Abstract
Disordered regions of proteins are highly abundant in various biological processes, involving regulation and signaling and also in relation with cancer, cardiovascular, autoimmune diseases and neurodegenerative disorders. Hence, recognizing disordered regions in proteins is a critical task. In this paper, we presented a new feature encoding technique built from physicochemical properties of residues selected as per the chaotic structure of related protein sequence. Our feature vector has been tested with various classification algorithms on an up-to-date data set and also compared to other methods. The proposed method shows better classification performance than many methods in terms of accuracy, sensitivity and specificity. Our results suggest that the new method that links the residues and their physicochemical properties using Lyapunov exponents is highly effective in recognition of disordered regions.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Although it is generally assumed that the three-dimensional (3D) structure of a protein determines its function, it does not hold for all proteins. That is, some proteins labelled as disorder regions, intrinsic disorders, intrinsically disordered regions or intrinsically unstructured proteins function without a defined, stable 3D structure under physiological conditions (Wright and Dyson 1999; Dunker et al. 2002).
Disordered regions in proteins hold critical function in related with cell cycle regulation, transcriptional and translational regulation, modulation of protein activity, assembly of other proteins, cell signaling, DNA recognition, protein-RNA recognition, membrane fusion and transport, regulation of nerve cell function (Tompa 2002; Stoffer and Volkert 2005). In this context, for structure-based rational drug design, disordered protein regions are considered to be of paramount importance for effective therapies (Cheng et al. 2006).
It is expensive and costly to determine disordered regions using wet lab methods such as X-ray crystallography, NMR spectroscopy, near ultraviolet circular dichroism (CD), far-ultraviolet CD (Ringe and Petsko 1986; Uversky et al. 2000). However, computational simulations can be used as an alternative in studying disordered regions in proteins. Over the last decade, some intensive techniques have been developed in silico environment such as DisEMBL (Linding et al. 2003), RONN (Yang et al. 2005), DISOPRED2 (Ward et al. 2004), VSL1 and VSL2 (Peng et al. 2006). But, the estimation error generated from these techniques still relatively high. Hence, there are significant motivations for developing new methods, which minimizes the estimation error.
There are many paradoxical and diverse dynamics that cause the disordered regions in a protein. Hence, we think that chaos theory can be an effective solution to predict the disordered regions. In the literature, there is just one study used chaos theory to encode the protein data in bioinformatics. That is, Gao et al. (2005) used three pseudo amino acid components: Lyapunov index, Bessel function, Chebyshev filter values to predict protein subcellular location. Authors used one Lyapunov index that cannot give homogeneous distribution of the LEs. Hence, their method cannot give exact recognition of chaotic behavior of protein structures.
In this paper, we present a technique that uses physicochemical properties of amino acids determined with respect to chaos theory to predict the disordered protein regions. Our prediction method links between residues and physicochemical properties from the point of chaotic structure of related protein.
Methods
Physicochemical Properties of Amino Acids
The amino acids, which are represented by characters as A, R, N, D, C, Q, E, G, H, I, L, K, M, F, P, S, T, W, Y and V, are the building blocks of proteins each having different characteristics in terms of the shape, the volume, and the chemical reactivity among others. Extensive experimental and theoretical research has been performed to express and derive these characteristic physicochemical properties of amino acids. Finally all these characteristic properties are gathered in a database termed AAindex (Kawashima and Kanehisa 2000) which represents physicochemical properties by amino acid indices, each has a set of 20 numerical values. AAindex currently contains 544 such indices.
Dataset
A total of 369 protein sequences, with pairwise sequence identity ≤25 %, were used. Structures with a resolution above 2 Å and an R-factor above 20 % are excluded. A residue is considered as disordered if it is present in the SEQRES but not in the ATOM field of the PDB file (Shimizu et al. 2007; Deiana and Giansanti 2010). These proteins includes disordered regions were extracted from two database: Disprot database (Vucetic et al. 2005) and Protein Data Bank (PDB) (Linding et al. 2003). In Disprot data, there are 96 proteins, minimum length of protein sequence is 49 residues and maximum length is 1861 residues. In PDB data, there are 273 proteins, minimum length is 43 residues and maximum length is 926 residues.
Lyapunov Exponents
LE is a quantitative measure for the divergence of nearby trajectories, the path that a signal vector follows through the phase space. The rate of divergence can be different for different orientations of the phase space. Thus, there is a whole spectrum of LEs—the number of them is equal to the number of dimension of the phase space. A positive exponent means that the trajectories are initially close to each other (divergence). The magnitude of a positive exponent determines the rate as to how rapidly they move apart. Similarly, for negative exponents, the trajectories move closer to each other (convergence) (Kennel et al. 1992).
The LE is calculated for each dimension for the phase space as
In Eq. 1, s(n) is the reference point and s(m) is the nearest neighbor of s(n) on a nearby trajectory. d(s(n), s(m)) is the initial distance between the nearest neighbors. d(s(n + 1),s(m + 1)) is the distance between s(n + 1) and s(m + 1) which are the next pair of neighbors on their trajectories (Abarbanel 1996).
Calculation of Lyapunov Exponents for a Protein Sequence
Whether a protein sequence has a chaotic structure according to a physicochemical property, numerical vectors that are built from physicochemical properties were constituted initially. Accordingly, for each residue, corresponding amino acid indices of every physicochemical properties were replaced from AAindex. For example, for a given AYCCEDEAKYYH protein, firstly, protein sequence was encoded using physicochemical-1 (alpha-CH chemical shifts) as follows;
Then, z-score normalization that is transformation of observed data to a mean of zero and a standard deviation of one (Hamann and Herzfeld 1991) was applied to pc1:
In next step, 10-D phase-space vectors was constructed from pc1z,
In our example;
Finally, nearest neighbors were accounted to use in Eq. 1 that gives LEs. These phase-space vectors were given to Tisean Package Program (Hegger et al. 1999) to calculate Lyapunov spectra, LEs from index 1,2,..,10 for each protein sequences.
Classification Algorithms
We used four classification algorithms: Bayesian network, Naïve Bayes, k-means and Sigmoid support vector machines (SVM). Bayesian networks are directed acyclic graphs that allow efficient and effective representation of the joint probability distribution over a set of random variables. Each vertex in the graph represents a random variable, and edges represent direct correlations between the variables. More precisely, the network encodes the following conditional independence statements: each variable is independent of its non-descendants in the graph given the state of its parents. These independencies are then exploited to reduce the number of parameters needed to characterize a probability distribution, and to efficiently compute posterior probabilities given evidence (Friedman et al. 1997).
Naïve Bayes is the simplest form of Bayesian network, in which all attributes are independent given the value of the class variable. This is called conditional independence. It is obvious that the conditional independence assumption is rarely true in most real-world applications. All Naïve Bayes classifiers assume that the value of a particular feature is independent of the value of any other feature, given the class variable (Zhang 2005).
K-means clustering is a method commonly used to automatically partition a data set into k groups. It proceeds by selecting k initial cluster centers and then iteratively refining them. Firstly, each instance is assigned to its closest cluster center. Secondly, each cluster center is updated to be the mean of its constituent instances. The algorithm converges when there is no further change in assignment of instances to clusters (Wagstaff et al. 2001). Prediction of disordered regions in protein is a binary classification problem. Hence, number of clusters (k) should be two for this problem.
SVM aims to find the maximum margin hyperplane to separate two classes of samples. Training vectors, x i , are mapped into to a higher dimensional space that allows a linear separation of classes which could not be linearly separated in the original space by the function \(\phi\). Furthermore, \(K(xi,xj) \equiv \phi (xi)\phi (xj)\) is called the kernel function. Though new kernels are being proposed by researchers, there are four basic kernels: linear, polynomial, radial bases and sigmoid (Hsu et al. 2003).
Proposed Feature Encoding Method
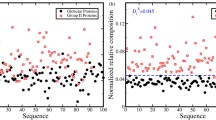
In this paper, we assigned physicochemical properties that ensure chaotic structure for the disordered protein sequences. In this manner, 6 physicochemical properties were obtained by investigating the phase space of protein sequences encoded with respect to each physicochemical property’s index values from AAindex. Working through, first of all, 10 LEs for each protein were obtained for 544 physicochemical properties as in (Kennel et al. 1992). In Table 1, LEs results for an example of a protein sequence of 3848 length are shown.
Number of positive and negative exponents can be enough to characterize the complete distribution. According to empirical results shown in Table 1, there are an equal distribution on positive and negative exponents, that is, five positive and negative LEs. This means that this protein has a chaotic structure for the physicochemical property tested and this physicochemical property’s index values can be used as for the feature vector to classify disordered residues. In this way, six physicochemical properties that shows chaotic structure for every protein in the dataset were determined shown in Table 2.
Ultimately, residues were encoded according to selected six-physicochemical properties as depicted in Fig. 1. Hence, each residue was encoded with 6 numerical index values further normalized using the z-score normalization method.
The proposed feature encoding technique were analyzed by various classification algorithms to generate the performance scores and then compared with the some effective methods.
Experimental Results
Tests were conducted on disordered region data sets assembled from up-to-date Disprot and PDB protein databases. tenfold cross validation (tenfold CV) testing protocol was used to evaluate the performance of proposed feature encoding method. In tenfold CV, the encoding scheme methods are trained using 90 % of the data and the remaining 10 % of the data are used for testing of the methods. This process is repeated 10 times so that each peptide in the data set is used once. Then the average performance score of each method over these10 turns are obtained.
The classification performance was assessed by the following scores:
TP, number of true positives (disordered residues that were classified correctly), FP, number of false positives (ordered residues that were classified as disordered), TN, number of true negatives (ordered residues that were classified correctly), and FN, number of false negatives (disordered residues that were classified as ordered) are derived from confusion matrix.
The performance of proposed feature encoding method with various classifier algorithms on the data set is shown in Table 3 in terms of accuracy, sensitivity, specificity and F-measure. In general, the results demonstrate that our method perform well in identifying disordered residues with the classifier algorithms.
Table 3 reports that Naïve Bayes algorithms obtained the best accuracy (75.2 %) and specificity (95.5 %) but worst sensitivity (0.06 %) scores. However, BayesNet show the best sensitivity (38.5 %) and F-measure (0.342) performance compared to other classifiers. Table 4 points out performance comparison with previously tested methods on Disprot and PDB data sets.
According to the results, VSL1 and VSL2 used sliding window technique in the protein sequences to build feature vectors outperform the other methods in terms of all performance metrics. While both VSL1 and VSL2 methods seek correlation from sequential windows, our method (LEs encoding) seeks correlation among phase vectors (disjoint windows) in the phase space. LEs encoding with Naïve Bayes classifier accuracy result, 75 %, is better than DisEMBL, RONN and DISOPRED2 methods. However, Naïve Bayes classifier’s sensitivity performance, 0.06 %, is the worst performance and specificity performance, 95.5 %, is the best performance compared to other methods shown in Table 4. It is revealed that although LEs encoding with Naïve Bayes classifier could not recognize the disordered regions, it discriminates effectively ordered regions in the protein sequences. Also, LEs encoding with BayesNet classifier’s sensitivity result, 38.5 %, is better than DisEMBL and RONN methods. It is brought out that our feature encoding method used 10 LEs for the selected 6-physicochemical properties make contribution untangling the underlying mechanism of disordered regions in proteins via chaotic approach.
Conclusion
Our feature encoding method based on LEs and physicochemical properties of amino acids is easy to implement and it shows better performance compared to some hitherto methods. Our encoding method that characterizes relationship among residues from the point of chaos theory can be used with other machine learning methods (e.g. ensemble of classifiers) to obtain higher performance and can be applied to any kind of protein sequence-based classification problems as well. This fact can initiate subsequent new studies for future researches. Ultimately, the development of robust feature encoding methods based on chaos theory and physicochemical properties of amino acids will open the door to more useful predictions.
References
Abarbanel HD (1996) Analysis of observed chaotic data, 1st edn. Springer, New York
Cheng Y, LeGall T, Oldfield CJ et al (2006) Rational drug design via intrinsically disordered protein. Trends Biotechnol 24(10):435–442
Deiana A, Giansanti A (2010) Predictors of natively unfolded proteins: unanimous consensus score to detect a twilight zone between order and disorder in generic datasets. BMC Bioinformatics 11(1):198
Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, Obradovic Z (2002) Intrinsic disorder and protein function. Biochemistry 41(21):6573–6582
Friedman N, Geiger D, Goldszmidt M (1997) Bayesian network classifiers. Mach learn 29(2–3):131–163
Gao Y, Shao S, Xiao X et al (2005) Using pseudo amino acid composition to predict protein subcellular location: approached with Lyapunov index, Bessel function, and Chebyshev filter. Amino Acids 28(4):373–376
Hamann IM, Herzfeld UC (1991) On the effects of preanalysis standardization. J Geol 99(4):621–631
Hegger R, Kantz H, Schreiber T (1999) Practical implementation of nonlinear time series methods: the TISEAN package. CHAOS 9:413
Hsu CW, Chang CC, Lin CJ (2003) A practical guide to support vector classification. http://www.csie.ntu.edu.tw/~Ecjlin/papers/guide/guide.pdf
Kawashima S, Kanehisa M (2000) AAindex: amino acid index database. Nucleic Acids Res 28(1):374
Kennel MB, Brown R, Abarbanel HD (1992) Determining embedding dimension for phase-space reconstruction using a geometrical construction. Phys Rev A 45(6):3403
Linding R, Jensen LJ, Diella F et al (2003) Protein disorder prediction: implications for structural proteomics. Structure 11(11):1453–1459
Peng K, Radivojac P, Vucetic S et al (2006) Length-dependent prediction of protein intrinsic disorder. BMC Bioinformatics 7(1):208
Ringe D, Petsko GA (1986) Study of protein dynamics by X-ray diffraction. Methods Enzymol 131:389–433
Shimizu K, Muraoka Y, Hirose S et al (2007) Predicting mostly disordered proteins by using structure-unknown protein data. BMC Bioinformatics 8(1):78
Stoffer DA, Volkert LG (2005) A neural network for predicting protein disorder using amino acid hydropathy values, computational intelligence. In: Proceedings of the 2005 IEEE symposium on in bioinformatics and computational biology, CIBCB’05, 1–8.
Tompa P (2002) Intrinsically unstructured proteins. Trends Biochem Sci 27(10):527–533
Uversky VN, Gillespie JR, Fink AL (2000) Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins 41(3):415–427
Vucetic S, Obradovic Z, Vacic V et al (2005) DisProt: a database of protein disorder. Bioinformatics, 21(1), 137–140
Wagstaff K, Cardie C, Rogers S, Schrödl S (2001) Constrained k-means clustering with background knowledge. In ICML, vol 1. pp. 577–584
Ward JJ, Sodhi JS, McGuffin LJ et al (2004) Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol 337(3):635–645
Wright PE, Dyson HJ (1999) Intrinsically unstructured proteins: reassessing the protein structure-function paradigm. J Mol Biol 293(2):321–331
Yang ZR, Thomson R, McNeil P, Esnouf RM (2005) RONN: the bio-basis function neural network technique applied to the detection of natively disordered regions in proteins. Bioinformatics 21(16):3369–3376
Zhang H (2005) Exploring conditions for the optimality of naive Bayes. Int J Pattern Recognit Artif Intell 19(02):183–198
Acknowledgments
This study was supported by Yalova University M.Sc. Project (Grant 2014/YL/037).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
Murat Gök, Osman Hilmi Koçal and Sevdanur Genç declare that they have no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human participants or animals performed by any of the authors.
Rights and permissions
About this article
Cite this article
Gök, M., Koçal, O.H. & Genç, S. Prediction of Disordered Regions in Proteins Using Physicochemical Properties of Amino Acids. Int J Pept Res Ther 22, 31–36 (2016). https://doi.org/10.1007/s10989-015-9481-9
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10989-015-9481-9