Abstract
The p38 protein kinase is a serine–threonine mitogen activated protein kinase, which plays an important role in inflammation and arthritis. A combined study of 3D-QSAR and molecular docking has been undertaken to explore the structural insights of pyrazolyl urea p38 kinase inhibitors. The 3D-QSAR studies involved comparative molecular field analysis (CoMFA) and comparative molecular similarity indices (CoMSIA). The best CoMFA model was derived from the atom fit alignment with a cross-validated r 2 (q 2) value of 0.516 and conventional r 2 of 0.950, while the best CoMSIA model yielded a q 2 of 0.455 and r 2 of 0.979 (39 molecules in training set, 9 molecules in test set). The CoMFA and CoMSIA contour maps generated from these models provided inklings about the influence of interactive molecular fields in the space on the activity. GOLD, Sybyl (FlexX) and AutoDock docking protocols were exercised to explore the protein–inhibitor interactions. The integration of 3D-QSAR and molecular docking has proffered essential structural features of pyrazolyl urea inhibitors and also strategies to design new potent analogues with enhanced activity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Mitogen activated protein kinases (MAP kinase) play crucial roles in the signal transduction pathways and are activated by various extracellular signals triggered by growth factors, physicochemical stress and cytokines [1, 2]. Four major cascades of MAP kinases have been reported namely, extracellular signal-regulated kinase (ERK), c-Jun N-terminal kinase or stress activated protein kinase (JNK/SAPK), ERK5 or big MAP kinase (BMK1) and p38 group of MAP kinases [3, 4]. The activation of MAP kinases involves dual phosphorylation of threonine and tyrosine residues in TXY motif that are located in the regulatory loop [5, 6].
Four isoforms of p38 kinase namely p38α (or p38), p38β, p38γ and p38δ, which differ in their tissue expression have been identified [7–10]. These homologues exhibit 40–70% sequence similarity among themselves and a common feature of presence of 12 amino acids in the activation loop, which comprises TGY motif [11]. A plethora of downstream substrates have been identified for p38 MAP kinase including downstream protein kinases (MAPKAPK-2 and PRAK), and several transcription factors (ATF-1 and 2, Elk-1, CREB, MEF2C and CHOP) [12, 13]. These substrates are in turn responsible for the biosynthesis of inflammatory proteins. Thus, p38 MAP kinase plays an important role in the biosynthesis of inflammatory proteins such as IL-1, IL-6, IL-8, TNF-α, and GM-CSF [14, 15].
The discovery of triaryl imidazole inhibitors (such as SB203580) having p38 kinase inhibitory activity as novel anti-inflammatory agents, has triggered the medicinal chemists with a relentless quest to search for selective p38 kinase inhibitors for the treatment of inflammatory diseases [16]. The reported structure activity relationship of archetypal pyridinylimidazole derivatives has revealed that pyridine ring at the 5th position is crucial for the activity, which may be attributed to the formation of hydrogen bond with Met109 in the ATP binding site [17–19]. Although, the representatives of pyridinylimidazole inhibitors have demonstrated encouraging in vitro p38 kinase inhibitory profile, they have suffered severe set back in clinical trials as they were found to inhibit the hepatic cytochrome P450 enzymes [20]. A number of new lead molecules have been identified that are distinct from pyridinyl/pyrimidinylimidazoles, viz diamides, benzophenones, indole amides, N-pyrazolyl ureas, dihydroquinazolinones, pyrido[3,2-d]pyrimidinones, pyridones and pyridylamino-quinazolines [21–23].
In the present study, we have selected a set of N-pyrazolyl urea derivatives that have been reported as potent and selective p38 kinase inhibitors in biochemical and cellular assays [24, 25]. These pyrazolyl urea inhibitors are structurally distinct from the traditional pyridinyl/pyrimidinyl imidazole inhibitors and have also been demonstrated the p38 kinase inhibition by a different mechanism. The X-ray crystallographic analysis (co-crystallized with BIRB 796, a potent pyrazolyl urea in phase II clinical trials) has revealed that the pyrazolyl urea inhibitors bind to the allosteric site rather than ATP binding site by inducing a large conformational change in the Asp168-Phe169-Gly170 (DFG) motif [26, 27]. Side effects that are caused by lack of target selectivity is found to be a major problem for many drugs, particularly those that target the protein kinases. In this regard the advantage of pyrazolyl urea inhibitors is that they target the unique allosteric site, as a result of which BIRB 796 has shown higher selectivity for p38 kinase over 11 other protein kinases [27].
Initially, 3D-QSAR studies were carried on reported N-pyrazolyl urea p38 kinase inhibitors (Table 1) in order to probe the biological affinity and to gain insight into the structure activity relationship. Further, to ascertain the binding interactions and conformations, all the molecules were docked in to the active site of p38 kinase. In this paper, we report the best predictive 3D-QSAR models derived by employing CoMFA [28] and CoMSIA [29, 30] techniques along with the overall binding interactions of N-pyrazolyl urea inhibitors with p38 kinase by using GOLD, Sybyl (FlexX) and AutoDock protocols. Based on the molecular field information of 3D-QSAR tools and molecular docking protocols, a few strategies were proposed to design new molecules with improved activity.
Materials and methods
Dataset and molecular modeling
A series of 48 pyrazolyl urea analogues have been selected from the literature to derive CoMFA and CoMSIA models, the structures and their p38 kinase inhibitory activities are listed in Table 1 [24, 25]. The activities of the dataset were distributed with in a range of 11–800 nM (approx. 70-fold). The inhibitors were randomly sorted out into training and test set containing 39 and 9 molecules, respectively. The p38 MAP kinase inhibitory activity (IC50) was expressed in nano molar and converted into pIC50 using the formula pIC50 = −log IC50.
All the molecular modeling studies were performed using Sybyl 6.9 version [31]. The crystal structures of p38 kinase with two reported inhibitors (1KV1 with I and 1KV2 with II, BIRB 796) were collected from Brookhaven Protein Databank (PDB). The conformation of the BIRB 796 (Scheme 1) was considered to model the most active molecule 28. Then the modeled structure was minimized using Tripos force field with distance dependent dielectric function and applying Gasteiger-Hückel partial atomic charges. Rests of the molecules were constructed using 28 as a template and minimized similarly.
Alignment
The crux of the 3D QSAR is the alignment of the inhibitors that defines the presumed pharmacophore of the series under the study. Three different alignment procedures were employed to test the sensitivity of CoMFA results. Among the dataset the common scaffold that depicted in Scheme 2 is used for the alignment.
The various alignment protocols that are employed in the study
Alignment 1. RMS fit alignment (atom fit) in which heavy atoms of the compound 28 were selected to superimpose rest of the molecules by minimizing the RMS distance (model I).
Alignment 2. Flexible alignment (multi fit), all the molecules were aligned with template by applying force (force constant 20 kcal/mol) and subsequent energy minimization (model II).
Alignment 3. Sybyl database alignment (model III).
For docking studies, between the two structures, KV1 and KV2, as the structural differences that are induced by the ligands are mostly trivial we have taken KV2 as a receptor. To the downloaded structure all the hydrogens were added after deleting the water and that was followed the Kollman_all partial atomic charges assigning. Using the Tripos force field minimization was performed up to 100 iterations. Then the active site was defined by considering all the important active site residues and further docking parameters were applied as mentioned in the methodology.
Results and discussion
CoMFA results
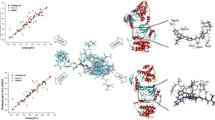
Three CoMFA models were built based on three different alignment protocols and the results are summarized in Table 2. CoMFA model derived from multifit alignment (model II) has resulted in a cross-validated q 2 value of 0.486 with six components, non-cross validated r 2 value of 0.948 and standard error of estimation (SEE) of 0.106. The steric and electrostatic contributions were found to be 83.5% and 16.5%, respectively. Comparatively, better correlation has been observed with database alignment (model III), which has shown a q 2 value of 0.536 with six components, r 2 value of 0.950 and SEE of 0.109 that was supported by 86% and 14% steric and electrostatic contributions, respectively. But the best CoMFA model was obtained from the atom fit alignment (model I) with q 2 value of 0.516 with six components, r 2 value of 0.950 and SEE of 0.108. The steric and electrostatic contributions were 84.5% and 15.5%, respectively. Figure 1 portrays the atom fit alignment of dataset molecules. Model I has been considered for further analysis due to its better r 2pred . Group cross validation was performed for 50 runs to substantiate the model and resulted in a mean group cross-validated r 2 was 0.513. Bootstrapping analysis for 100 runs was resulted with an r 2bs of 0.975 and a deviation of 0.011 (Table 3).
CoMSIA results
The atom fit model of CoMFA was further used to develop CoMSIA models and nine different models with varying CoMSIA field combination were generated. The results of all different contributions are listed in Table 4. A model with the combination of steric, hydrophobic and hydrogen bond acceptor fields has exhibited q 2 and r 2 value of 0.455 and 0.979, respectively with considerably good F-value, 179.72 from seven components. The steric, hydrophobic and hydrogen bond acceptor field contributions were 34.9, 31.4 and 33.7, respectively. This model was further subjected to group cross validation for 50 runs and correlation of 0.431 was obtained. The robustness of this model was ascertained by boot strapping for 100 runs as summarized in Table 3. The activity predicted by CoMFA and CoMSIA for both training and test sets are listed in Tables 5 and 6 and the graphs of experimental activity against the CoMFA and CoMSIA predicted activities of both training and test sets are presented in Fig. 2.
Docking results
The inhibitor (I) in the crystal structure 1KV1 (source: human) occupies new allosteric binding pocket, which is different from ATP binding pocket. Furthermore, the inhibitor orients ∼60° to ATP and no overlap has been observed between the inhibitor and ATP. It has also been reported that for appropriate binding interactions a large conformational change is required in the conserved region of the active site (Asp168-Phe169-Gly170) residues. To facilitate the required interactions, the side chain of Phe169 moves away by ∼10 Å to new position (DFG out conformation) so that one face of Phe169 side chain shields the inhibitor while other side is exposed to the solvent. This conformational change in Phe169 creates a large hydrophobic pocket that accommodates the t-butyl pyrazole moiety of I [27]. With regards to hydrogen bond interactions, Glu71 forms two hydrogen bonds with urea hydrogens and Asp168 forms one hydrogen bond with urea oxygen. On the other hand, inhibitor II (BIRB 796) containing a tolyl group on pyrazole ring has demonstrated hydrophobic interactions with the side chain of Glu71 in helix αC. A slight change has been noticed in the conformation of the Glu71 which leads to form a single hydrogen bond between Glu71 and urea hydrogen and this new conformation of Glu71 is favorable for the hydrophobic interaction with the tolyl group and thus contributing for the activity [27]. In addition to above interactions, another hydrogen bond between morpholine oxygen and backbone NH of Met109 also enhances the activity.
Three docking protocols, GOLD, Sybyl (FlexX) and AutoDock, are employed in order to get an overall picture of binding conformations and inhibitor–protein interactions. After careful examination of these three results, a complete overview of receptor–inhibitor binding interactions is presented herewith. For better understanding of the inhibitor structure, it is partitioned as portrayed in the Scheme 3.
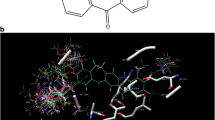
The urea group (U) was observed lying in allosteric pocket and encircled by Asp168 on one side, side chain of Leu167 on top, side chain of Leu75 on rear, side chains of Glu71 and Lys53 from the underside. As expected, the Glu71 and Asp168 residues were involved in hydrogen bond formation with urea hydrogen and oxygen atoms, respectively. The t-butyl pyrazolyl moiety (P1) was positioned in a lipophilic pocket where the pyrazolyl ring stacked between His148 ring and side chain of Leu75 and t-butyl group pierced deeply in a large lipophilic pocket created by Leu74, Val83, Ile84, Ile141, Ile146, Met78 and Ile166. The urea phenyl ring (P2) was stacked between Ile84, Val38, Ala51 and Lys53, and also exhibiting T-shape π–π interaction with Phe169 (∼5.4 Å) as depicted in Fig. 3. The substituents of N1-pyrazolyl ring (P1S) were exposed to solvent region. The substituents on phenyl ring were observed to force the phenyl ring to orient in such a fashion that it could display cation–π interaction with side chain NH +3 of Lys53 with a distance ranging between 6–7 Å in addition to the lipophilic interactions with side chain of Glu71. Also, there was a possibility for meta and para groups to form hydrogen bonds with near by residues, Lys53, Arg67 and Glu71. The residues Leu167, Lys165, Ala157, Met109, Thr106 and Val38 were found around the P2S substituents.
Docking interactions of 28 (green) with active site residues and for the purpose of comparison the X-ray structure of BIRB 796 (red) is also shown. Protein tertiary structure is displayed in magenta color helices, yellow color β-sheets and cyan color coils. The ball & stick models represent the important active site residues whereas other active site residues are shown in lines. The non-bonded interactions are displayed in dotted lines
CoMFA and CoMSIA contour maps
Figure 4 depicts the CoMFA steric and electrostatic maps. In CoMFA contour maps, three green steric favorable regions have been observed. A green contour is found around meta/para position of aryl group of the P1S, a big green contour is on the brink of P2S and another contour is seen next to the ortho position of P2. Four sterically unfavorable yellow regions are noticed, a yellow contour map is located near P1S, a couple of yellow contours is close to the meta position of P2 and the another yellow contour in proximity to P2S are spotted. A red and a blue contour are located near P2S and another blue contour encircling P1S is also seen.
In CoMSIA, only one big green contour near P2S and a big yellow contour above P2S are found. Out of two orange contours, one is found near U and ortho position of P2 and another one is noticed at L. A total of three white regions are found and these three regions are distributed along these lines, one big white region near P1S and other two on either side of P2 meta position. Only one magenta contour near P2S and two cyan contours around P1S are stippled. All these CoMSIA maps have shown in Fig. 5a and b.
(a) CoMSIA steric and hydrophobic fields wherein the orange color represents for favorable hydrophobic region and white color represents for unfavorable region. (b) CoMSIA hydrogen bond acceptor contour map where magenta color represents favorable hydrogen bond acceptor and cyan color represents the unfavorable acceptor region
Analysis
The presence of green, blue, white and cyan regions suggests steric, electropositive favorable, less hydrophobic (hydrophilic) and hydrogen bond acceptor unfavorable fields around P1S region. Strong evidence for this speculation can be gained concomitantly from docking studies wherein, the orientation of P1S substituents towards the solvent region support the hydrophilic environment and presence of Lys53, Arg67 and Glu71 residues that render steric as well as hydrogen-bonding interactions. The aryl ring in P1S region is reported to exhibit π–CH2 interaction with side chain of Glu71 and stabilizes its orientation [24]. In addition to this we have noticed another interaction with Lys53 where the side chain NH +3 demonstrated a cation–π interaction with the phenyl ring (6–7 Å). Within the available SAR, compounds 27–30 have exhibited potent activity, compounds 26, 35, 40–46 and 48, showed considerably good activity, whereas rest of the molecules, 31–34, 36–39 and 47 have displayed reduced activity. The inhibitor 27 containing phenyl ring at P1S position found occupying this pocket and making lipophilic π–CH2 interaction with Glu71. When compared to inhibitor 35 (H) and 26 (CH3), 27 has shown an augmented activity perhaps due to increased lipophilic interactions with Glu71 of the kinase. Likewise, substituents of meta and para position of phenyl ring have shown wide-ranging activity because of varying interactions with the nearby residues. The presence of NO2 at meta position as in 28 has drastically increased the activity where the NO2 group involved in hydrogen bonding interactions with side chain NH +3 of Lys53 (2.2–2.7 Å). The presence of same NO2 group at para position (43) resulted in decreased activity due to the absence of these hydrogen-bonding interactions. The same analysis could be extended to molecules 29 and 30 and also for 40 and 42. In compound 42, the oxygen atom of –OCH3 is involved in hydrogen bond formation (2.5 Å) with NH +3 of Lys53 while such interaction is absent in 40 because the –OCH3 group is in para position. To perceive the contribution of these molecules towards 3D maps, quiet a few models were developed by excluding the above-discussed molecules. One CoMSIA model that was built by ignoring 28, 29, 33 and 44 resulted in contour plots without the white contour (hydrophilic favorable) near P1S. Closer look at the binding orientations of these molecules have revealed that the meta and para substituents of the phenyl ring are oriented in a less hydrophobic (hydrophilic) environment. At the same time, the compounds (31, 36–38) with polar substituents have exhibited comparatively lower activities. Thus we can infer from this analysis that the P1S pocket possesses two different types of environments, liphophilic and hydrophilic and only those groups which utilize these two regions effectively add to the activity and results in potent inhibitors. Amongst the dataset, only ring substituted (at P1S) compounds appear to satisfy this criterion where the phenyl group comfortably occupied the pocket and as a result the meta and para groups could effectively interact with hydrophilic part of Lys53 and Arg67, respectively. On the other hand, inhibitors with flexible aliphatic chains probably do not utilize this pocket very effectively thus leading to decrease in the potency. The phenyl ring of 48 containing a methyl substitution at ortho position is oriented differently from that of 28.
The P2 part of the inhibitor reflects the need of less bulky flat aromatic ring systems. The active site analysis of corresponding P2 part has revealed a big hydrophobic pocket where Phe169 surrounds one side of the P2. Yellow contour at 5th position of P2 indicates unfavorable steric interactions and the one compound (32) with 5-bromo substituent has been found to oriented close to Phe169 and thus leading to unfavorable steric interactions and poor activity. Moreover, the docking interactions of this molecule indicated that the U part was not oriented as found in other unsubstituted molecules. The change in the orientation of U led to formation of a hydrogen bond between urea hydrogen and backbone carbonyl group of Asp168 leading to an increase in the distance between Glu71 side chain and urea hydrogen by 3.9 Å and subsequent loss in the usual hydrogen bond between them. Another sterically unfavorable yellow contour has been located closer to 3rd position of P2 aromatic ring. Although this P2 part was positioned in a big hydrophobic pocket, bulky groups in this region still extend unfavorable steric interaction with the protein. For instance, a phenyl group substituted (biphenyl type) at this position as in 34 has led to disorientation and resulted in exposing this phenyl group in to a polar region of the protein and also influencing other binding interactions. To ascertain these features by knowing the size and shape of the active site, the protein has been subjected for the site ID and molcad analysis in Sybyl as displayed in Fig. 6. The surface analysis of this part reveals that the active site is became narrow at the corresponding P2 region and gradually widened into the equivalent P2S region. Hence, the biphenyl group of 34 is experienced unfavorable interactions in P2 region and turned out as the least active molecule. However, the crystal structure of BIRB 796 indicates that the flat ‘naphthyl’ group in P2 region was placed appropriately in this pocket and exhibited potent activity.
The P1S and P2 show distinct maps with regard to the substituents, whereas P2S has wide-ranging flexible substituents with multiple conformational poses. Accordingly, four types of the CoMFA contour maps, green, yellow, blue and red have resulted in P2S region and similarly different types of CoMSIA maps have resulted. With such type of extensive map information, it is not undemanding to classify these maps with respect to groups and so the collective analysis of these maps has been presented here with. The presence of steric and electrostatic maps rendered some indications in support of the presence of residues, Leu167, Lys165, Ala157, Met109, Thr106 and Val38. The non-polar part of molecules displays steric interactions with side chains of these amino acid residues. Groups like, anilides (5, 13 and 17) and carbamates (8 and 9) with bulky alkyl side chains have demonstrated a better activity and this may be attributed to the increase of electronegative potential of the ‘–NHCO–’ group. Increase in the activity with respect to the alkyl side chain has been noticed in 4, 5 and 6 with oxygen linker and 15, 16, 17 and 13 with CH2 linker. Although, the P2S groups interact effectively with the active site, as the P2S pocket found comparatively small and less conformational flexibility, the orientation of long chain P2S groups (3, 18) shoving U and P1 moieties from their original place led to the distortion of the groups resulting in the loss of hydrogen bond interactions.
An orange contour in the CoMSIA model at the linker region indicates favorable for hydrophobic interactions. Among the molecules with different linkers (X = CH2, S, O, NH and extended groups), the molecules with sulfur exhibited greater potency indicating favorable lipophilic contribution by this linker. In addition, the sulfur linker is probably facilitating the influential π–π interactions between P2 phenyl ring and aromatic side chain of Phe169. In inhibitor 19 the phenyl ring of P2 is displaced (5.29 Å) away from the side chain of Phe169 when compared to that seen in inhibitor 20 (4.85 Å) so that the π–π interaction was prominent in 20. A few molecules with extended linker (21 and 22) have illustrated hydrogen bonds with Met109 and Gly110.
The foremost aspiration of any QSAR and docking studies is to suggest new strategies toward design inhibitors with improved activity. In the present data series none of the inhibitors were able to optimally utilize both P1S and P2S. Thus, it occurred to us that the inhibitors, which are able to access both the pockets efficiently might result in enhanced activity. Hence, we propose that new hybrid inhibitors can be designed which may access both the pockets efficiently. A cursory look at the crystal structure (1KV2) reveals that the inhibitor’s (BIRB 796) pendant tolyl group is positioned in P1S while the ethoxy morpholine is situated in the P2S pocket. Hence, the P1S aryl rings have to be preserved and few modifications can be done in the P2. For example, the 4th position of P2 can be substituted with bulky groups that could creep into the distant lipophilic pocket. Docking studies on the molecule 20 indicate that the thiopyridine ring was found to access the corresponding pocket well. The P1S aryl ring can be consider for further substitution at meta position with NO2 or NH2 groups in order that they may exhibit hydrogen bonds with Lys53. Also, there is a possibility that the thiopyridine ring may enable nitro or amino group of P1S to exhibit hydrogen bond interactions with Lys53.
Methodology
CoMFA and CoMSIA field calculations
In CoMFA, a 3D cubic lattice was defined automatically by extending at least 4 Å beyond all the investigated molecules in all the three axes (X, Y, Z directions) with 2.0 Å grid spacing. The steric and electrostatic interaction fields were calculated using Lennard-Jones and Columbic potentials, respectively at each lattice intersection. The interaction energies were calculated using standard Tripos force field with distant dependent dielectric constant ε = ε0 R ij with ε0 =1.0. An sp3 carbon atom with a radius of 1.52 Å bearing +1 charge was employed as a probe atom to calculate the CoMFA fields. Steric and electrostatic cutoffs were applied to truncate the contribution up to ±30 kcal/mol and CoMFA standard scaling option was applied.
The CoMSIA analysis introduced by Klebe et al. calculates five similarity descriptors namely steric, electrostatic, hydrophobic, hydrogen bond donor and acceptor [27]. A distant dependent Gaussian type function was used to calculate similarity indices at all the grid points. A default attenuation factor (α) of 0.3 was employed as a smoothening function. The probe atom with charge +1, hydrophobicity +1, and hydrogen bond donor and acceptor property of +1 was used. The steric indices were related to third power of the radii, partial atomic charges were used to derive the electrostatic fields. Atom based parameters developed by Viswanadhan et al. were employed to compute hydrophobic descriptors and rule based method was used to get the hydrogen bond donor and acceptor fields [32].
Partial least square analysis
Partial least square analysis was employed to derive the correlation between biological activity and descriptors calculated by CoMFA and CoMSIA [33]. Leave one out (LOO) method was initially carried out to check the predictivity of the model and also to determine the optimum number of components with minimum standard error of prediction (SEP). The optimum number of components obtained from the LOO method was further applied to derive the final non-cross validated correlation r 2. To get the confidence intervals (mean and standard deviation) and to assess the robustness of the model, group cross validation and bootstrapping were performed. The group cross validation was conducted for 50 runs in which the training set molecules were randomly split into groups and subjected to the correlation. The mean of 50 cross-validation runs was considered as correlation coefficient, r 2cv . The bootstrapping (r 2bs ) analysis for 100 runs was performed with optimum number of components, which provides an estimate of the variability of the parameters in a final model [34].
Molecular docking
The binding interactions can be ascertained by docking the inhibitors in the active site. GOLD 2.2 [35] docking program was used to calculate the docking modes of pyrazolyl urea inhibitors into p38 active site. GOLD employs genetic algorithm in which the information about the ligand conformation and hydrogen bonding is encoded in chromosome. GOLD considers complete ligand flexibility and partial protein flexibility and the energy functions partly based on conformational and non-bonded interactions. Several types of scoring functions viz. GoldScore, ChemScore and User-defined score are available. The ligand and water molecules in the p38 kinase crystal structure were removed and after that hydrogens were added. The protein was assigned with Kollmann partial atomic charges and local minimization of hydrogens was carried to relieve the unwanted contacts. Following default genetic algorithm parameters were used: 100 population size, 1.1 for selection, 5 number of islands, 100,000 number of genetic operations and 2 for the niche size. The active site was defined with 10 Å radius from Asp168 and GOLD score was selected to rank order the docked conformations.
FlexX molecular docking program interfaced with Sybyl 6.9 was employed to find the docking interactions. FlexX is an automated docking program based on incremental construction [36, 37]. FlexX considers ligand flexibility by changing the conformations of the ligand in the active site while making the protein rigid. Active site was defined around 6.5 Å and certain care was taken to cover the important residues like, Asp168 and Glu71. The default FlexX score was used to rank order the docked conformations and the best-docked conformations were then used to analyze the binding interactions.
The AutoDock program is an automated grid-based docking procedure of ligands with biomacromolecular targets that employs Monte Carlo simulated annealing/Genetic algorithm techniques for conformational exploration with a rapid energy evaluation using molecular affinity potentials [38, 39]. The AutoGrid settings with 60 × 60 × 60 grid size and a grid spacing of 0.375 Å were used for preparing the grid. The Lamarckian genetic algorithm, with 250,000 energy evaluations and 27,000 generations with step sizes of 0.2 for translation and 5.0 for quaternion and torsion, respectively, has been employed. The best-docked conformations in each of the cases were taken for further analysis.
Conclusions
The two well known 3D-QSAR tools, CoMFA and CoMSIA studies were intended to develop statistically significant prognostic models of pyrazolyl urea derivatives and to bring out structural requirements sought for the potent activity. In CoMFA, a model with atom fit alignment has demonstrated an excellent conventional correlation, and consistent statistical relationships have been observed as reflected in the group cross validation and bootstrap correlation coefficient. Comparatively, a higher steric contribution has been observed in this model. A CoMSIA model with steric, hydrophobic and hydrogen bond acceptor descriptor fields have been derived using the best predictive CoMFA model. The steric contour maps of both CoMFA and CoMSIA were found complementing each other, apart from other informative contour maps.
Docking studies have been employed to explore binding interactions and conformations, and further to explain the relative difference in the biological activity of the inhibitors. All the pyrazolyl urea inhibitors were found docked in the active site exhibiting much needed interactions for the activity and the interactions were consistent with the reported crystal structures. The important observation is that the hydrophobic properties of the inhibitor are found to be very important, as is the effective utilization of both P1S and P2S regions. In P1S, groups that could explore the lipophilic interactions with Glu71 and hydrogen bond interactions with nearby amino acid residues demonstrated potent activity. Similarly, groups that could pierce deep lipophilic P2S pocket support for the potent activity.
References
Pearson G, Robinson F, Beers T, Xu E, Berman K, Cobb M (2001) Endocr Rev 22:153
Kulkarni RG, Achaiah G, Sastry GN (2006) Curr Pharm Des 12:2437
Ono K, Han J (2000) Cell Sign 12:1
Zarubin T, Han J (2005) Cell Res 15:11
Schaeffer H, Weber M (1999) Mol Cell Biol 19:2435
English J, Pearson G, Wilsbacher J, Swantek J, Karandikar M, Xu S, Cobb MH (1999) Exp Cell Res 253:255
Behr TM, Berova M, Doe CP, Ju H, Angermann CE, Boehm J, Willette RN (2003) Cur Opin Invest Drugs 4:1059
Jiang Y, Chen C, Li Z, Guo W, Gegner A, Lins S, Han J (1996) J Biol Chem 271:17920
Li Z, Jiang Y, Ulevitch J, Han J (1996) Biochem Biophys Res Comm 228:334
Jiang Y, Gram H, Zhao M, New L, Gu J, Feng L, Padova F, Ulevitch R, Han J (1997) J Biol Chem 272:30122
Hanks SK, Hunter T (1999) FASEB J 9:576
New L, Jiang Y, Zhao M, Liu K, Zhu W, Flood LJ, Kato Y, Parry G, Han J (1998) EMBO J 17:3372
Newton R, Holden N (2003) Biodrugs 17:113
Jackson R, Bolognese B, Hillegass L, Kassis S, Adams J, Griswold DE, Winkler JD (1998) J Pharmacol Exp Ther 284:687
Lee JC, Kumar S, Griswold DE, Underwood DC, Votta BJ, Adams JL (2000) Immunopharmacology 47:185
Lee C, Laydon J, McDonnell P, Gallagher TF, Kumar S, Green D, McNulty D, Blumenthal MJ, Heys JR, Landvatter SW (1994) Nature 372:739
Gallagher TF, Susan M, Thomson SM, Garigipati RS, Sorenson ME, Smietana JM, Lee D, Bender PE, Lee JC, Laydon JT, Griswold DE (1995) Bioorg Med Chem Let 5:1171
Wang Z, Canagarajah J, Boehm JC, Kassisa S, Cobb MH, Young PR, Abdel-Meguid S, Adams JL, Goldsmith EJ (1998) Structure 6:1117
Young PR, McLaughlin MM, Kumar S, Kasis S, Doyle ML, McNultry D, Gallagher TF, Fisher S, McDonnell PC, Carr SA, Huddleston MJ, Seibel G, Porter TG, Livi GP, Adams JL, Lee JC (1997) J Biol Chem 272:12116
Laufer S, Wagner G, Kotschenreuther A, Albrecht W (2003) J Med Chem 46:3230
Cirillo F, Pargellis C, Regan J (2002) Curr Top Med Chem 2:1021
Stelmach E, Liu L, Patel B, Pivnichny V, Scapin G, Singh B Hop CECA, Wang Z, Strauss JR, Cameron PM, Nichols EA, O’Keefe SJ, O’Neill EA, Schmatz DM, Schwartz CD, Thompson CM, Dennis M, Zaller DM, Doherty JB (2003) Bioorg Med Chem Lett 13:277
Lee M, Dominguez C (2005) Cur Med Chem 12:2979
Dumas J, Hatoum-Mokda H, Siblety R, Riedl B, Scott J, Monahan MK, Lowinger TB, Brennan C, Natero R, Turner T, Johnson JS, Schoenleber R, Bhargava A, Wilhelm SM, Housley TJ, Ranges GE, Shrikhande A (2000) Bioorg Med Chem Letts 10:2051
Dumas J, Hatoum-Mokda H, Siblety R, Riedl B, Scott J, Khire U, Lee W, Wood J, Wolanin D, Cooley J, Bankston D, Redman AM, Schoenleber R, Caringal Y, Gunn D, Romero R, Osterhout M, Paulsen H, Housley TJ, Wilhelm SM, Pirro J, Chien D, Ranges GE, Shrikhande A, Muzsi A, Bortolon E, Wakefield J, Ostravage CG, Bhargava A, Chaub T (2002) Bioorg Med Chem Letts 12:1559
Regan J, Pargellis CA, Cirillo PF, Gilmore T, Hickey ER, Peet GW, Proto A, Swinamer A, Moss N (2003) Bioorg Med Chem Letts 13:3101
Pargellis C, Tong L, Churchill L, Cirrillo F, Gilmore T, Graham AG, Grob PM, Hickey ER, Moss N, Pav S, Regan J (2002) Nat Struct Biol 9:268
Cramer D, Patterson E, Bunce D (1988) J Am Chem Soc 110:5959
Klebe G, Abraham U, Mietzner T (1994) J Med Chem 37:4130
Klebe G, Abraham U (1999) J Comp Aided Mol Des 13:1
Sybyl 6.9, Tripos Inc., 1699 S. Hanley Rd., St Leuis, MO 63144 USA
Viswanadhan VN, Ghose AK, Revankar GR, Robins RK (1989) J Chem Inf Comput Sci 29:163
Geladi P (1998) J Chemon 2:231
Cramer RD, Bunce D, Patterson E (1988) Quant Struct Act Relat 7:18
GOLD, version 2.2, Cambridge Crystallographic Data Centre: Cambridge, U.K
Rarey M, Kramer B, Lengauer T, Klebe G (1996) J Mol Biol 261:470
Kramer B, Rarey M, Lengauer T (1999) Proteins: Str Func Genet 37:228
Goodsell DS, Olson AJ (1990) Proteins: Str Func Genet 8:195
Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ (1998) J Comput Chem 19:1639
Acknowledgements
RG thanks AICTE for providing the financial assistance and PS thanks CSIR for a senior research fellowship. The Director of IICT is thanked for his continued encouragement.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kulkarni, R.G., Srivani, P., Achaiah, G. et al. Strategies to design pyrazolyl urea derivatives for p38 kinase inhibition: a molecular modeling study. J Comput Aided Mol Des 21, 155–166 (2007). https://doi.org/10.1007/s10822-006-9092-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-006-9092-9