Abstract
Dunaliella salina is known as the most salinity-tolerant unicellular eukaryote. To explore its molecular response mechanisms to high salinity concentrations, D. salina transcriptomes, small RNA groups and degradomes were analyzed under salinity stress conditions, by high throughput sequencing. A total of 1008 microRNA (miRNA) sequences were identified, including 998 known conserved miRNAs and 10 novel miRNAs. Further analysis of miRNA expression in D. salina under salinity stress found that 49miRNAs showed significant differences in expression. For the first time in D. salina, 745 target genes, regulated by 194 miRNAs, were validated by degradome sequencing. Gene ontology (GO) enrichment analysis and KEGG analysis showed that these miRNA target genes are involved in a variety of molecular biological regulation processes, such as signal transduction, material transport, transcriptional regulation and protein processing. In combination with transcriptome sequencing results, 14 differentially expressed miRNAs and 87 differentially expressed target genes were found to negatively correlate in expression. Further analysis showed that mmu-miR-466, dme-miR-2493, mmu-mir-669h, dre-mir-29a and dme-mir-9388 play an important role in osmoregulation in response to high salinity stress in D. salina. These results enrich existing hypotheses, while providing new insights into the molecular mechanism of salinity tolerance in D. salina.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Dunaliella salina is widely distributed green alga, commonly found in saline lakes. Dunaliella salina cells have no cell wall and cells are small and varied in shape, with two equal length flagella located at the front of the cell enabling swimming. Dunaliella salina is a high-salinity-tolerant unicellular eukaryote able to grow and reproduce in culture media containing 0.05–5.0 mol L−1 NaCl (Borowitzka 2013), making it an ideal model organism to study physiological and molecular mechanisms of plant salinity tolerance. Better understanding of the molecular mechanisms of D. salina adaptation to high-salinity environments is of scientific significance and has high application value. The osmotic regulation of D. salina in adaptation to high-salinity environments has been extensively studied (Weiss and Pick 1990; Li et al. 2006; Chen et al. 2011; Zhao et al. 2013), in addition to cloning of D. salina salinity-tolerant genes (He et al. 2007; Jia et al. 2009) and proteome (Liska et al. 2004; Katz et al. 2007). However, D. salina molecular response mechanisms to salinity stress has not been clearly established.
MicroRNAs (miRNAs) are sequences of about 20–25 nucleotides and are a class of endogenous non-coding RNA found in eukaryotes. Mature miRNA is composed of primary transcripts (pri-miRNA), which after a series of shear processing steps are then assembled into the multi-protein RNA-induced silencing complex (RISC). Via complementary base pairing with target mRNAs, miRNAs degrade target mRNA or inhibit translation of the target mRNA (Park et al. 2005; Jones-Rhoades et al. 2006). Studies have identified miRNA responses to environmental stress such as drought (Wang et al. 2016), extreme temperatures (Li et al. 2016), metals (Han et al. 2016), and salinity exposure (Yu et al. 2016). Therefore, analysis of miRNA is of interest to study the molecular mechanisms of D. salina stress responses.
In this study, the D. salina transcriptome, small RNA, and degradome were systematically analyzed under salinity stress conditions, using high throughput sequencing technology to identify changes in miRNAs and corresponding target genes. Results provide important information for elucidating the molecular mechanisms of the D. salina response to high-salinity environments.
Materials and methods
Culture and salinity stress treatment conditions for D. salina
The Dunaliella salina strain CCAP 19/3 was provided by the Aquatic Biology Laboratory of Dalian Ocean University, Dalian, China, and cultured in Conway medium (Walne 1966) containing 1 mol L−1 NaCl, under the culture conditions of 25 °C, 22.5 μmol photons m−2 s−1 light, and a 12-h light (L) and 12 h dark (D) photoperiod. These conditions were maintained until the logarithmic growth period, when solid NaCl was added to the culture solution at a final concentration of 3 mol L−1. Total RNA was extracted from algal cells exposed to salinity stress for 0 h (as control group (Con)), 1 h (S_1), and 6 h (S_6), each with three biological repeats, using a Total RNA Purification Kit-RNAiso Plus (TaKaRa, Japan), and the quality and purity of the total RNA extract was analyzed using an Agilent 2100 (Agilent Technologies, USA) and a Nanodrop 2000 (NanoDrop, USA).
Construction of transcriptome library and sequencing
The transcriptome library was constructed according to the instructions of the Illumina Kit and after a quality and purity test, mRNA with Poly (A) were purified from the total RNA using oligo(dT) magnetic beads and then fragmented using a fragmentation reagent. First-strand cDNA was synthesized using six random hexamer primers and a template of short fragments, with second-strand cDNA synthesized using the buffer, dNTPs, RNase H, and DNA polymerase I. The products were then purified using AMPure XP beads and the cohesive ends were modified into blunt ends using T4 DNA polymerase and Klenow DNA polymerase. In addition, poly-adenylated caps were added to the 3′ ends. The final sequencing library was prepared by PCR enrichment, where after the quality test, the transcriptome library was sequenced using an Illumina Hiseq2000/2500 (Illumina, USA).
Small RNA sequencing and miRNA identification
Nine small RNA libraries were prepared using the TruSeq SmallRNA Sample Prep Kits. The constructed libraries were sequenced using an Illumina Hise-q2000/2500. The original sequence was evaluated by Illumina Fast QC for data quality, to ensure data quality scores of > Q30. A series of data processing steps were used to remove impure sequences produced by sample preparation, sequencing joints, and atypical miRNA feature sequences, as well as those smaller than 18 nt or larger than 25 nt. The remaining sequences were aligned to multiple RNA databases: mRNA, RFam (excluding miRBase), and Repbase. The unmatched sequences were mapped to specific species’ precursors, as listed in miRBase 21.0 to identify known miRNAs and novel 3p- and 5p-derived miRNAs.
Degradome sequencing and target gene identification
Triplicate exposures performed for each group (Con, S_1 and S_6) were mixed separately to construct separate degradome libraries for each exposure group. The three constructed libraries were then sequenced using an Illumina Hiseq2000/2500. The data obtained by sequencing was processed for further analysis and a degradome density file was generated by aligning the obtained sequences to a cDNA database of selected species. Targetfinder target gene prediction software was used to predict the target gene mRNA sequences according to miRNA sequences, which were then compared to the degradome density file, to identify common mRNAs (miRNA target genes). Differential expression analysis and functional annotation of these transcripts was then performed.
qRT-PCR validation
To validate the high throughput sequencing results, ten differentially expressed miRNA samples (five from 1 h group and five from 6 h group) were randomly selected to carry out real-time fluorescence quantitative PCR (qRT-PCR) analysis. Reverse transcription was performed on RNA samples, using a SYBR PrimeScript miRNA RT-PCR Kit (TaKaRa, Japan). Specific miRNA primers were designed according to their respective miRNA sequences, with D. salina 18S rRNA genes used as the endogenous genes. miRNA names, sequences, and forward primers used for qRT-PCR analysis are listed in Appendix S1 and universal reverse primer is provided in the kit. The SYBR PrimeScript miRNA RT-PCR kit was used to carry out qRT-PCR using 96-well plates, with each PCR reaction repeated in triplicate.
Results
Construction of D. salina small RNA library and small RNA sequencing
Nine separate small RNA libraries were constructed, for each triplicate exposure repeat, for the control group (CON), 1 h salinity stress group (S_1), and 6 h salinity stress group (S_6) samples. Libraries were constructed according to TruSeq Small RNA Sample Prep kit instructions and the constructed libraries were sequenced using an Illumina Hiseq2000/2500. The obtained data are shown in Table 1. The numbers of raw reads from the three exposure groups (Con, S_1, and S_6), were 11260671, 10253264, and 11032749, respectively. After removing 3′ joint sequences, any sequences with a base length no longer in the range of 18-24 nt, sequences with high single base content and pollution sequences, 7922103, 5341924, and 6457750 valid reads were obtained, respectively. In total, the numbers of repetitive sequences were 238470, 225933, and 282209, accounting for 35.17, 35.29, and 38.69% of the unique raw reads, respectively.
The number of raw reads (total and unique) and valid reads (total and unique) obtained from the three exposure groups were mostly within the same order of magnitude, with slight differences observed due to the variation in salinity stress duration. Among the three exposure groups, the number of reads obtained from group S_6 was significantly higher than the number of reads for the Con group, while the number of reads obtained from group S_1 was slightly less than that the Con group. All valid triplicate reads were then compared with the Rfam database, to identify non-coding small RNA, including tRNA, rRNA, snoRNA, and snRNA. The number and proportion of small RNA identified for each exposure group are shown in Table 1.
Based on the statistical analysis, we performed length distribution analysis on the total number of filtered valid reads. As shown in Fig. 1, most data obtained were distributed between 20 and 22 nt, which is consistent with the miRNA length of 18–24 nt and also conforms to the typical endoribonuclease Dicer enzyme cutting characteristics.
Identification of known and novel miRNAs in D. salina
miRNAs identified in different groups are shown in Table 2. In total, 2225 known miRNA precursors and 2151 unique miRNA sequences were identified. Among them, the number of known miRNA sequences identified in the S_6 group was significantly higher than that of both the Con group and the S_1 group, while the number of known miRNA sequences in group S_1 were slightly less than in the Con group. These findings are in accordance with the distribution of unique sequences previously identified as varying between the exposure groups.
In total, 998 known conserved miRNA and 10 novel miRNA were identified in D. salina, with the names and sequences of the novel miRNAs shown in Table 3.
Salinity stress responsive miRNAs in D. salina
To identify miRNAs in D. salina that respond to salinity stress, differential miRNA expression was compared for the nine separate libraries, using the read counts generated by high-throughput sequencing. Following 1 h of salinity stress, 15 miRNAs presented with significant differences in expression (p < 0.05), with 10 miRNAs significantly downregulated and 5 miRNAs significantly upregulated (Fig. 2). After 6 h of salinity stress, 28 miRNAs were significantly downregulated and 12 miRNAs were significantly upregulated. This finding suggests that the differential expression of these miRNAs was induced by salinity stress, which may play an important role in cellular responses to salinity stress.
To verify the accuracy of the variation in miRNA expression, established by high throughput sequencing analysis, real-time fluorescence quantitative PCR (qRT-PCR) was performed, using 10 randomly selected and differentially expressed miRNAs. The changes in expression of these 10 selected miRNAs were found to be consistent with the results of high-throughput sequencing, indicating that high throughput sequencing is a reliable method for the analysis of miRNA expression.
Target prediction of the known and novel miRNAs by degradome sequencing
In order to explore the function of the identified salinity stress responsive miRNAs, degradome sequencing was performed. Analysis using Cleaveland software showed that in the mixed degradome samples, 194 known miRNAs and 745 target genes were identified, with 10 novel miRNAs found to have 54 target genes. 132 miRNAs were found to have multiple target genes, indicating that these miRNAs have a variety of functions in cellular responses to salinity stress. In addition, 235 target genes were found to be regulated by multiple miRNAs. It is of note that the same outcome has also been reported in degradome analysis of Arabidopsis (Addoquaye et al. 2009), suggesting that these miRNA target genes may be involved in a variety of molecular biological regulation mechanisms.
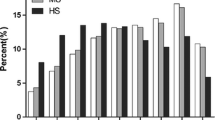
To better understand the potential biological functions of the miRNAs identified as being modified by salinity stress in D. salina, GO and KEGG analysis of the associated target genes were performed. GO analysis identifies gene molecular functions, cellular components, and biological processes, with the GO functional results showing that the D. salina miRNA target genes have a variety of biological functions. The most frequent categories in the biological process are transport, protein folding, and transcription (Fig. 3). Fifteen categories are in the cellular component, with the most abundant being the membrane, nucleus, and cytoplasm. Of the molecular function categories, the most highly represented were ATP binding, metal ion binding, and serine/threonine protein kinases (STPK).
KEGG metabolic pathway analysis showed that the miRNA target gene expression products were mainly involved in bio-synthetic metabolism pathways, including energy metabolism and the bio-synthesis of amino acids, nucleotides, vitamins and co-factors, carbohydrates, and lipids (Fig. 4). Gene products were also found to be involved in the transmission of genetic information, including replication and repair, transcription, translation, folding, and degradation of proteins. These results indicate that anabolism, genetic information transmission, and signal transduction play important roles in the response of D. salina to high levels of salinity stress, providing a novel understanding of the salinity tolerance mechanisms of D. salina.
Correlation analysis of D. salina miRNAs and target genes
In plants miRNA achieves post transcriptional regulation of its target gene, via degradation of the target gene mRNA. Therefore, identifying miRNAs and target genes with negatively correlated expression profiles is key to identifying the induction of miRNAs and the regulation of target genes during salinity stress. In the present study changes in gene expression were analyzed in transcripts from all three exposure groups (Con, S_1, and S_6), as reported by high throughput sequencing. Results showed that after 1 h of salinity stress conditions, there were 394 differentially expressed target genes (p < 0.05), of which 159 were significantly upregulated and 235 were significantly downregulated. Following 6 h of salinity stress, there were 2242 differentially expressed target genes (p < 0.05), 1563 of which were significantly upregulated and 679 were significantly downregulated. Moreover, a joint analysis of transcriptome sequencing results and degradome sequencing results, found that 15 miRNAs exhibited significant differences in expression, corresponding to significant variation in expression of 159 target genes. Among all target genes identified, 53 had functional annotations in the GO database.
Further analysis found that some miRNAs regulate multiple target genes. For example, mmu-miR-466ihas 88 target genes and was significantly downregulated, resulting in 59 target genes being upregulated. Dre-mir-29a and mse-mir-932 were both found to be significantly downregulated, resulting in the significant upregulation of nine target genes each. Cte-mir-2696 was significantly upregulated and significantly downregulated two target genes. The remaining 10 miRNAs corresponded to a single target gene, while 14 miRNAs and 87 target genes were found to display negatively correlated expression profiles. Figure 5 shows negatively correlated miRNAs and target genes, with these miRNAs potentially playing an important role in response to salinity stress in D. salina.
Expressions levels of negatively correlated miRNAs (a) and their target genes (b) in D. salina at three different salinity treatment duration, each with three biological repeats. Red indicates upregulated; green indicates downregulated. Con: control group; S_1: 1-h salinity stress group; S_6: 6-h salinity stress group
Discussion
miRNA is essential for life activities of flora and fauna, by regulating the expression of genes. Although miRNA has been reported to have been involved in biological regulation in Arabidopsis under salinity stress conditions (Park et al. 2005), it has not previously been reported in D. salina.
Dunaliella salina has a relatively simple structure, but a strong ability to tolerate high salinity levels, suggesting that D. salina has regulation mechanisms associated with target gene expression, in response to salinity stress. In this study, the D. salina transcriptome, small RNA, and degradome profile were systematically analyzed under salinity stress conditions using high throughput sequencing technology. As shown in Tables 2 and 3, 1008 miRNAs were identified for the first time in D. salina, including 998 known conserved miRNAs and 10 novel miRNAs. The D. salina miRNA sequence length was mainly 21 nt, which is in accordance with the results reported previously for unicellular alga C. reinhardtii (Zhao et al. 2007). Further analysis of D. salina miRNA expression under salinity stress found that 49 D. salina miRNAs showed significant differences in expression, including miR2916 (Qin et al. 2015), miR398 (Jagadeeswaran et al. 2009), miR397 (Sunkar and Zhu 2004), miR393 (Gao et al. 2011), miR396 (Gao et al. 2010), miR156 (Liu et al. 2008), miR160 (Zhao et al. 2009), and miR169 (Zhao et al. 2009). These have been previously reported to be involved in the regulation processes in response to salinity stress, although the mechanisms remain unknown.
Typically, miRNA is partially or completely matched with the target gene, causing endonuclease digestion of genes or translation inhibition, to negatively regulate target gene expression. Therefore, finding the differentially expressed target genes is highly valuable for elucidating the gene biological functions. In this study, new degradome sequencing technology, which has high flux and high coverage, was shown to significantly improve the probability of capturing target miRNA fragments. All sequences were analyzed by against each possible miRNA target gene transcript, and a statistically significant number of miRNA target gene degradation sites could be obtained by degradome sequencing. Therefore, degradome signals could be distinguished from those associated with other mechanisms, ensuring the reliability of degradome sequencing results. For these reasons, degradome sequencing has been widely used in the study of plant miRNA target genes, resulting in a high number of validated miRNA target genes being known (Pantaleo et al. 2010; Li et al. 2011). As shown in Figs. 3 and 4, 745 target genes were identified and through GO and KEGG analysis, these miRNA target genes were found to possibly be involved in a variety of molecular biological regulatory mechanisms.
Through adjustment of cell volume, ion transport and mass synthesis of glycerol, D. salina successfully adapt to changes in osmotic pressure. However, it is still unclear how cells sense external salinity change, how they activate the synthesis of glycerol, and what the regulation mechanisms are for gene expression in glycerol synthesis.
To adapt to high-salinity environment D. salina cells lose water, which results in cell shrinkage (Weiss and Pick 1990; Borowitzka 2018). The results of the present study show that mmu-miR-466f, dme-miR-2493, and mmu-mir-669h co-regulate actin beta/gamma and plasma membrane H+-ATPase. In eukaryotic cells, microfilaments are composed of actin and are a key component of cytoskeleton maintaining the cell shape. Under high salinity stress conditions, ATP is hydrolyzed by H+-ATPase to form Pi. The rate of H+ secretion increases rapidly, reaching a maximum at 3 min (Chen et al. 1991). At the same, the pH change activates Na+/H+ antiporter membrane proteins, causing an influx of Na+ and an efflux of H+(Weiss and Pick 1990; Pick and Avron 1992). Na+ is extruded through an Na+-ATPase and Na+ pump membrane protein activated by the high cell concentration of Na+, maintaining the Na+ balance (Weiss and Pick 1990; Katz and Pick 2001; Popova et al. 2005). Jones and Sattelle (2008) showed that in Caenorhabditis elegans, Na+/H+ antiporters release protons as “transmitters” to activate cys-loop ligand-gated ion channels, depolarizing the cells and inducing enteric muscle contraction. Such ion channels have also been found in C. reinhardtii and are involved in adaptation to pH change (Merchant et al. 2007; Mukherjee 2015). In the transcriptome library established in the present study, similar sequences were also found with several conserved domains. We presume that under high salinity stress, D. salina cells release protons through H+-ATPase and Na+/H+ antiporters, serving as transmitters to activate cys-loop ligand-gated ion channel, and therefore causing cell shrinkage.
Calcium plays an important role in cellular osmotic stress response (Issa 1996). Under hyperosmotic stress, the cell concentration of Ca2+ increased due to the influx of Ca2+ through Ca2+ channels (Borowitzka 2018), although the mechanisms of Ca2+ channels remain unclear.
Mmu-miR-466i regulates multiple target genes including STPK, ion channel protein, molecular chaperone, 26S lysosome, guanosinecyclase, and transcription factors, among others. It activates guanylatecyclase, which can activate the cGMP signaling system. CGMP will bind to and open the CNGC calcium channel of the intimal system, causing calcium ions to enter the cytoplasm, produce calcium, and then activate calcium-dependent protein kinase (CDPK). CDPK is unique to plants, algae, and some protozoans and has been found to regulate many substrates including transcription factors, metabolic enzymes, ion channels, ion pumps, and skeleton proteins, among others (Harmon 2000). In our previous study, a halotolerant-related CDPK gene was cloned showing that its expression was induced by high salinity using qRT-PCR (Zhang 2013). Mmu-miR-466i also regulates calmodulin-dependent kinase (CaMKK) which activates calmodulin-dependent protein kinase (CaMK). CaMK may regulate cell cycles and rapid growth in the logarithmic phase of unicellular algae (Means 1994). Moreover, the data shows that the target gene of mmu-miR-466f and dme-miR-2493, calmodulin, regulates Na+/H+ antiporters(Emmer et al. 1989). Mmu-miR-466f and mmu-miR-466i belong to the same miRNA family. Based on these findings, we may presume that mmu-miR-466, dme-miR-2493, and mmu-mir-669h co-regulate the rapid volume change and ion transport of D. salina under high salinity stress.
Glycerol is the most important substance in D. salina osmoregulation with the synthesis of glycerol starting from dihydroxyacetone phosphate (DHAP), via glycerol cycle (Borowitzka 2018). DHAP for glycerol synthesis is derived mainly from starch catabolism, as photosynthesis is inhibited under high salinity stress (Goyal 2007a, b). It was observed that 1 h after salinity stress was imposed, the light-harvesting complex II chlorophyll a/b binding protein gene was significantly downregulated by mmu-miR-466f, indicating that photosynthesis was inhibited. Conversely, 6 h after salinity stress, the gene was upregulated by mmu-mir-669 h (not expressed differentially in the 1 h group). It may be that with the extension of salinity stress exposure time, starch stored in D. salina cells is depleted and photosynthesis is gradually enhanced.
Under high salinity stress conditions, the ATP concentration decreases while the concentration of Pi increases. High concentrations of Pi are able to induce the activity of starch phosphorylase, one of the key enzymes for glycerol synthesis in D. salina (Degani et al. 1985; Bočvar 2008). Belmans and Van Laere (2010) also reported that the reduction in ATP concentration induces glycerol. One starch phosphorylase gene in D. salina was successfully cloned and expressed in E. coli (GenBank Accession No. KF061044). Starch is converted into glycerol 1-phosphate by starch phosphorylase, then into DHAP, which is converted into glycerol 3-phosphate by a DHAP/glycerol 3-phosphate dehydrogenase. Glycerol 3-phosphate is then dephosphorylated by a specific phosphatase to produce glycerol (Borowitzka 2018). Glycerol 3-phosphate dehydrogenase (GPDH) is a key enzyme in glycerol synthesis, responsible for glycerol synthesis and accumulation (Sun et al. 2007; Saito and Posas 2012), although the regulation mechanism for GPDH gene expression remains still unclear. These findings show that GPDH is regulated by dre-mir-29a, and MAPK is regulated by dme-mir-9388 and dme-miR-2493. Previous studies have reported cloning of the DsMAPK gene with its expression proven to be induced by high salinity stress using qRT-PCR. Under high salinity stress, its expression was upregulated, reaching a maximum at 1 h (Zhang 2013). Zhao et al. (2015) proposed that DtMAPK regulates the expression of DtGPDH and glycerol synthesis.
Further work is required to fully establish the relationship between miRNAs and their predicted target genes.
In conclusion, the D. salina transcriptomes, small RNA groups, and degradomes were analyzed under salinity stress conditions, by high throughput sequencing. A total of 1008 microRNA (miRNA) sequences were identified, with 49 miRNAs showing significant differences in expression. For the first time in D. salina, 745 target genes, regulated by 194 miRNAs, were validated by degradome sequencing. These target genes are involved in a variety of molecular biological regulation processes, such as signal transduction, material transport, transcriptional regulation, and protein processing. In combination with transcriptome sequencing results, 14 differentially expressed miRNAs and 87 differentially expressed target genes were found to negatively correlate in expression. miRNAs such as mmu-miR-466, dme-miR-2493, mmu-mir-669h, dre-mir-29a, and dme-mir-9388 may play an important role in osmoregulation in response to high salinity stress in D. salina. These results enrich and develop existing theories, while providing new insights into the molecular mechanism of salinity tolerance in D. salina.
References
Addoquaye C, Miller W, Axtell MJ (2009) CleaveLand: a pipeline for using degradome data to find cleaved small RNA targets. Bioinformatics 25:130–131
Belmans D, Van Laere A (2010) Glycerol cycle enzymes and intermediates during adaption of Dunaliella teriolecta cells to hyperosmotic stress. Plant Cell Environ 10:185–190
Bočvar DA (2008) Regulation of salt-induced starch degradation in Dunaliella tertiolecta. J Plant Physiol 127:77–96
Borowitzka MA (2013) Dunaliella: biology, production, and markets. In: Richmond A, Hu Q (eds) Handbook of microalgal culture. John Wiley & Sons, Ltd, London, pp 359–368
Borowitzka MA (2018) The ‘stress’ concept in microalgal biology—homeostasis, acclimation and adaptation. J Appl Phycol. https://doi.org/10.1007/s10811-018-1399-0
Chen Z, Jiao XZ, Liu H (1991) Role of the plasma membrane H+-ATPase during the osmoregulation of the alga Dunaliella salina under hypertonic stress. Plant Physiol J 17:333–341
Chen H, Lao YM, Jiang JG (2011) Effects of salinities on the gene expression of a (NAD+)-dependent glycerol-3-phosphate dehydrogenase in Dunaliella salina. Sci Total Environ 409:1291–1297
Degani H, Sussman I, Peschek GA, Avron M (1985) 13C- and 1H-NMR studies of osmoregulation in Dunaliella. Biochim Biophys Acta Mol Cell Res 846:313–323
Emmer E, Rood RP, Wesolek JH, Cohen ME, Braithwaite RS, Sharp GWG, Murer H, Donowitz M (1989) Role of calcium and calmodulin in the regulation of the rabbit ileal brush-border membrane Na+ /H+ antiporter. J Membr Biol 108:207–215
Gao P, Bai X, Yang L, Lv D, Li Y, Cai H, Ji W, Guo D, Zhu Y (2010) Over-expression of Osa-MIR396c decreases salt and alkali stress tolerance. Planta 231:991–1001
Gao P, Bai X, Yang L, Lv D, Pan X, Li Y, Cai H, Ji W, Chen Q, Zhu Y (2011) Osa-MIR393: a salinity- and alkaline stress-related microRNA gene. Mol Biol Rep 38:237–242
Goyal A (2007a) Osmoregulation in Dunaliella, part I: effects of osmotic stress on photosynthesis, dark respiration and glycerol metabolism in Dunaliella tertiolecta and its salt-sensitive mutant (HL 25/8). Plant Physiol Biochem 45:696–704
Goyal A (2007b) Osmoregulation in Dunaliella, part II: photosynthesis and starch contribute carbon for glycerol synthesis during a salt stress in Dunaliella tertiolecta. Plant Physiol Biochem 45:705–710
Han X, Yin H, Song X, Zhang Y, Liu M, Sang J, Jiang J, Li J, Zhuo R (2016) Integration of small RNAs, degradome and transcriptome sequencing in hyperaccumulator Sedum alfredii uncovers a complex regulatory network and provides insights into cadmium phytoremediation. Plant Biotechnol J 14:1470–1483
Harmon AC (2000) CDPKs-a kinase for every Ca2+ signal? Trends Plant Sci 5:154–159
He Q, Qiao D, Bai L, Zhang Q, Yang W, Li Q, Cao Y (2007) Cloning and characterization of a plastidic glycerol 3-phosphate dehydrogenase cDNA from Dunaliella salina. J Plant Physiol 164:214–220
Issa AA (1996) The role of calcium in the stress response of the halotolerant green alga Dunaliella bardawil Ben-Amotz et Avron. Phyton 36:295–302
Jagadeeswaran G, Saini A, Sunkar R (2009) Biotic and abiotic stress down-regulate miR398 expression in Arabidopsis. Planta 229:1009–1014
Jia Y, Xue L, Liu H, Li J (2009) Characterization of the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene from the halotolerant alga Dunaliella salina and inhibition of its expression by RNAi. Curr Microbiol 58:426–431
Jones AK, Sattelle DB (2008) The cys-loop ligand-gated ion channel gene superfamily of the nematode, Caenorhabditis elegans. Invertebr Neurosci 8:41–47
Jones-Rhoades MW, Bartel DP, Bartel B (2006) MicroRNAS and their regulatory roles in plants. Annu Rev Plant Biol 57:19–53
Katz A, Pick U (2001) Plasma membrane electron transport coupled to Na+ extrusion in the halotolerant alga Dunaliella. Biochim Biophys Acta Bioenerg 1504:423–431
Katz A, Waridel P, Shevchenko A, Pick U (2007) Salt-induced changes in the plasma membrane proteome of the halotolerant alga Dunaliella salina as revealed by blue native gel electrophoresis and nano-LC-MS/MS analysis. Mol Cell Proteomics 6:1459–1472
Li Z, Meng X, Liu C, Yu L, Chen X (2006) Effects of osmotic stress on intracellular glycerol content and enzyme activity in Dunaliella salina. Chinese Bull Bot 23:145–151
Li B, Qin Y, Hui D, Yin W, Xia X (2011) Genome-wide characterization of new and drought stress responsive microRNAs in Populus euphratica. J Exp Bot 62:3765–3779
Li SP, Dong HX, Yang G, Wu Y, Su SZ, Shan XH, Liu HK, Han JY, Liu JB, Yuan YP (2016) Identification of microRNAs involved in chilling response of maize by high-throughput sequencing. Biol Plant 60:251–260
Liska AJ, Shevchenko A, Pick U, Katz A (2004) Enhanced photosynthesis and redox energy production contribute to salinity tolerance in Dunaliella as revealed by homology-based proteomics. Plant Physiol 136:2806–2817
Liu HH, Tian X, Li YJ, Wu CA, Zheng CC (2008) Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA 14:836–843
Means AR (1994) Calcium, calmodulin and cell cycle regulation. FEBS Lett 347:1–4
Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Maréchal-Drouard L (2007) The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318:245–250
Mukherjee A (2015) Computational analysis of a cys-loop ligand gated ion channel from the green alga Chlamydomonas reinhardtii. Mol Biol 49:742–754
Pantaleo V, Szittya G, Moxon S, Miozzi L, Moulton V, Dalmay T, Burgyan J (2010) Identification of grapevine microRNAs and their targets using high-throughput sequencing and degradome analysis. Plant J 62:960–976
Park MY, Wu G, Gonzalez-Sulser A, Vaucheret H, Poethig RS (2005) Nuclear processing and export of microRNAs in Arabidopsis. Proc Natl Acad Sci U S A 102:3691–3696
Pick U, Avron M (1992) Modulation of Na+/H+ antiporter activity by extreme pH and salt in the halotolerant alga Dunaliella salina. Plant Physiol 100:1224–1229
Popova LG, Shumkova GA, Andreev IM, Balnokin YV (2005) Functional identification of electrogenic Na+-translocating ATPase in the plasma membrane of the halotolerant microalga Dunaliella maritima. FEBS Lett 57922:5002
Qin Z, Chen J, Jin L, Duns GJ, Ouyang P (2015) Differential expression of miRNAs under salt stress in Spartina alterniflora leaf tissues. J Nanosci Nanotechnol 15:1554–1561
Saito H, Posas F (2012) Response to hyperosmotic stress. Genetics 192:289–318
Sun XF, Huang F, Liang X, Zhang FW, Yang WG, Bai LH, Qiao DR, Cao Y (2007) Expression of GPD gene from Dunaliella salina treated with different stress and glycerol synthesis of the cells. J Sichuan Univ 44:433–438
Sunkar R, Zhu JK (2004) Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16:2001–2019
Walne PR (1966) Experiments in the large scale culture of the larvae of Ostrea edulis. Fish Investig 2:1–53
Wang Y, Li L, Tang S, Liu J, Zhang H, Zhi H, Jia G, Diao X (2016) Combined small RNA and degradome sequencing to identify miRNAs and their targets in response to drought in foxtail millet. BMC Genet 17:57
Weiss M, Pick U (1990) Transient Na+ flux following hyperosmotic shock in the halotolerant alga Dunaliella salina : a response to intracellular pH changes. J Plant Physiol 136:429–438
Yu Y, Wu G, Yuan H, Cheng L, Zhao D, Huang W, Zhang S, Zhang L, Chen H, Zhang J (2016) Identification and characterization of miRNAs and targets in flax (Linum usitatissimum) under saline, alkaline, and saline-alkaline stresses. BMC Plant Biol 16:124
Zhang XL (2013) Cloning and bioinformatic analysis of CDPK gene of Dunaliella salina. J Nuclear Agric Sci 27:418–424
Zhao T, Li G, Mi S, Li S, Hannon GJ, Wang XJ, Qi Y (2007) A complex system of small RNAs in the unicellular green alga Chlamydomonas reinhardtii. Genes Dev 21:1190–1203
Zhao B, Ge L, Liang R, Li W, Ruan K, Lin H, Jin Y (2009) Members of miR-169 family are induced by high salinity and transiently inhibit the NF-YA transcription factor. BMC Mol Biol 10:1–10
Zhao LN, Gong WF, Chen XW, Chen DF (2013) Characterization of genes and enzymes in Dunaliella salina involved in glycerol metabolism in response to salt changes. Phycol Res 61:37–45
Zhao R, Ng DHP, Fang L, Chow YYS, Yuan KL (2015) MAPK in Dunaliella tertiolecta regulates glycerol production in response to osmotic shock. Eur J Phycol 18:243–248
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 31472260). D. salina cells were provided by the Aquatic Biology Laboratory of Dalian Ocean University, Dalian, China.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Gao, X., Cong, Y., Yue, J. et al. Small RNA, transcriptome, and degradome sequencing to identify salinity stress responsive miRNAs and target genes in Dunaliella salina. J Appl Phycol 31, 1175–1183 (2019). https://doi.org/10.1007/s10811-018-1612-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10811-018-1612-1