Abstract
A series of mIDH1 inhibitors derived from 3-pyrimidine-4-oxazolidin-2-ketone derivatives were studied by QSAR model to explore the key factors that inhibit mIDH1 activity. The generated model was cross-verified and non-cross-verified by Topomer CoMFA and HQSAR methods; the independent test set was verified by PLS method; the Topomer search technology was used for virtual screening and molecular design; and the Surflex-Dock method and ADMET technology were used for molecular docking, pharmacology and toxicity prediction of the designed drug molecules. The Topomer CoMFA and HQSAR cross-validation coefficients q2 are 0.783 and 0.784, respectively, and the non-cross-validation coefficients r2 are 0.978 and 0.934, respectively. Ten new drug molecules have been designed using Topomer search technology. The results of molecular docking and ADMET show that the newly designed drug molecules are effective. The docking situation, pharmacology and toxicity prediction results are good. The model can be used to predict the bioactivity of the same type of new compounds and their derivatives. The prediction results of molecular design, molecular docking and ADMET can provide some ideas for the design and development of novel mIDH1 inhibitor anticancer drugs, and provide certain theoretical basis of the experimental verification of new compounds in the future.
Graphic abstract
Newly designed molecules after docking with corresponding proteins in the PDB library, it can explore the targets of drug molecules acting with large proteins and the related force, which is very helpful for the design of new drugs and the mechanism of drug action.

Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Isocitrate Dehydrogenase 1 (IDH1) is a rate-limiting enzyme in the tricarboxylic acid cycle, which is involved in life activities such as glutamine metabolism, phospholipid metabolism, fat synthesis, insulin secretion and cell reactive oxygen regulation. It plays an important role in the process and is also closely related to the occurrence of tumors [1].
With the deepening of IDH1 and tumor research, it has been found that IDH1 is closely related to the occurrence, development, metastasis and prognosis of many tumors. IDH1 mutations are heterozygous and usually involve amino acid substitutions in the active site of the enzyme at codon 132 [2]. As studies have found, changes in IDH1 expression level and mutations in 132 amino acids are closely related to the occurrence and development of many tumors, such as glioma [3], gastric cancer and colorectal cancer [4], esophageal cancer [5], lung cancer [6] and other cancers.
In general, IDH1 catalyzes the conversion to isocitrate to α-ketoglutaric acid (α-KG), and produces non-mitochondrial nicotinamide adenine dinucleotide phosphate (NAPDH). Mutated Isocitrate Dehydrogenase 1(mIDH1) enables it to acquire new catalytic activity by using NAPDH to further convert α-KG to R-2-hydroxyglutarate (R-2HG) [7, 8]. High levels of R-2HG induce hypermethylation of histones and chromatin, and competitively inhibit several α-KG-dependent dioxygenases, eventually leading to the occurrence of tumors [9]. Although the connection between mIDH1 and tumors was first discovered in gliomas [10], with the deepening of research, it was found that it has a certain correlation between many tumors. Therefore, mIDH1 is likely to become a new target for tumor treatment.
So far, many mIDH1 inhibitors have been reported at home and abroad. Among them, Novartis Institute of Biomedical Research [11] reported the use of 3-pyrimidin-4-yl-oxazolidin-2-one derivatives as IDH1R132H inhibitors and has made certain progress. Especially, IDH305 has entered clinical trials for the treatment of patients with advanced malignant tumors containing IDH1R132H mutations [12]. However, the safety and effectiveness of new mIDH1 inhibitors is still a big problem. Therefore, it is necessary to use molecular modeling methods to conduct detailed studies on the relationship between their structure and activity. (Fig. 1 shows one of the compounds IDH305 synthesized by Novartis Institute of Biomedical Research).
In this study, the relationship between structure and activity of 3-pyrimidin-4-yl-oxazolidin-2-one derivatives as mIDH1 inhibitors was studied. Topomer CoMFA and HQSAR were used to study the quantitative structure–activity relationship (QSAR), and the generated model was cross-verified and non-cross-verified to determine its predictive activity; Virtual screening based on R groups was carried out in ZINC database using Topomer Search technology. New mIDH1 inhibitors with higher activity were designed through recombination of screened small molecular fragments and molecular fragments with relatively high contribution value in molecular concentration, and their activity was predicted by the QSAR model established. Then, molecular docking technology was used to dock these newly designed compound molecules with macromolecular proteins to explore the mechanism of action between them, explore the binding mode and target between ligands and protein receptors, and find the amino acid residues that form hydrogen bond interactions between ligands and crystal structures. Finally, ADMET prediction was used to test the safety and safety of 10 designed drug molecules to ensure that the designed molecules have high safety and good pharmacological effects.
Materials and methods
Molecular modeling and data set
In this study, the molecular structure of all compounds in the molecular construction was constructed by using the SKETCH MOLECULE in SYBYL2.0-X. All the molecules were loaded with Gasteiger-Huckel charge, and the standard Tripos molecular force field and Powell energy gradient algorithm were used to optimize the energy of all small molecules. The maximum number of iterations was 1000, the energy convergence limit was set as 0.005 kcal·mol−1, and the default values of the rest parameters were adopted [13, 14]. Energy minimization keeps compounds in a stable state of low energy.
The 3-pyrimidin-4-yl-oxazolidin-2-ones and the effective concentration reduction in the half maximal inhibitory concentration (IC50 mol·L−1) were selected from four patents of Novartis (WO2013046136, WO2014141104, WO2014141153 and WO2014147586) and literature [15]. For the convenience of calculation, these IC50 values were converted into corresponding pIC50 values to represent their biological activity and used as a dependent variable in Topomer CoMFA and HQSAR models, and the equivalent relation is: pIC50 = -lgIC50. The chemical structure and activity of 41 3-pyrimidin-4-yl-oxazolidin-2-one derivatives are shown in Table 1. The data set was divided into two parts, namely the training set for QSAR model generation (30 compounds, accounting for about 3/4) and the test set for external validation of the model (11 compounds, accounting for about 1/4).
Construction of the QSAR model
Quantitative structure–activity Relationship (QSAR) is a common research method to explore the relationship between molecular structure and its biological activity. Mathematical models are usually used to study the quantitative relationship between molecular structure and molecular biological activity. Molecular structure parameters are taken as independent variables and molecular biological activity as dependent variables to establish the quantitative relationship expression between activity and structure [16, 17]. Three-dimensional Quantitative structure–activity Relationship (3D-QSAR) is the study of QSAR based on the 3D structure of compounds and biomacromolecules. Compared with the traditional QSAR method, 3D-QSAR added a 3D conformation of the properties of bioactive molecules, introduces and bioactive molecules in the QSAR of 3D structure information about parameter as a variable, so it can more accurately reflect the reality of the bioactive molecules and receptors, more profoundly illustrates the interaction between drug and the receptor mechanism, to provide certain theoretical basis to the research and development of new drugs [18].
Topomer CoMFA analysis
Comparative molecular field analysis (CoMFA) is one of the most commonly used methods for studying the 3D-QSAR between drugs and receptors. CoMFA can fully consider the 3D structure information of molecules, characterize the 3D field and static electric field by molecular structure, and then obtain the relationship between these 3D characteristic information and compound activity by partial Least Squares [19] (PLS). By constructing the molecular structure, the lowest energy conformation of the molecule was obtained, the common skeleton was selected for molecular superposition, the molecular force field around the compound was calculated according to the probe atoms, and the relationship between the structure and activity of the drug was analyzed. Particularly, the results of molecular superposition have a great impact on the accuracy of model prediction [20]. Therefore, to obtain more accurate results often needs to try different ways of molecular overlap, and it takes a lot of time.
Topomer CoMFA, proposed by Cramer [21] in 2004, is a new 3D-QSAR tool that can predict the biological activity or properties of compounds. Unlike CoMFA, Topomer CoMFA is based on the joint technology of Topomer and CoMFA. It compensates for the shortcomings of the traditional CoMFA approach of manually superposed molecules, using 3D-QSAR to correlate the biological activity of a range of molecules with their spatial and static electric fields. Standard Tripos force field method and SP3 carbon probe atom with + 1 charge were used for calculation of space field and static electric field [22]. Space and electrostatic descriptors were used as independent variables, pIC50 value was used as dependent variable, and partial least square regression was used to establish the Topomer CoMFA model. In addition, Topomer CoMFA has the advantage of high repeatability. Topomer CoMFA can be used to quickly establish a prediction model for analysis and evaluation, providing a theoretical basis for the structural optimization of the same type of small-molecule inhibitors [23].
Topomer CoMFA does not need skeleton based 3D structure superposition. Generally, the operation process of Topomer CoMFA is as follows: the 3D model of Topomer is established according to the molecular structure fragment of the training set, and the fracture mode of the compound structure is confirmed by the structural characteristics of the compound. Topomer CoMFA uses topological molecular technology to cut the ligand molecules into two or more small fragments, while preserving the common skeleton as much as possible. It automatically builds a standard 3D topological model for each fragment, and generates a set of space and electrostatic fields for each group of topological bodies. Finally, taking the obtained parameters as the independent variables and the biological activity value as the dependent variables, the relationship between the compound activity and molecular field characteristics was established by the partial least squares method, and the 3D-QSAR model was generated.
HQSAR study
Hologram Quantitative structure–activity Relationship (HQSAR) is a 2D-QSAR technology, which is different from the general 3D-QSAR research method. It can determine the relationship between biological activities and structural fragments without the need for molecular superposition, selection of 3D Structure and active conformation. It relies on 2D chemical database storage and search techniques that rely on linear symbols that define chemical structures, a process that involves generating hashes into arrays of fragments called molecular holograms [24]. The ability to achieve molecular arrangement and conformational specification by transforming the chemical structure representation of molecules into corresponding molecular holograms [25]. According to the principle of hologram, the influence of any group or atom in the drug molecule on the drug activity can be accurately determined [26], and the operation process is relatively fast, saving time.
The HQSAR method usually consists of three main steps:①Cut the molecules under study into segments of appropriate size. The distinction is determined by two parameters: Fragment size and Fragment distinction;②The encoded molecular fragments are converted into molecular holograms;③After obtaining the molecular hologram, partial least squares regression was used to establish the quantitative relationship model between the molecular hologram and the properties of the compound. In this study, the atoms that hold these fragments are encoded in different colors in the hologram to reflect their contribution to biological activity. Negative contributions, intermediate contributions, and positive contributions are shown in red, white, and green, respectively. Each fragment is defined by its unique fragment type parameters, consisting of atoms (A), chemical bond type (B), atomic connections (C), hydrogen atoms (H), chirality (CH), and participation as donor or acceptor (DA). All the characteristic fragments of each molecule are mapped into a certain length of integer string, namely molecular holography. The length of integer string is called the length parameter of molecular holography. In the HQSAR module, 12 prime numbers (53, 59, 61, 71, 83, 97, 151, 199, 257, 307,353 and 401) are provided as holographic lengths. Through the combination of different parameters, multiple HQSAR models with different prediction ability can be obtained.
Validation of the QSAR model
The LOO cross-validation method is adopted as the internal validation. The cross-validation coefficient q2 is calculated by formula (1) [27, 28]: (\(\widehat{{y_{i} }}\) and \(y_{i}\) are the predicted value and the experimental value of the test set, respectively, and \(\overline{y} \) and \(\hat{y} \) are the average activity values of the experimental value and the predicted value of the training set respectively. "n" is the number of molecules in the test set, and "i" is the ordinal number of molecules in the test set.)
After cross-validation, the conventional correlation coefficient r2 is usually used for non-cross-validation analysis, as shown in Formula (1). Meanwhile, the Standard Error of Estimate (SEE) and F value of the predicted value were obtained [26].
A good internal validation showed only a high q2 in the training set of compounds, but it did not indicate the high predictive ability of the established models, therefore external validation was essential. The predictive ability of QSAR models was validated by calculating biological activities of the compounds which were not included in the training set and used as a test set. External test set validation is an effective method to evaluate the predictive power of the corrected model. Usually, the data set is randomly divided into training set and test set. The training set establishes a correction model, and the test set is used as an independent subset to test the predictive ability of the model. The external test set compares the predicted value with the experimental value to make an accurate evaluation of the predictive ability of the model [29]. The QSAR model can be verified and evaluated by the following parameters, and the predictive ability of QSAR model can be further verified by calculating the biological activity of compounds in the test set [30, 31].
Prediction correlation coefficient \({\text{q}}_{pred}^{2}\) based on test set, formula (3):
\(\mathop \sum \nolimits_{i = 1}^{n} \left( {y_{i} - \widehat{{y_{i} }}} \right)^{2}\) is the sum of squares of the actual molecular activity of the test set and the mean molecular activity of the training set (SD); \(\mathop \sum \nolimits_{i = 1}^{n} \left( {\widehat{{y_{i} }} - y_{i} } \right)^{2}\) is the sum of squares of the deviation between the predicted value of the test set and the actual activity value (PRESS).
Root mean square error (RMSE), mean absolute error (MAE) and concordance correlation coefficient (CCC) were used to evaluate the performance of Topomer CoMFA and HQSAR regression models [31]. Formulae (4)-(6):
In order to obtain the best prediction model for the test set, additional validation is performed on the model, where \(r\) and \(r_{0}\) are regression correlation coefficients between the actual activity value and the predicted activity value, and \(k\) and \(k^{\prime}\) represent the slope of the model. The parameters are as follows [32], formula (7)–(10):
Another validation statistic parameter \(r_{m}^{2}\) and \(\Delta r_{m}^{2}\) is used to further evaluate the model [31] [36], formula (11)–(13):
Virtual screening
Virtual screening is a good method of finding leading compounds in drug research and development. It plays a great role in the identification and optimization of early drug research and development, and can effectively reduce the cost of drug research and development and improve the speed of research and development [33]. In this study, the Topomer search technology in SYBYL2.0-X was used for molecular virtual screening. Topomer Search is a ligand based virtual screening method that can be used for the optimization and skeleton transition of lead compounds, and its screening speed is faster than molecular docking and pharmacophore based virtual screening [34]. Topomer search technology can be used in combination with receptor-based molecules or as a preliminary screening tool when there is no receptor structure [35]. Its basic principle is to use the R group in the Topomer CoMFA model as the question form, search for molecular fragments with high similarity in the compound database [36] through the Topomer similarity comparison, and select the fragments with high activity contribution value. Through the reasonable combination of the obtained fragments and the basic skeleton, compounds with high biological activity were obtained [37].
In this study, compound 11 with the highest activity was used as a template molecule for cutting to obtain the corresponding molecular fragments, and then the Topomer similarity and threshold comparison score were used to predict its contribution to the activity. Then, the Topomer search technology was used to conduct virtual screening based on R group in ZINC database, and the similarity between the searched compound fragments and the database compound fragments was evaluated based on the Topomer distance. The maximum distance of Topomer is 185, and other parameters are the default values. The molecular fragments with high similarity and high contribution value are screened out in the database. Finally, the search fragments are combined with the basic skeleton reasonably to design new compounds.
Molecular docking
The act of molecular docking is a method of drug design based on the characteristics of the receptor and the interaction between the receptor and drug molecules. It mainly studies the interaction between molecules (such as ligands and receptors), and is a theoretical simulation method to predict the binding pattern and affinity of small molecular ligands and large molecular proteins through geometric and energy matching and recognition [38], which is an important technology in the field of computer-aided drug research. Surflex-dock is a semi-flexible docking with rigid protein structure. It connects ligand pairs to the binding site of the receptor based on the Protomol method. The Surflex-Dock [39] module in SYBYL2.0-X software package is selected to perform simulated docking, avoiding the tedious process of searching for active sites. This process compiles specific docking methods, extracts ligands bound at the original sites to generate docking protocols, and then connects target ligands to the previous specific sites. In addition, the conformation of the generated ligand molecule compared with the original ligand was used as a reference to determine the suitability of the docking method [40]. The docking results of the original ligand can also be compared with the potential ligand molecules to further screen drugs that are more suitable as inhibitors.
The protease crystal used in molecular docking in this paper is from PDB database [41], ID: 5TQH. In this study, 5TQH macromolecular protein was pretreated before molecular docking, the required small molecule ligands were extracted from the macromolecular complex, the unnecessary small molecule ligands and all water molecules were deleted, the protein was hydrogenated and Gasteiger-Huckel charge was added [42]. The docking region was determined by analyzing the interaction between ligands and active residues, and the active site of the docking was determined according to the small molecular ligands. The docking effect was evaluated by the values of the compound's scoring functions total-score, Crash and Polar [43]. The Score function value of total-score indicates the affinity between the small molecular ligand extracted from the large molecular protein and the receptor. The higher the value, the higher the affinity. Crash represents the imbalance between ligands and receptors extracted from macromolecular proteins, and the closer to zero the better. Polar has the score of polarity function. When the binding site is on the molecular surface, the greater the value, the better; Inside the molecule, the smaller the value, the better. In this study, we mainly used the total-Score value Score to screen the best configuration, and generally the total-score value should be greater than 5.0.
ADMET prediction
Current drug research and development is a high-risk investment, which often faces some unexpected and even catastrophic failures at different stages of drug discovery [44]. One of the main reasons for the failure of research and development is the lack of efficacy and safety, which is related to the absorption, distribution, metabolism and excretion (ADME) characteristics and various toxicity (T) of drugs in the human body. Therefore, rapid ADMET evaluation is urgently needed to reduce the failure in drug discovery [45]. ADMET (Absorption, Distribution, Metabolism, Excretion and Toxicity) is a prerequisite, and molecular properties play a key role in the prodromal clinical stage, so it is necessary to test the ADMET properties of the designed compounds in advance to ensure the drug's adaptability to the human body [46]. The attribute of ADMET obtained in this paper comes from ADMET-lab [47].
Absorption parameters
Absorption is the process of drugs entering the human circulatory system from the site of drug administration, which can be found in various epithelial cell membranes such as oral cavity, stomach and intestine.
HIA (human intellectual absorption) [48]: for oral drugs, the intestinal tract is the most important absorption site, so the absorption of oral drugs in the human intestinal tract is a necessary prerequisite for its obvious curative effect. F (30% bioavailability) [49] for any drug administered by oral route, oral bioavailability is one of the most important pharmaceutical parameters, because it is an indicator of the efficiency of drug delivery to systemic circulation.
Distribution parameters
Drug distribution is a transport process between blood and tissue. After the drug is absorbed into the blood from the drug delivery site, the circulatory system will act as a transporter to deliver the drug to its target organ, target tissue and target site.
PPB (plasma protein binding) [50]: one of the main mechanisms of drug absorption and distribution is through PPB, so the combination of drugs and plasma proteins has a great impact on its pharmacodynamic behavior. Moreover, PPB can directly affect oral bioavailability, because the free concentration of the drug is at risk when the drug binds to serum protein in the process. BBB (blood – brain barrier) [51]: blood brain barrier is an important pharmacokinetic characteristic of a drug, which means that it can or cannot penetrate the blood brain barrier. BBB penetration is important for drugs targeting brain receptors.
Metabolic parameters
Metabolism is the symbol of the life system. It enables organisms to create a feasible environment in which complex biochemical transformation can be carried out to maintain homeostasis.
CYP450 3A4, CYP450 2D6 and CYP450 2C9 [52, 53]: the metabolic system is very complex and adaptable. In this process, a large number of different enzyme families are involved, which can be divided into two categories: microsomal enzymes, such as cytochrome P450 (CYP) enzymes, which are important for most drugs, and non-microsomal enzymes, which are important for a few drugs. Therefore, the recognition of CYP450 enzyme substrates or inhibitors is very important in drug development.
Excretion parameters
Excretion is the elimination process of drugs or their metabolites in vivo. The excretion characteristics of molecules can affect drug efficiency and the corresponding drug side effects.
T1/2 (half-life) and Cl (clearance) [54]: drug clearance is an important pharmacokinetic parameter, which determines the half-life together with the distribution volume, thus determines the drug delivery frequency.
Toxicity parameters
We should not only understand the pharmacological effects, but also attach great importance to the toxic effects.
Ames (the Ames test for carcinogenicity) [55]: mutagenic effect has a close relationship with carcinogenicity. At present, the ames test is the most widely used method to test the mutagenicity of compounds. Carcinogenicity (mouse) [56]: it is an important task to determine the carcinogenicity of mammals (such as rats or mice). It is helpful for the safety test of drugs.
Results and discussion
HQSAR results and analysis
The HQSAR model can be optimized by changing parameters, including holographic length, fragment size, and fragment characteristic parameters. In this article, all 12 hologram lengths (53, 59, 61, 71, 83, 97, 151, 199, 257, 307, 353, and 401) are selected and the fragment size is set as the default (4–7). The optimal model was determined as the model with minimum standard error, and different HQSAR models were established for 30 compounds in the training set using different fragment types. The results of selecting 10 models with better data are listed in Table 2
As shown in Table 2, the best model can be obtained when the Fragment Size is set to "C/CH/DA" (1–1), with its main parameters q2 = 0.750, r2 = 0.933, N = 5, HL = 71. Then, based on the results in Table 2, set the Fragment Distinction as "C/CH/DA", and use different Fragment sizes to establish 8 HQSAR models (Table 3). The result shows the best Fragment size for the Fragment size of "7–10". Its main parameters q2 = 0.784, r2 = 0.934, PCs = 5, HL = 83. In addition, its predictive correlation coefficient \(r_{pred}^{2}\) was calculated by formula (3), and the result was 0.865. Therefore, when the Fragment distinction is "C/CH/DA" and the Fragment size is "7–10", the model established is the best HQSAR model (2–6), and the remaining parameters are shown in Table 3.
Topomer CoMFA results and analysis
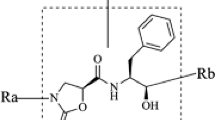
The most active compound 11 was used as the template, and all the compounds were divided according to the segmentation method shown in Table 4. The molecules were divided into three parts. The molecules that were not automatically segmented were manually cut, and the cut compounds were automatically divided into Ra(red), Rb(blue) and common skeleton (green).
Using compound 11 as template and using Topomer CoMFA method, the relationship between structure and activity of 3 pyrimidine-4-oxazolidin-2-one derivatives as mIDH1 inhibitors was studied as follows:
Generally, the larger the cross-validation coefficient q2, the larger the non-cross-validation coefficient r2 and the smaller the SEE, which means the better the correlation and the stronger the prediction ability of the model; q2 stderr and r2 stderr are the standard errors of q2 and r2. The smaller the value is, the higher the model quality is. F value is mainly used to judge whether there is a significant difference between samples. The larger the value is, the more significant the difference is and the smaller the error is (F > 100). When the cross-validation coefficient q2 is greater than 0.5 and the non-cross-validation coefficient r2 is greater than 0.6, and the difference between r2 and q2 is better than 0.3, it can be proved that the established model has a high predictive ability [26, 57].
It can be seen from Table 4 that the principal components "N" of the three models are 5, 5 and 3, respectively, q2 is 0.783, 0.667 and 0.671, respectively, which are greater than 0.5, r2 is 0.978, 0.983 and 0.927, respectively, which are greater than 0.6. The results show that the QSAR models constructed by the three cutting methods have good fitting and prediction ability. By analyzing the results of the three models, we found that Model 3–1 not only has good prediction ability, but also retains the core skeleton of the parent compound in the cutting mode, which is more conducive to the selection of core skeleton and R group. Therefore, we used Model 3–1 in the subsequent Topomer CoMFA research for structure–activity analysis.
The data show that the best model of q2 is greater than 0.5, r2 is greater than 0.6, r2—q2 < 0.3, q2pred is 0.792 (> 0.6), F is 282.660 (F > 100), which indicates that the model established by this method is an ideal Topomer CoMFA model, and its statistical results have high predictive ability. In addition, the predicted values of SEE, q2 stderr and r2 stderr are 0.138, 0.440 and 0.140, respectively, which indicates that the error of the model is small, and further indicates that the established model has high reliability.
Comparative analysis of HQSAR model and Topomer CoMFA model
The HQSAR model can discover the potential influence of debris and atoms on the activity with the help of the model color code map, thus helping to reveal the molecular mechanism affecting the activity of compounds. By analyzing the color code map, we can determine which fragments or atoms may be key contributors to the mIDH1 inhibitor activity value.
The HQSAR contribution diagram of several compounds in this paper is shown in Fig. 2, and the blue part is the common skeleton. There are many green and yellow fragments in Fig. 2a and c, which indicates that these fragments or atoms have positive contribution to biological activity. These groups should be retained when synthesizing compounds that may have better biological activity. Some of the fragments in Fig. 2b and d are orange and red, indicating that these fragments or atoms have adverse contributions to biological activity. When synthesizing compounds with better biological activity, these groups should be removed or replaced. The white atoms of other substituents in the compounds indicate that they have a neutral contribution to biological activity and may be replaced by substituents that can produce stronger inhibitory effect.
Figure 3 shows the 3D contour map of the Topomer CoMFA model of template compound 11. Figure 3a and b are the steric and electrostatic contour maps of Ra groups respectively. Figure 3c and d are, respectively, steric and electrostatic contour maps of Rb group. In Fig. 3a and b, the green part indicates that increasing the volume of substituent is beneficial to the improvement of compound activity, while the yellow part indicates that decreasing the volume of substituent is beneficial to the improvement of compound activity. In Fig. 3c and d, the red region indicates that increasing the electronegativity of groups is conducive to the improvement of compound activity, while the blue region indicates that decreasing the electronegativity of groups is conducive to the improvement of compound activity.
As shown in Figs. 3 and 4, Fig. 3a shows the steric contour map of Ra groups of template molecules. There are green groups at the positions of O-14, C-15 and F-16, indicating that the selection of large groups at these positions is beneficial to the improvement of activity; yellow groups are found inside the N-9 and O-12 positions, indicating that the volume of substituents at these positions needs to be reduced to increase the activity of the compound. For example, compared with compound 16 (pIC50 = 5.54), compound 15 (pIC50 = 7.14) increased the substituent at C-15 and F-16 positions, and the activity was significantly improved. Figure 3b shows the electrostatic contour map of the Ra group of template molecule. There are red groups at O-14, indicating that the introduction of high electronegative substituents at this position is conducive to the improvement of the activity of the compound. There are blue groups at C-10 and C-15 positions, indicating that the introduction of low electronegativity substituents at these positions is conducive to improving the activity of the compound. For example, compound 19 introduces a highly electronegative substituent at the C-15 position compared to compound 13, resulting in reduced activity.
Figure 3c shows the steric contour map of the Rb group of template molecule. There are large green groups at C-26, C-27, C-28 and Cl-30, indicating that the selection of large groups at these positions is conducive to improving the activity; There are yellow groups at the O-19, C-21, N-22 and C-25 positions, so it is necessary to introduce substituents of small groups at these positions to improve the activity of the compound. For example, compared with compound 19 (pIC50 = 5.46), compound 20 (pIC50 = 6.23) increased the volume and activity of substituents at the positions of C-26, C-27, C-28 and Cl-30. Figure 3d shows the electrostatic contour map of the template molecule Rb group. C-25 and C-26 positions have blue groups, indicating that the introduction of low negative substituents at these positions is conducive to improving the activity of the compound. There are a large number of red groups in the inner and outer positions near C-27, C-28, C-29 and O-19. Therefore, high negative substituents should be introduced at these positions to improve the activity of the compound. For example, compound 29 (pIC50 = 7.44) had a lower electronegative activity near the O-19 site than compound 31 (pIC50 = 7.80).
Compare the color code map of HQSAR model (Fig. 2) with the 3D contour map of Topomer CoMFA model (Fig. 3). In the 3D contour map of Topomer CoMFA model near C-27 (Fig. 3c), there are large green groups. In the color code map of HQSAR model, the same position of molecule 11 (pIC50 = 8.30) (Fig. 2a) shows yellow and green (favorable contribution), while the same position of molecule 7 (pIC50 = 4.95) (Fig. 2b) shows red (unfavorable contribution), which corresponds to Topomer CoMFA, In the same position of the 3D contour map of the model (Fig. 3c), the rule that the substituent of green group decreases and the volume activity decreases is given. Near C-15 of the atomic distribution number diagram, the 3D equipotential diagram of the Topomer CoMFA model (Fig. 3a) gives the suggestion of increasing the volume and enhancing the activity (green group), which corresponds to the favorable contribution (green and yellow) of compound 14 (PIC50 = 8.10) in the same part of the color code diagram of HQSAR model (Fig. 2c); In the position of C-18 and C-20, compound 12 (pIC50 = 5.56) used the unfavorable orange and favorable contribution yellow, respectively, in the HQSAR model color code diagram (Fig. 2d) to correspond to the yellow group with reduced volume activity and the red group with enhanced negative rising activity in the same position as the Topomer CoMFA model (Fig. 3c, d). Combined with the color code map of HQSAR model (Fig. 2) and the 3D contour map of Topomer CoMFA model (Fig. 3), the two methods get consistent results, which fully shows the accuracy and good prediction ability of the model established in this study (Table 5).
Based on the results of the training set, we set the Fragment Size to 7–10, and set the Fragment Size to "C/Ch/DA" to perform HQSAR validation on the training set, and predict the pIC50 value of the test set (Table 6). Finally, regression analysis and residual analysis are carried out for the obtained HQSAR model prediction data. Figure 5 is the linear regression correlation diagram and the residual analysis diagram between the experimental and predicted values of the training set and the test set of the HQSAR model and the Topomer CoMFA model. It is shown in Fig. 5 that all the samples in the linear regression analysis chart are uniformly distributed around the 45° line (Fig. 5a and c), and the data in the residue analysis chart are also mainly concentrated around the zero-scale line (Fig. 5b, d), which proves that both models have good predictive ability.
By comparing the color code map of the HQSAR model (Fig. 2) and the 3D contour map of the Topomer CoMFA model (Fig. 3), the 3D contour map of Topomer CoMFA model near C-27 has large green groups. In the color code map of HQSAR model, the molecules of compound 11 (pIC50 = 8.30) at the same position are shown in yellow and green (favorable contribution), while the molecules of compound 7 (pIC50 = 4.95) at the same position are shown in red (unfavorable contribution), at the same time corresponding to the Topomer CoMFA model of 3D contour of green groups in the same position in the reduced in volume by the substituent activity rules. The results obtained by the two methods are consistent, which fully demonstrates the accuracy and good predictive ability of the proposed model.
Verification results of QSAR model
In order to further verify the predictive power of QSAR models, independent test sets generated by non-models are needed for verification. Table 7 lists the results of the external validation parameters. r2pred is the predictive correlation coefficient of the compound test set. The external validation parameters of Topomer CoMFA and HQSAR are 0.792 and 0.865, respectively, indicating that the external prediction ability of the two models is reliable. \({\text{r}}_{0}^{2}\) and \(r_{0}^{2\prime }\) determine the square correlation coefficient of the regression line through the origin between the predicted activity value and the experimental activity value. The value of k and k´ are the slope of their models, respectively. It can be seen from the data in the table that the regression analysis values of the two models are also very good. Other parameters are shown in Table 7:
Through comparative analysis of the results of the HQSAR model and the Topomer CoMFA model (Tables 6 and 7), regression and residual analysis (Fig. 5, Table 6), the values of the two models are very close to each other, and the data of the models basically reach the standard range. This indicates that the fragment interval and fragment size in the HQSAR model established in this study and the choice of cutting mode in the Topomer CoMFA model are the best choices. Therefore, the model can be used to predict the bioactivity of these chemical types, new compounds and their derivatives more accurately.
Molecular design results
The results of HQSAR and Topomer CoMFA model were used as reference. Based on the template compound 11, the Topomer search technology was used to search the conformation in the ZINC database, and the structures of Ra and Rb were used to search and screen in the ZINC database. Among the structural fragments, the Topomer distance is close to 185 and the activity contribution value is higher than that of Ra group (contribution value is 0.97) and Rb group (contribution value is 1.37). Finally, 1 Ra group and 8 Rb groups were selected from ZINC database. In addition, Rb groups of compounds 14 and 41 in training set were extracted (contribution values were 1.41 and 1.69). 1 Ra group and 10 Rb groups were obtained to replace the Ra group and Rb group of the template compound 11 respectively for molecular design, and finally 10 new mIDH1 inhibitors were designed. According to the above method, a new molecular structure was constructed in SYBYL-X 2.0, and the molecular structure was optimized and named by the same method. The activity of compound 11 was predicted by Topomer CoMFA model. The structures and predicted activity values of the 10 newly designed compounds (pIC50) are shown in Table 8.
As shown in Table 8, the predicted activity values (pIC50) of the newly designed 10 molecules were all above the activity values (pIC50 = 8.30) of the template molecules. The structural analysis of the newly designed compound molecule showed that –OH replaced –CH2F electronegativity of template molecule Ra group, and the activity of -CH2F decreased. For Rb group, 4–06 and 4–07 replaced the -H of the substituted template molecule with the more negative -Cl at the 29 position, and the activity was enhanced; 4–03, 4–04 and 4–10 increased the substitutions near the C-26, C-27 and C-28 positions, leading to increased volume activity. The data results conform to the analysis results of the Topomer CoMFA model.
Molecular docking results analysis
Prior to molecular docking, the large proteins were pretreated by hydrogenation, charge addition, extraction of original ligands, removal of water molecules, other residues and terminal residues, etc. The molecules after treatment were shown in Fig. 6a (gray area represents prototype molecules). Then protein eutectic of macromolecular ligand to withdraw from the crystal structure, the eutectic with weight through docking technology new access in the crystal structure, and to the original ligands for reference, to verify the reliability of the docking, as shown in Fig. 6b (red bar back together ligands, green rod benchmark ligand) said. It is shown in Fig. 6b that the conformation of the crystal ligand almost completely overlaps with that of the ligand after docking (the similarity is 0.912), and their rotation trend is basically similar. This indicates that the conformation of ligand molecules before and after docking is basically unchanged, and the docking method is reasonable and reliable.
The template molecules were docked with the newly designed 10 molecules, and the template molecules were used as the control. The values of the scoring functions total-score, Crash and Polar and the number of hydrogen bonds formed were used as evaluation criteria. The higher the total score value is, the more the docking results can meet the requirements of the analysis results. The results are shown in Table 9.
The plane force analysis diagram of molecular docking is shown in Fig. 7, in which the ball stick model is a small molecule ligand, the spherical shape represents the amino acid residues that form the force, the hydrogen bonding action is shown as a green dotted line, and the hydrophobic action is shown as a pink dotted line. Hydrogen bonding is the main force to maintain protein and ligand molecules, which makes the binding between them more stable.
Figure 7a shows the docking analysis of the ligand extracted from 5TQH protein crystal. It is shown in Fig. 7a that the ligand mainly forms four hydrogen bonds with A/ILE128, A/LEU120 and A/SER278 residues in the protein crystal, and forms hydrophobic interaction with amino acid residues such as A/TRP124, A/ALA111 and A/VAL281. Total score, crash and polar were 9.4819, -1.4541 and 3.4803, respectively. It can be seen that the selection of ligands and protein crystals is more appropriate, and the docking method is reasonable and reliable.
Figure 7b presents the docking analysis diagram of compound 11 as a template molecule with 5TQH protein crystals in the training set. As shown in Fig. 7b, compound 11 mainly formed hydrogen bonding interactions with B/ILE128 residues in the crystal structure and hydrophobic interactions with amino acid residues such as B/TRP124, B/VAL281 and B/ALA111, with total score, crash, and polar of 7.1685, −1.9240, and 3.4635, respectively; Fig. 7c shows that the newly designed compound 4–02 formed a total of six hydrogen bonding interactions with residues B/LYS126, B/GLN277, B/TYR272 and B/SER278 and hydrophobic interactions with amino acid residues such as B/ILE130, B/VAL255 and B/ALA111, with a total score, crash, and polar of 7.0208, −4.0453, and 3.6820, respectively. Figure 7d shows that the newly designed compounds 4–05 formed a total of seven hydrogen bonding interactions with residues B/ALA111, B/GLN277, B/SER278, B/SER287 and B/LEU120, and formed hydrophobic interactions with amino acid residues such as B/VAL281, B/ILE130 and B/TRP124, with total score, crash, and polar of 7.1048, -2.7462, and 3.8948, respectively.
The results showed that the newly designed compounds formed strong hydrogen bonding interactions with amino acid residues such as B/LEU120, B/GLN277, B/SER287, and these interactions enhanced the binding strengths of the ligands and receptors, so the docking results of the designed compounds were reliable and beneficial.
ADMET prediction result analysis
Results as shown in Table 10, it can be seen that all the values of the designed drugs are basically good without the carcinogenicity of rats, indicating that the designed drugs have high molecular safety and obvious pharmacological effects. Moreover, all 10 molecules have mutagenicity, which is of great help to understand the carcinogenic mechanism of mIDH1.
Conclusion
In this paper, the Topomer CoMFA method of the second generation CoMFA and the fast and simple HQSAR method were used to establish QSAR models for 41 3-pyrimidin-4-yl-oxazolidin-2-one derivatives as mIDH1 inhibitors. According to the statistical verification results, the model with high predictive ability was finally obtained. Then, based on the results of Topomer CoMFA and HQSAR, some novel 3-pyrimidine-4-yl-oxazolidin-2-one derivatives mIDH1 inhibitors were designed through virtual screening, and 10 new molecules were finally determined. External validation showed that the activity values of these new molecules were all higher than the original template molecules. Then molecular docking was used to explore the binding mode and target between ligands and protein receptors, and amino acid residues were found to form hydrogen bond interaction between ligands and crystal structure. It was proved that the newly designed compound formed strong hydrogen bonds with amino acid residues B/LEU120, B/GLN277, B/SER287, etc., and these interactions enhanced the binding strength of ligands and receptors. Including AMES prediction, the results show that all 10 drug molecules are mutagenic, which is of great help to understanding the carcinogenic mechanism of mIDH1. In addition, AMES prediction was carried out, and the results showed that all 10 drug molecules are mutagenic, which is very helpful for understanding the carcinogenic mechanism of mIDH1. This research provides a certain idea for the design and development of new mIDH1 inhibitor anticancer drugs, helps to better understand its inhibitory mechanism, and provides a certain theoretical basis for future experimental verification of new compounds.
Data availability
All data generated or analyzed during this study are included in the article and supplementary materials.
References
Popovici-Muller J, Lemieux RM, Artin E et al (2018) Discovery of AG-120 (Ivosidenib): a first-in-class mutant IDH1 inhibitor for the treatment of IDH1 mutant cancers. ACS Med Chem Lett 9(4):300–305. https://doi.org/10.1021/acsmedchemlett.7b00421
Kessler J, Güttler A, Wichmann H et al (2015) IDH1R132H mutation causes a less aggressive phenotype and radiosensitizes human malignant glioma cells independent of the oxygenation status. Radiotherapy Oncol 116(3):381–387. https://doi.org/10.1016/j.radonc.2015.08.007
Molenaar RJ, Verbaan D, Lamba S et al (2014) The combination of IDH1 mutations and MGMT methylation status predicts survival in glioblastoma better than either IDH1 or MGMT alone. Neuro-Oncology 16(9):1263–1273. https://doi.org/10.1093/neuonc/nou005
Li J, Huang J, Fang H et al (2016) Decreased expression of IDH1-R132H correlates with poor survival in gastrointestinal cancer. Oncotarget 7(45):73638–73650
Xuan C, Li Q, Cong W et al (2016) Prognostic and diagnostic potential of isocitrate dehydrogenase 1 in esophageal squamous cell carcinoma. Oncotarget 7(52):86148–86160
Sun N, Chen Z, Tan F et al (2013) Isocitrate dehydrogenase 1 is a novel plasma biomarker for the diagnosis of non-small cell lung cancer. Clin Cancer Res 19(18):5136–5145. https://doi.org/10.1158/1078-0432.CCR-13-0046
Gross S, Cairns RA, Minden MD et al (2010) Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2 mutations. J Experiment Med 207(2):339–344. https://doi.org/10.1084/jem.20092506
Dang L, White DW, Gross S et al (2009) Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462(7274):739–744. https://doi.org/10.1038/nature08617
Xu W, Yang H, Liu Y et al (2011) Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell 19(1):17–30. https://doi.org/10.1016/j.ccr.2010.12.014
Parsons DW, Jones S, Zhang X et al (2008) An integrated genomic analysis of human glioblastoma multiforme. Science 321(5897):1807–1812. https://doi.org/10.1126/science.1164382
Levell JR, Caferro T, Chenail G et al (2017) Optimization of 3-Pyrimidin-4-yl-oxazolidin-2-ones as allosteric and mutant specific inhibitors of IDH1. ACS Med Chem Lett 8(2):151–156. https://doi.org/10.1021/acsmedchemlett.6b00334
Cho YS, Levell JR, Liu G et al (2017) Discovery and evaluation of clinical candidate IDH305, a brain penetrant mutant IDH1 inhibitor. ACS Med Chem Lett 8(10):1116–1121. https://doi.org/10.1021/acsmedchemlett.7b00342
Tong J-B, Luo D, Feng Y et al (2021) Structural modification of 4, 5-dihydro-[1, 2, 4] triazolo [4, 3-f] pteridine derivatives as BRD4 inhibitors using 2D/3D-QSAR and molecular docking analysis. Molecul Divers. https://doi.org/10.1007/s11030-020-10172-5
Gasteiger J, Marsili M (1980) Iterative partial equalization of orbital electronegativity—a rapid access to atomic charges. Tetrahedron 36(22):3219–3228. https://doi.org/10.1016/0040-4020(80)80168-2
Yao K, Liu P, Liu H et al (2019) 3D-QSAR, molecular docking and molecular dynamics simulations study of 3-pyrimidin-4-yl-oxazolidin-2-one derivatives to explore the structure requirements of mutant IDH1 inhibitors. J Mol Struct 1189(5):187–202. https://doi.org/10.1016/j.molstruc.2019.03.092
Verma J, Khedkar V et al (2010) 3D-QSAR in drug design - a review. CTMC 10(1):95–115. https://doi.org/10.2174/156802610790232260
JitenderMarshall RG et al (1988) Three-dimensional structure-activity relationships. Trends Pharmacol Sci 9(8):285–289. https://doi.org/10.1016/0165-6147(88)90012-0
Panwar U, Singh SK (2021) Atom-based 3D-QSAR, molecular docking, DFT, and simulation studies of acylhydrazone, hydrazine, and diazene derivatives as IN-LEDGF/p75 inhibitors. Struct Chem 32(1):337–352. https://doi.org/10.1007/s11224-020-01628-3
Tenenhaus M, Vinzi VE, Chatelin YM et al (2005) PLS path modeling. Comput Stat Data Anal 48(1):159–205. https://doi.org/10.1007/s11224-020-01628-3
Shirota Y, Luo H, Qin W et al (2002) Hepatitis C virus (HCV) NS5A binds RNA-dependent RNA Polymerase (RdRP) NS5B and modulates RNA-dependent RNA Polymerase Activity*. J Biol Chem 277(13):11149–11155. https://doi.org/10.1074/jbc.M111392200
Cramer RD, Cruz P, Stahl G et al (2009) ChemInform abstract: virtual screening for R-groups, including predicted pIC50 contributions, within large structural databases. Using Topomer CoMFA ChemInform 40(15):2180–2195. https://doi.org/10.1021/ci8001556
Ferreira MMC (2002) Multivariate QSAR. J Brazilian Chem Soc 13(6):742–753. https://doi.org/10.1590/S0103-50532002000600004
Cramer RD (2003) Topomer CoMFA: a design methodology for rapid lead optimization. J Med Chem 46(3):374–388. https://doi.org/10.1021/jm020194o
Jiao L, Zhang X, Qin Y et al (2016) Hologram QSAR study on the electrophoretic mobility of aromatic acids. Chemometrics Intell Lab Syst 157(15):202–207. https://doi.org/10.1016/j.chemolab.2016.06.020
Cheng Y, Mei Z, Tung CH et al (2010) Studies on two types of PTP1B inhibitors for the treatment of type 2 diabetes: hologram QSAR for OBA and BBB analogues. Bioorg Med Chem Lett 20(11):3329–3337. https://doi.org/10.1016/j.bmcl.2010.04.033
Waller CL (2004) A comparative QSAR study using CoMFA, HQSAR, and FRED/SKEYS paradigms for estrogen receptor binding affinities of structurally diverse compounds. J Chem Inform Comput Sci 44(2):758–765. https://doi.org/10.1021/ci0342526
Pérez-Areales FJ, Betari N, Viayna A et al (2017) Design, synthesis and multitarget biological profiling of second-generation anti-Alzheimer rhein–huprine hybrids. Future Med Chem 9(10):965–981. https://doi.org/10.4155/fmc-2017-0049
Abdizadeh R, Hadizadeh F, Abdizadeh T (2020) QSAR analysis of coumarin-based benzamides as histone deacetylase inhibitors using CoMFA, CoMSIA and HQSAR methods. J Mol Struct 1199(5):126961. https://doi.org/10.1016/j.molstruc.2019.126961
Verma J, Khedkar VM, Coutinho EC (2010) 3D-QSAR in drug design–a review. Curr Top Med Chem 10(1):95–115. https://doi.org/10.2174/156802610790232260
Busch KW, Swamidoss IM, Fakayode SO et al (2004) Determination of the enantiomeric composition of some molecules of pharmaceutical interest by chemometric analysis of the UV spectra of cyclodextrin guest–host complexes. Analytica Chimica Acta 525(1):53–62. https://doi.org/10.1016/j.aca.2004.07.066
Ojha PK, Mitra I, Das RN et al (2011) Further exploring rm2 metrics for validation of QSPR models. Chemometrics Intell Lab Syst 107(1):194–205. https://doi.org/10.1016/j.chemolab.2011.03.011
Rácz A, Bajusz D, Héberger K (2015) Consistency of QSAR models: correct split of training and test sets, ranking of models and performance parameters. SAR QSAR Environ Res 26(7):1–18. https://doi.org/10.1080/1062936X.2015.1084647
Golbraikh A, Tropsha A (2002) Beware of q2! J Mole Graphics Model 20(4):269–276. https://doi.org/10.1016/S1093-3263(01)00123-1
Walters WP, Stahl MT, Murcko MA (1998) Virtual screening—an overview. Drug Discovery Today 3(4):160–178. https://doi.org/10.1016/S1359-6446(97)01163-X
Doman TN, McGovern SL, Witherbee BJ et al (2002) Molecular docking and high-throughput screening for novel inhibitors of protein Tyrosine Phosphatase-1B. J Med Chem 45(11):2213–2221. https://doi.org/10.1021/jm010548w
Yuhong X, Jia S, Zhuoyong Z (2014) Topomer CoMFA and virtual screening studies of azaindole class renin inhibitors. Combinatorial Chem High Throughput Screen 17(5):458–472. https://doi.org/10.2174/1386207317666140107094708
Sterling T, Irwin JJ (2015) ZINC 15–Ligand discovery for everyone. J Chem Inf Model 55(11):2324–2337. https://doi.org/10.1021/acs.jcim.5b00559
Tong JB, Bai M, Zhao X (2016) 3D-QSAR and docking studies of HIV-1 protease inhibitors using R-group search and Surflex-dock. Med Chem Res 25(11):2619–2630. https://doi.org/10.1007/s00044-016-1701-0
Liu J, Yan L, Zhang HX et al (2012) Studies of H4R antagonists using 3D-QSAR, molecular docking and molecular dynamics. J Mol Model 18(3):991–1001. https://doi.org/10.1007/s00894-011-1137-x
Pearlman DA, Case DA, Caldwell JW et al (1995) AMBER, a package of computer programs for applying molecular mechanics, normal mode analysis, molecular dynamics and free energy calculations to simulate the structural and energetic properties of molecules. Comput Phys Commun 91(1):1–41. https://doi.org/10.1016/0010-4655(95)00041-D
Cleves AE, Jain AN (2015) Knowledge-guided docking: accurate prospective prediction of bound configurations of novel ligands using Surflex-Dock. J Comput Aided Mol Des 29(6):485–509. https://doi.org/10.1007/s10822-015-9846-3
Sussman JL, Lin D, Jiang J et al (1998) Protein data bank (PDB): database of three-dimensional structural information of biological macromolecules. Acta Crystallographica Sect D 54(61):1078–1084. https://doi.org/10.1107/S0907444998009378
Jain AN (2007) Surflex-Dock 2.1: Robust performance from ligand energetic modeling, ring flexibility, and knowledge-based search. J Comput Aided Mol Des 21(5):281–306
Xu C, Ren Y (2015) Molecular modeling studies of [6,6,5] Tricyclic Fused Oxazolidinones as FXa inhibitors using 3D-QSAR, Topomer CoMFA, molecular docking and molecular dynamics simulations. Bioorg Med Chem Lett 25(20):4522–4528. https://doi.org/10.1016/j.bmcl.2015.08.070
Khanna I (2012) Drug discovery in pharmaceutical industry: productivity challenges and trends. Drug Discovery Today 17(19–20):1088–1102. https://doi.org/10.1016/j.drudis.2012.05.007
Cheng F, Li W, Liu G et al (2013) In silico ADMET prediction: recent advances, current challenges and future trends. Current Topics Med Chem 13(11):1273–1289. https://doi.org/10.2174/15680266113139990033
Hodgson J (2001) ADMET—turning chemicals into drugs. Nat Biotechnol 19(8):722–726. https://doi.org/10.1038/90761
Dong J, Wang N-N, Yao Z-J, et al. (2018) ADMETlab: a platform for systematic ADMET evaluation based on a comprehensively collected ADMET database 10(1):29. http://europepmc.org/abstract/MED/29943074https://doi.org/https://doi.org/10.1186/s13321-018-0283-xhttps://europepmc.org/articles/PMC6020094https://europepmc.org/articles/PMC6020094?pdf=render.
Artursson P, Karlsson J (1991) Correlation between oral drug absorption in humans and apparent drug permeability coefficients in human intestinal epithelial (Caco-2) cells. Biochem Biophys Res Commun 175(3):880–885. https://doi.org/10.1016/0006-291X(91)91647-U
Tian S, Li Y, Wang J et al (2011) ADME evaluation in drug discovery prediction of oral bioavailability in humans based on molecular properties and structural fingerprints. J Mol Pharm 8(3):841–851
Trainor GL (2007) The importance of plasma protein binding in drug discovery. Exp Opinion Drug Discovery 2(1):51–64. https://doi.org/10.1517/17460441.2.1.51
Clark DE (2003) In silico prediction of blood–brain barrier permeation. Drug Discovery Today 8(20):927–933. https://doi.org/10.1016/S1359-6446(03)02827-7
Li J, Schneebeli ST, Bylund J et al (2011) IDSite: an accurate approach to predict P450-mediated drug metabolism. J Chem Theory Comput 7(11):3829–3845. https://doi.org/10.1021/ct200462q
Sheridan R, Korzekwa K, Torres R et al (2007) Empirical regioselectivity models for human cytochromes P450 3A4, 2D6, and 2C9. J Med Chem 50(14):3173–3184. https://doi.org/10.1021/jm0613471
Wang Y, Xing J, Xu Y et al (2015) In silico ADME/T modelling for rational drug design. Quart Rev Biophys 48(4):488–515. https://doi.org/10.1017/S0033583515000190
Aronov AM (2005) Predictive in silico modeling for hERG channel blockers. Drug Discovery Today 10(2):149–155. https://doi.org/10.1016/S1359-6446(04)03278-7
Gerald L, Kennedy et al (1986) Estimation of acute oral toxicity in rates by determination of the approximate lethal dose rather than the LD50. J Appl Toxicol 6(3):145–148. https://doi.org/10.1002/jat.2550060302
Roy PP, Leonard JT, Roy K (2008) Exploring the impact of the size of training sets for the development of predictive QSAR models. Chemom Intell Lab Sys Chemometrics Intell Lab Syst 90(1):31–42
Funding
This project was funded by the National Natural Science Foundation of China (21475081), the Natural Science Foundation of Shaanxi Province (2019JM -237), and the Graduate Innovation Fund of Shaanxi University of Science and Technology.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation and data collection were performed by Jian-Bo Tong, Shuai Bian, Xing Zhang, and Ding Luo. Data analysis was performed by Jian-Bo Tong. The first draft of the manuscript was written by Shuai Bian and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Tong, JB., Bian, S., Zhang, X. et al. QSAR analysis of 3-pyrimidin-4-yl-oxazolidin-2-one derivatives isocitrate dehydrogenase inhibitors using Topomer CoMFA and HQSAR methods. Mol Divers 26, 1017–1037 (2022). https://doi.org/10.1007/s11030-021-10222-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11030-021-10222-6