helgeson 1994b; Thieme et al. 1999; Gavrilenko et al. 2003). Resistances to PVY, PLRV, and green peach aphid derived from S. etuberosum, also were identified in the BC1 and BC2 progeny of somatic hybrids (Novy et al. 2002), and PLRV resistance was shown to be expressed in the BC3 generation as well (Novy et al. 2007).
The stable transmission of PLRV resistance following three generations of backcrossing to cultivated potato indicate that genomic differences between S. etuberosum and cultivated potato have not detrimentally impacted the introgression of this trait. Segregation for PLRV resistance in the two BC3 populations used in this present study most closely fit a gene model whereby resistance was conferred by a single dominant gene inhibiting the systemic spread of PLRV from infected foliage to tubers (Novy et al. 2007). Localization of the PLRV resistance gene to one genomic region also supports this gene model (Gillen and Novy 2007).
Potato leafroll virus resistance conferred by a single gene demonstrated as heritable following successive backcrossing to cultivated potato would be advantageous to potato breeders. Efforts in developing PLRV-resistant potato varieties have been hampered due to polygenic inheritance of resistance or difficulty in introgressing identified monogenic resistance from donor wild species (Jansky 2000; Solomon-Blackburn and Barker 2001; Taliansky et al. 2003). Of the 13 most widely grown potato cultivars in North America, none of them are resistant to PLRV (Corsini and Brown 2001), even though this virus is considered among the most problematic of the potato viruses on a world-wide scale (Solomon-Blackburn and Barker 2001).
The objective of the present study was to map the location of PLRV resistance in the BC3 progeny of a S. etuberosum somatic hybrid. Currently, selecting for PLRV resistance is done under field conditions and is a time and labor-intensive process. Marker-assisted selection for PLRV resistance would speed this process and would be a great help to potato breeding programs. An earlier study localized the PLRV resistance of S. etuberosum to chromosome 4 (Gillen and Novy 2007). This study reports on mapping the location of a PLRV resistance gene derived from S. etuberosum, represented by the designation Rlr etb . New information also is presented regarding genomic structural differentiation between the E-genome of S. etuberosum and the A-genome of S. tuberosum identified during mapping of Rlr etb .
Materials and methods
Plant material
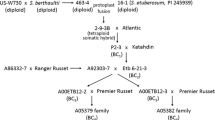
Somatic hybrids were produced by protoplast fusion of the diploid (2n = 2x = 24) S. etuberosum (PI 245939) (clone designation: 16-1) with a diploid S. tuberosum subsp. tuberosum haploid-wild species hybrid [US-W730 × S. berthaultii (PI 265857)] designated 463-4 (Novy and Helgeson 1994a). BC1 individuals were produced by crossing somatic hybrid 2-9-3B with the potato cultivar Atlantic, and BC2 individuals were produced by crossing the BC1 individual P2-3 with the cultivar Katahdin. The BC2 individual Etb 6-21-3 was used as the PLRV resistant parent in crosses with the advanced breeding clone A92303-7 to produce a BC3 family of four individuals designated A00ETB12, and with the potato cultivar GemStar Russet to produce a family of 35 individuals designated as AO1687 (Table 1).
Screening for PLRV resistance
Plant material was previously screened and assessed for response to PLRV infection in the field as reported by Novy et al. (2007), with ELISA testing of harvested daughter tubers conducted to ensure veracity in the classification of response to PLRV infection. The cultivar Liu was used as a PLRV resistant control and the cultivars Russet Burbank and Ranger Russet were used as PLRV susceptible controls.
DNA extraction and marker analysis
Young leaf tissue of potato plants grown in the field or greenhouse was used for DNA extraction as described by Gillen and Novy 2007. The protocol involved either a modification of a CTAB extraction procedure (Doyle and Doyle 1987) or a modification of a nuclei extraction procedure (Bernatsky and Tanksley 1986).
RFLP analysis using probe TG443 was carried out following the procedure described by Gillen and Novy (2007). Probe labeling and detection was carried out using Gene Images™ labeling and detection systems (Amersham Biosciences, Piscataway, NJ, USA). A polymorphism specific to S. etuberosum and 6.5 kb in size was scored from genomic DNA digested with the restriction enzyme EcoRV.
Cleaved amplified polymorphism (CAP) markers were amplified using primers obtained from tomato or potato (Table 2) (Chen et al. 2001; Frary et al. 2005; Wu et al. 2006). Most were COSII markers (second generation conserved ortholog set markers) (Wu et al. 2006), the exceptions being cLEC7B23 (Frary et al. 2005), ANTL (a known function gene available at http://sgn.cornell.edu/), and Dpe-P (Chen et al. 2001). All markers had been previously placed on the Solanum lycopersicum (LA925) × S. pennellii (LA716) high-density map (tomato map) (Frary et al. 2005; Fulton et al. 2002; Wu et al. 2006) with the exception of Dpe-P, which was mapped in potato (Chen et al. 2001). The tomato map was used as a reference to identify markers mapped to tomato linkage group 4 that likely were syntenic with Rlr etb, and therefore would be useful in establishing genetic linkages to this resistance gene. Due to the length of many PCR fragments (often greater than 1,000 bp), an extended PCR protocol was used as follows: 1 cycle of 94°C for 3 min, an annealing temperature of 55°C for 2 min, 72°C for 1 min 30 sec; followed by 39 cycles of 94°C for 45 sec, an annealing temperature of 55°C for 1 min 30 sec, 72°C for 1 min 30 sec; then a final extension step for 10 min at 72°C. Amplified fragments were screened for both amplicon and restriction site polymorphisms (Table 2). Restriction enzymes were used at a concentration of 0.05 U ml−1 with digestion reactions conducted at the enzyme-specific recommended temperatures for 3 h, followed by 20 min of heat inactivation. PCR and restriction digest products were analyzed by gel electrophoresis using 0.7% Seakem® LE Agarose (Cambrex Bio Science Rockland, Inc., Rockland, ME, USA) and 1.15% Synergel™ (Diversified Biotech, Boston, MA, USA). Only markers unique to the S. etuberosum parent were analyzed.
S. etuberosum 16-1: S1 progeny bulk analysis
The original S. etuberosum parent, 16-1, which does not produce tubers, and which had previously been maintained in tissue culture was lost in 2004. However, S. etuberosum and the other two diploid species within sect. Etuberosum are unique among diploid potato species in that they are self fertile and have a high level of genetic homozygosity (Spooner et al. 1992, 1996). Prior to its loss, greenhouse plants of 16-1 had flowered and were selfed to produce S1 progeny. To obtain DNA samples that closely reconstituted the genetic identity of the original parent, S1 plants were grown in the greenhouse and leaves bulked together for DNA extraction. Leaf tissue was collected and bulked from 65 S1 plants; from 131 g of bulked tissue, 15 g was used for a large-scale DNA extraction using a modification of a nuclei extraction procedure (Bernatsky and Tanksley 1986; Gillen and Novy 2007).
To verify that this bulk of S1 plants provided a DNA sample that approximated the 16-1 parent, two CAPs markers from each of the 12 chromosomes in the tomato map were evaluated for polymorphisms between the progeny bulk and remnant DNA retained of the original 16-1 parent. Each of the markers was evaluated with 12 restriction enzymes (data not shown). The number of enzymes that cut amplified fragments ranged from 5 to 11 out of 12 enzymes with an average of 8.1 enzymes per marker, resulting in a total of 195 restriction sites evaluated. From among these, no polymorphism was detected between the 16-1 parent and the progeny bulk DNA, indicating that the full genome of clone 16-1 was represented in its 65 S1 progeny. The S1 bulk DNA was used in the place of the 16-1 parent in this study.
Mapping of RFLP and CAPs markers
Molecular markers were scored in BC3 individuals as well as parental clones to determine whether polymorphisms were unique to S. etuberosum. Only marker fragments unique to S. etuberosum were evaluated, as S. tuberosum specific markers are present in all progeny of this backcross population. Such S. etuberosum-specific markers were expected to be simplex in the BC2 parent and to segregate for presence/absence in a 1:1 ratio in the BC3 population. A goodness-of-fit test was performed to determine whether markers fit the expected ratios.
A genetic map was constructed using the statistical program TetraploidMap (Hackett and Luo 2003). Markers were grouped using the cluster analysis function of the program and compared against the tomato or potato map they originated from using a LOD threshold of 3.0. Marker order was determined using a two-point linkage analysis and simulated annealing (Hackett and Luo 2003).
Results
Initial genomic localization studies for PLRV resistance were carried out using a combination of RFLP and SSR markers on six BC2 and four BC3 breeding clones of the A00ETB12 family (Gillen and Novy 2007). The marker TG443, mapped to linkage group 4 of tomato and potato, was identified as co-segregating with the PLRV resistance gene, Rlr etb (Gillen and Novy 2007). An additional 35 BC3 clones from family AO1687 were screened with TG443, and among the 39 BC3 clones (total across both BC3 families), TG443 was present in 18 (Fig. 1). The segregation of TG443 with PLRV resistance is outlined in Table 3. Observed segregation for the four classes in Table 3 were tested against a model in which TG443 was unlinked to Rlr etb , with an expected 25% of BC3 individuals present in each class. A chi-square test using Excel software rejected at the 5% level of significance that TG443 was unlinked to Rlr etb , with a calculated P value of 0.01—corroboration of the linkage of TG443 with Rlr etb . On the basis of the frequency of recombination between TG443 and Rlr etb (represented by the TG443(+)/Susceptible and TG443(−)/Resistant classes in the BC3), genetic distance between the two is calculated to be 24 cM.
Ideogram of S. etuberosum chromosome 4 in 39 BC3 progeny of a somatic hybrid between S. etuberosum and a S. tuberosum haploid × S. berthaultii hybrid determined with CAPs and RFLP markers. Each chromosome is marked with nine markers, placed in the order that minimized the number of recombinants. See Fig. 2 to see how this order differs from that of the published tomato map of these markers. Markers present in the respective BC3 are represented by black bars over gray areas, and absent markers are represented by black bars behind white areas. The score for PLRV resistance of each BC3 clone is indicated by (R) for resistant clones and (S) for susceptible clones following each clonal designation. The exception being clone AO1687-32 for which its response to infection by PLRV remains undetermined
Confirmation of the linkage of TG443 with Rlr etb , allowed the selection of CAPs markers published on the SOL Genomics Network (http://sgn.cornell.edu/) that could be used to saturate chromosome 4 and allow mapping of Rlr etb (Mueller et al. 2005, 2008). Of the 84 CAPs markers that were amplified using PCR in this study, 79 had primers that amplified a fragment near the expected size from S. etuberosum. Of these, 62 were tested for amplicon or restriction fragment polymorphisms specific to S. etuberosum, and 45 produced polymorphisms that were scored in 39 BC3 clones. The two markers C2_At1g35720 and C2_At4g09010 produced secondary amplicon polymorphisms that mapped to locations other than the expected synteny groups along with polymorphisms that mapped to expected locations. Of the 5 markers that did not amplify well from S. etuberosum, 2 of them were COSII markers (C2_At1g43580 and C2_At3g17210) and the other three used primers designed from tomato sequences (TG223, Brix 9-2-5, and U168526).
When analyzed with the goodness-of-fit test, the majority of markers did not deviate from the expected 1:1 segregation ratio. Exceptions include the following: four from chromosome 3 that formed a linkage group (C2_At2g01770, C2_At5g62390, TG324, and C2_At4g14570) had a lower than expected number that scored positive for the markers from S. etuberosum; and three markers, C2_At2g20860, C2_At2g42810, & C2_At3g15290, from linkage group 7 of the tomato map that scored positive for these markers from S. etuberosum in 38, 38, and 37 out of 39 BC3 clones, respectively.
Localization of Rlr etb
Nine markers unique to S. etuberosum from linkage group 4 of the tomato map were identified as being linked with Rlr etb (Fig. 2). The scores for the 39 BC3 clones with these nine markers are displayed in Fig. 1. The order of the markers in the figure is arranged to minimize the number of recombinants. All nine markers co-segregated with Rlr etb in 26 out of 38 clones, as measured by the presence of all nine in PLRV resistant clones and their absence in susceptible clones. Potential recombinants include three resistant clones that scored negative for all nine markers, one susceptible clone scored positive for all nine markers, and eight clones that scored positive for some markers and negative for others (Fig. 1).
Linkage maps generated by TetraploidMap of S. etuberosum synteny groups 4 and 9 in the BC3 progeny of a somatic hybrid between S. etuberosum and a S. tuberosum haploid × S. berthaultii hybrid determined with CAPs and RFLP markers. The published tomato map of these markers is included for comparison (Wu et al. 2006) and to show the proposed structural differences within the S. etuberosum chromosomes. The tomato map of chromosome 4 is inverted compared to the published map (Wu et al. 2006). The proposed location of the PLRV resistance gene Rlr etb is shown on the map. Circled markers were mapped to chromosome 4 in the tomato map, but formed a separate synteny group that aligned with markers from chromosome 9 of tomato
Linkage analyzes of these nine markers indicate that Rlr etb is located outside the limits of this map at a potential distance of 13.6 cM from the marker C2_At1g42990 (Fig. 2). The order that minimized the number of recombinants was not conserved with the tomato map (Fig. 2) (Wu et al. 2006). cLEC7B23 and C2_At3g03990 switched places in the marker order of BC3 progeny compared to the tomato map, while C2_At1g76080, which should have been most tightly linked to Rlr etb , given the tomato map marker order, was instead closer to the other end of the synteny group.
E-genome structure
Markers from across linkage group 4 were analyzed to determine whether one could be located that was more tightly linked to Rlr etb . Instead, 8 markers that mapped beyond C2_At3g03990 across the remainder of linkage group 4 of the tomato map formed a separate synteny group (Fig. 2—markers referenced are circled). ANTL, which mapped 1.5 cM from C2_At3g03990 on group 4 of the tomato map, did not show linkage with this marker in the etuberosum-derived BC3. In all, 17 markers mapped to linkage group 4 of the tomato map were scored. Markers most closely linked to Rlr etb were at the extreme end of the published tomato map (C2_At1g42990 and C2_At1g79600). At the other end of the published tomato map of chromosome 4, C2_At3g51010 is located 19.7 cM from the beginning of the map. The 17 markers are spaced an average of 7.3 cM apart across the tomato map.
Markers from chromosomes other than 4 that had been identified by Gillen and Novy (2007) as present in the resistant parent, Etb 6-21-3, were used to screen the BC3 population. This effort was undertaken to identify translocated regions of S. etuberosum now associated with the section of chromosome 4 linked to Rlr etb . Based on previous segregation analysis (Gillen and Novy 2007), and on the identification of TG10, a marker from linkage group 9, as being linked to markers from linkage group 4 of tomato and potato in the published map for the E-genome (Perez et al. 1999), chromosome 9 was selected as a potential candidate for this translocated region. Twelve markers unique to S. etuberosum distributed across linkage group 9 of the tomato map formed a synteny group with the 8 markers at the end of chromosome 4 of the tomato map that had not previously integrated with markers of linkage group 4 of the BC3 population (Fig. 2). Linkage analysis with TetraploidMap indicates ANTL from chromosome 4 is 26.2 cM from C2_At3g24010 of chromosome 9 in the BC3 progeny (Fig. 2).
Additional markers unique to S. etuberosum from chromosomal regions other than 9 that were present in the resistant parent, Etb 6-21-3, also were analyzed for possible linkage to Rlr etb . Marker selection focused on chromosomal regions known to contain resistance genes in Solanaceous species (Grube et al. 2000) according to the ‘SOLanaceae Function Map for Pathogen Resistance’ compiled by Christiane Gebhardt (https://gabi.rzpd.de/) (Gebhardt and Valkonen 2001; Meyer et al. 2005). In addition to chromosomes 4 and 9, this included regions of chromosomes 1, 3, 5, 6, and 7 (Table 2). No additional markers linked to Rlr etb were identified. However, synteny groups were formed for linkage groups 3 and 6 with markers in chromosomal regions that previously were separate linkage groups in the published map of the E-genome (Perez et al. 1999) (Fig. 3). These results indicate that these unconsolidated sections of chromosomes 3 and 6 of the E-genome may in fact be from the same chromosomes as expected based on the tomato and potato maps. Other than those results already mentioned, markers located on the same linkage groups in the tomato map were grouped into predicted synteny groups in the BC3 population in this study (chromosomes 1, 5, and 7- data not shown) (Table 2) with the exception of C2_At1g14790, which did not segregate with the other two markers from chromosome 5 and was not grouped by TetraploidMap with any other scored markers.
Linkage maps generated by TetraploidMap of S. etuberosum synteny groups 3 and 6 in the BC3 progeny of a somatic hybrid cross between S. etuberosum and a S. tuberosum haploid × S. berthaultii hybrid (Novy and Helgeson 1994b) determined with CAPs markers. The maps generated by this study are on the left, with the published tomato map of these markers (Wu et al. 2006) in the center, and the E-genome map (Perez et al. 1999) on the right. Comparison between the maps indicates potential consolidation of unlinked sections of chromosomes 3 and 6 in the E-genome (Perez et al. 1999). C2_At2g01770, which is from the tomato map of chromosome 4, produced a secondary amplicon polymorphism that segregated with markers from chromosome 3
Discussion
COSII markers (Wu et al. 2006) were used for the majority of the marker analysis of the current study. A previous study with this material had used mainly RFLP markers and SSR markers developed in tomato (Gillen and Novy 2007). In the current study, PCR-generated markers were emphasized due their advantages over RFLP markers with respect to ease of use and applicability to marker assisted selection, with lesser amounts of DNA required relative to RFLPs—an advantage when extracting DNA from breeding populations having large numbers of individuals. SSR markers were initially evaluated, but many of them did not amplify well from the S. etuberosum parent. Unlike the SSRs evaluated in this and the previous study (Gillen and Novy 2007), the primers for COSII markers were designed based on a consensus sequences across several species (Wu et al. 2006). This design strategy likely enabled the high level of success attained at evaluating S. etuberosum specific polymorphisms with COSII markers in the current study. The size of the S. etuberosum-derived, BC3 population used in this study did not appear to have impacted the grouping and ordering of the COSII markers relative to tomato (Figs. 2, 3). With few exceptions, the majority of markers not associated with putative translocations showed good concordance in both grouping and order with the tomato map—such concordance would not have been expected if the population size used for mapping Rlr etb had been inadequate for this purpose.
The loss of S. etuberosum specific marker fragments in the BC3 population indicates that recombination is taking place between S. etuberosum and S tuberosum chromosomes supporting previous reports of recombination in the BC2 by Gillen and Novy (2007). This is in agreement with previous studies of S. etuberosum and S. palustre crosses with tuberous Solanum species that indicate recombination takes place in spite of a bias against homeologous pairing (McGrath et al. 1996; Ramanna and Hermsen 1982; Williams et al. 1993). Williams et al. (1993) observed a lack of recombination in chromosomes 4 and 9 of S. palustre with S. tuberosum. However, despite the observed structural differences between chromosomes 4 and 9 of S. etuberosum and S. tuberosum in the present study, the ideograms for chromosome 4 of the BC3 in Fig. 1 depict single and multiple cross-over events are occurring between the distinctly different E- and A-genomes of the two species. This suggests that Rlr etb can be integrated into the genome of cultivated potato, and that there is potential for marker assisted selection of Rlr etb in potato breeding programs. However, observed discrepancies from a 1:1 ratio in chromosomes 3 and 7 and may indicate a potential bias toward loss of chromosome 3 and retention of chromosome 7 of S. etuberosum.
Translocations between chromosomes 4 and 9 of the E-genome were not reported in the published map of the E-genome (Perez et al. 1999). However, only markers from the center of the tomato map of chromosome 9 were evaluated in that study, which may have been too distant to detect this translocation event. Also, unlike the present study, markers used in chromosomes 3 and 6 of the published E-genome map were not able to be consolidated into single groups (Perez et al. 1999). A reduced level of recombination between chromosomes 3 and 6 of S. etuberosum and S. tuberosum may have allowed detection of these groups in the present study. Perez et al. (1999) reported greater recombination values in the E-genome compared to the A-genome which would make linkage between distant markers more difficult to detect. Also, the use of COSII markers, not yet identified and mapped at the time when the E-genome map was published, also provided additional saturation of chromosomes 3 and 6 allowing consolidation of previously separate synteny groups.
This study has identified a COSII marker, C2_At1g42990, at a genetic distance of 13.6 cM from Rlr etb —an improvement in linkage relative to RFLP marker TG443 at a distance of 24 cM. Our attempts at identifying markers more closely linked to Rlr etb for use in marker assisted selection led to the identification of translocations among chromosomes 4 and 9 of the E-genome of S. etuberosum relative to the A-genome of S. tuberosum. Translocations between chromosomes 4 and 9 of the E-genome had not previously been reported and support previous reports of structural differentiation between the two genomes (Perez et al. 1999). Translocations between chromosomes 4 and 9 have confounded our efforts to develop molecular markers more tightly linked to Rlr etb . However, a larger population of 115 BC4 individuals has been developed and was screened for response to infection by PLRV in 2008. This BC4 population with its larger number of individuals will be characterized using additional mapped markers from candidate chromosomes thought to be associated with chromosome 4 of S. etuberosum via translocations. This approach should allow for the further mapping of the location of Rlr etb and the identification of molecular markers applicable for use in marker assisted selection for this unique source of PLRV resistance.
References
Bernatsky R, Tanksley SD (1986) Genetics of actin-related sequences in tomato. Theor Appl Genet 72:314–321. doi:10.1007/BF00288567
Chen X, Salamini F, Gebhardt C (2001) A potato molecular-function map for carbohydrate metabolism and transport. Theor Appl Genet 102:284–295. doi:10.1007/s001220051645
Corsini D, Brown CR (2001) Important potato cultivars. In: Loebenstein G, Berger PH, Brunt AA, Lawson RH (eds) Virus and virus-like diseases of potatoes, production of seed potatoes. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 39–52
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemistry Bull 19:11–15
Frary A, Xu Y, Liu J, Mitchell S, Tedeschi E, Tanksley S (2005) Development of a set of PCR-based anchor markers encompassing the tomato genome and evaluation of their usefulness for genetics and breeding experiments. Theor Appl Genet 111:291–312. doi:10.1007/s00122-005-2023-7
Fulton TM, Van der Hoeven R, Eannetta NT, Tanksley SD (2002) Identification, analysis, and utilization of conserved ortholog set markers for comparative genomics in higher plants. Plant Cell 14:1457–1467. doi:10.1105/tpc.010479
Gavrilenko T, Thieme R, Heimback U, Thieme T (2003) Fertile somatic hybrids of Solanum etuberosum (+) dihaploid Solanum tuberosum and their backcrossing progenies: relationships of genome dosage with tuber development and resistance to potato virus Y. Euphytica 131:323–332. doi:10.1023/A:1024041104170
Gebhardt C, Valkonen JPT (2001) Organization of genes controlling disease resistance in the potato genome. Annu Rev Phytopathol 39:79–102. doi:10.1146/annurev.phyto.39.1.79
Gillen AM, Novy RG (2007) Molecular characterization of the progeny of Solanum etuberosum identifies a genomic region associated with resistance to potato leafroll virus. Euphytica 155:403–415. doi:10.1007/s10681-006-9342-x
Grube RC, Radwanski ER, Jahn M (2000) Comparative genetics of disease resistance within the Solanaceae. Genetics 155:873–887
Hackett CA, Luo ZW (2003) TetraploidMap: construction of a linkage map in autotetraploid species. J Hered 94:358–359. doi:10.1093/jhered/esg066
Jansky SH (2000) Breeding for disease resistance in potato. In: Janick J (ed) Plant breeding reviews. Wiley, New York, pp 69–155
Matsubayashi M (1991) Phylogenetic relationships in the potato and its related species. In: Tsuchiya T, Gupta P (eds) Chromosome engineering in plants: genetics, breeding, evolution. Elsevier, Netherlands, pp 93–118
McGrath JM, Wielgus SM, Helgeson JP (1996) Segregation and recombination of Solanum brevidens synteny groups in progeny of somatic hybrids with S. tuberosum: intragenomic equals or exceeds intergenomic recombination. Genetics 142:1335–1348
Meyer S, Nagel A, Gebhardt C (2005) PoMaMo-a comprehensive database for potato genome data. Nucleic Acids Res 33:D666–D670. doi:10.1093/nar/gki018
Mueller LA, Solow TH, Taylor N, Skwarecki B, Buels R, Binns J, Lin C, Wright MH, Ahrens R, Wang Y, Herbst EV, Keyder ER, Menda N, Zamir D, Tanksley SD (2005) The SOL Genomics Network. A comparative resource for Solanaceae biology and beyond. Plant Physiol 138:1310–1317. doi:10.1104/pp.105.060707
Mueller LA, Mills AA, Skwarecki B, Buels RM, Menda N, Tanksley SD (2008) The SGN comparative map viewer. Bioinformatics 24:422–423. doi:10.1093/bioinformatics/btm597
Novy RG, Helgeson JP (1994a) Somatic hybrids between Solanum etuberosum and diploid, tuber bearing Solanum clones. Theor Appl Genet 89:775–782
Novy RG, Helgeson JP (1994b) Resistance to potato virus Y in somatic hybrids between Solanum etuberosum and S. tuberosum × S. berthaultii hybrid. Theor Appl Genet 89:783–786
Novy RG, Nasruddin A, Ragsdale DW, Radcliffe EB (2002) Genetic resistances to potato leafroll virus, potato virus Y, and green peach aphid in progeny of Solanum etuberosum. Am J Potato Res 79:9–18
Novy RG, Gillen AM, Whitworth JL (2007) Characterization of the expression and inheritance of potato leafroll virus (PLRV) and potato virus Y (PVY) resistance in three generations of germplasm derived from Solanum etuberosum. Theor Appl Genet 114:1161–1172. doi:10.1007/s00122-007-0508-2
Perez F, Menendez A, Dehal P, Quiros CF (1999) Genomic structural differentiation in Solanum: comparative mapping of the A- and E-genomes. Theor Appl Genet 98:1183–1193. doi:10.1007/s001220051183
Ramanna MS, Hermsen JGTH (1981) Structural hybridity in the series Etuberosa of the genus Solanum and its bearing on crossability. Euphytica 30:15–31. doi:10.1007/BF00033655
Ramanna MS, Hermsen JGT (1982) Gene transfer from non-tuberous to tuberous Solanum species: evidence from meiosis in hybrids. Euphytica 31:565–572. doi:10.1007/BF00039194
Solomon-Blackburn RM, Barker H (2001) Breeding virus resistant potatoes (Solanum tuberosum): a review of traditional and molecular approaches. Heredity 86:17–35. doi:10.1046/j.1365-2540.2001.00799.x
Spooner DM, Hijmans R (2001) Potato systematics and germplasm collecting, 1989–2000. Am J Potato Res 78:237–268
Spooner DM, Douches DS, Contreras MA (1992) Allozyme variation within Solanum sect. Petota, series Etuberosa (Solanaceae). Am J Bot 79:467–471. doi:10.2307/2445161
Spooner DM, Tivang J, Nienhuis J, Miller JT, Douches DS, Contreras MA (1996) Comparison of four molecular markers in measuring relationships among the wild potato relatives Solanum section Etuberosum (subgenus Potatoe). Theor Appl Genet 92:532–540. doi:10.1007/BF00224555
Taliansky M, Mayo MA, Barker H (2003) Potato leafroll virus: a classic pathogen shows some new tricks. Mol Plant Pathol 4:81–89. doi:10.1046/j.1364-3703.2003.00153.x
Thieme RT, Gavrilenko T, Thieme T, Hemibach U (1999) Production of potato genotypes with resistance to potato virus Y (PVY) by biotechnological methods. In: Altman A, Ziv M, Izhar S (eds) Proceedings of the 9th international congress IAPTC and biotech on “plant biotechnology and in vitro biology in the 21st centrury”, Jerusalem, Israel. Kluwer Academic Publishers, The Netherlands, pp 557–560
USDA, ARS, National Genetic Resources Program. Germplasm Resources Information Network-(GRIN). [Online Database] National Germplasm Resources Laboratory, Beltsville, MD. Available: http://www.ars-grin.gov/cgi-bin/npgs/html/obs.pl?1189587 (21 Aug 2008)
Valkonen JPT, Brigneti G, Pehu E (1992a) Resistance to Myzus persicae (Sulz.) in wild potatoes of the series Etuberosa. Acta Agric Scand Sect BSoil Plant Sci 42:118–127
Valkonen JPT, Brigneti G, Salazar LF et al (1992b) Interactions of the Solanum spp. of the Etuberosa group and nine potato-infecting viruses and a viroid. Ann Appl Biol 120:301–313. doi:10.1111/j.1744-7348.1992.tb03427.x
Williams CE, Wielgus SM, Haberlach GT, Guenther C, Kim-Lee H, Helgeson JP (1993) RFLP analysis of chromosomal segregation in progeny from an interspecific hexaploid somatic hybrid between Solanum brevidens and Solanum tuberosum. Genetics 135:1167–1173
Wu F, Mueller LA, Crouzillat D, Petiard V, Tanksley SD (2006) Combining bioinformatics and phylogenetics to identify large sets of single-copy orthologous genes (COSII) for comparative, evolutionary and systematic studies: a test case in the Euasterid plant clade. Genetics 174:1407–1420. doi:10.1534/genetics.106.062455
Acknowledgments
The authors wish to acknowledge and thank Mark Fristad, Darren Hall, Charlene Miller, Brian Schneider, Penny Tubbs, and Steve Wheeler for their contributions to this research. Special thanks to Isabel Vales, Oregon State University, for her review of this manuscript and her helpful comments. Partial funding for this research was supported by the ARS-State Partnership Potato Program.
Disclaimer
Mention of trade names or commercial products is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kelley, K.B., Whitworth, J.L. & Novy, R.G. Mapping of the potato leafroll virus resistance gene, Rlr etb , from Solanum etuberosum identifies interchromosomal translocations among its E-genome chromosomes 4 and 9 relative to the A-genome of Solanum L. sect. Petota . Mol Breeding 23, 489–500 (2009). https://doi.org/10.1007/s11032-008-9251-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11032-008-9251-x