Abstract
Systemic lupus erythematosus (SLE) is a chronic autoimmune disease which involves many organs and presents with various symptoms. It has been shown that genetic and environmental factors play a major role in this disease and may affect the onset, activity, damage, and mortality of the disease. According to recent studies, methyl-CpG-binding protein 2 (MECP2) has been associated with SLE in various populations. Herein, we studied MECP2 polymorphism in Iranian lupus patients and controls. The study included a total of 884 samples of Iranian ancestry (492 independent SLE patients and 392 unrelated healthy controls). Healthy controls were gender-, ethnic-, and age-matched with the patients. Patient and control samples were genotyped for rs1734787, rs1734791, rs1734792, and rs17435 by applying the Allelic Discrimination Real-Time PCR System. Our results showed a significant association between rs1734787 and rs1734791 SNPs and the risk of SLE in the Iranian population (p = 0.028, p = 0.028), but did not show any significant association with rs1734792 and rs17435 SNPs (p = 075, p = 0.75). The rs1734787 C and the rs1734791 T allele frequencies in the patients were significantly higher than the control group (p = 0.014, p = 0.012). In addition, a significant CTAT haplotype frequency was observed in cases with SLE (p = 0.012), and a significant AAAT haplotype frequency was observed in the control group (p = 0.0003). However, there was no significant association between genotype frequencies and SLE patients. Also, there was no significant association between these SNPs and clinical features. The result of this study suggests that polymorphism in the MECP2 locus is associated with the susceptibility of Iranian SLE patients.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
INTRODUCTION
Systemic lupus erythematosus (SLE) is a chronic autoimmune disease which is linked to the connective tissue. It may involve many organs of the body such as the kidneys, skin, lungs, heart, the hematopoietic system, and the brain [1, 2]. This disease mostly affects women between 20 and 40 years of age and occurs nine times more often in women than in men [3]. SLE patients produce various pathogenic autoantibodies, most of them are nonspecific. Two kinds of the produced autoantibodies are almost specific; anti-dsDNA and anti-phospholipid [4]. The SLE etiology is partially known; nonetheless, it has been shown that genetic and environmental factors play an important role in this disease [5]. Several genes impact the probability of developing lupus when actuated by the environmental factors. The most important genes, which are associated with SLE, are located in the HLA region on chromosome 6 [6]. Several genes which contain the risk variant for SLE are IRF5, PTPN22, STAT4, ITGAM, BLK [7], TNFSF4, and BANK1 [8]. Another gene, which contains the risk variant for SLE according to numerous studies, is Methyl-CpG-binding protein 2 (MECP2). The major role of this gene is it’s binding to methylated cytosine in CpG islands and recruits histone deacetylase complexe to promoter sequences of target genes, which leads to the deacetylation of histone residues. The consequence of this process is the increment of the interaction between DNA and histone proteins, which induces heterochromatin formation that is inaccessible for the transcription factors and results in the suppression of gene expression [5]. Also, MECP2 has an ability to recruit DNA methyltransferase1 (DNMT1), which causes inactivation of gene expression. Single nucleotide polymorphisms (SNPs) of the MECP2 gene have been associated with SLE pathogenesis in some studies [9, 10]. A study on individuals of European descent showed an association between lupus and variants within the MECP2 gene [11]. Another study regarding gene association in SLE concluded that the susceptibility of genes including MECP2 was correlated with SLE in individuals of European ancestry [10]. A study of the Korean population confirmed that there were significant associations between the variants of SNPs within MECP2 in Korean patients and systemic lupus erythematous [5]. Due to the important role of MECP2 in this disease, our aim was to investigate whether SNPs of MECP2 are associated with susceptibility and clinical features of SLE in Iranian patients.
MATERIALS AND METHODS
Patients and Controls
SLE patients were recruited from the outpatient rheumatology clinic of Shariati Hospital, Tehran, Iran, from February 2012 to April 2014. The study included a total of 884 samples of Iranian ancestry (492 independent SLE patients and 392 unrelated healthy controls). The patient group had a mean age of 37.5 ± 12.2 years and included 428 females and 64 males. The mean age of the healthy controls was 38.5 ± 13 years (324 females and 68 males) and they had neither clinical evidence nor family history of any type of autoimmune disorder. The healthy controls were gender-, ethnic-, and age-matched with the patients (Table 1). All cases fulfilled the revised criteria of the American College of Rheumatology for the classification of SLE [12]. All patients were seen by a rheumatologist for the diagnosis of SLE. Informed consent was obtained from all patients and controls. In order to exclude intra-family correlation, neither two SLE patients nor two controls from the same family were chosen. Five milliliters of blood samples were obtained from all subjects for extraction of genomic DNA. Clinical features and laboratory tests for the classification of the disease were performed in SLE patients. The correlation between various genotypes of each SNP with clinical features of the disease was evaluated. The Human Research Ethics Committee of Tehran University of Medical Sciences approved this study.
Genotyping
The genomic DNA was extracted from the whole blood using the phenol–chloroform method [13]. The extracted DNA samples were stored at −20 °C. Approximately 30 ng of the genomic DNA of each sample was used for genotyping. Amplification was performed in 10-μl reaction volumes, containing 5 μl of the TaqMan Genotyping master mix (PN, 4371355), 0.25 μl of TaqMan Genotyping assay mix (PN, 4351376), 0.25 μl of distilled water, and 4.5 μl of genomic DNA. Genotyping of MECP2 was performed using the MGB-TaqMan Allelic Discrimination method (Applied Biosystems, Foster City, CA, USA). Patient and control samples were genotyped for rs1734787, rs1734791, rs1734792, and rs17435 by applying the StepOnePlus Real-Time PCR System (Applied Biosystems). The PCR reaction was done in 96-well plates on a programmable thermal cycler. The PCR cycle includes a denaturation phase at 95 °C for 10 min, followed by 40 cycles at 95 °C for 15 s and at 60 °C for 1 min.
Statistical Analysis
Genotype and allele distribution between case and control groups were evaluated by the Chi-Square test. The Pearson Chi-Square test was applied in order to determine the association between genotype and allele frequencies, and risk of SLE. An alpha of 0.025 was regarded as the significant level. All p values were two-tailed. The odds ratio (OR) and 95 % confidence interval (CI) were also calculated. The genotype distributions of rs1734787, rs1734791, rs1734792, and rs17435 were tested for deviation from the Hardy–Weinberg equilibrium in cases and controls. The Bonferroni correction method was used to correct multiple comparisons, controlling the false discovery rate (FDR) [14]. Statistical analyses were carried out using SPSS for Windows (version 22.0, IBM SPSS Inc., USA). SHEsis online software was used for haplotype, genotype analysis, linkage disequilibrium, and also Hardy–Weinberg equilibrium [15].
RESULTS
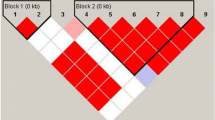
We genotyped four SNPs (rs1734787, rs1734791, rs1734792, and rs17435) in 492 independent SLE patients and 392 unrelated healthy controls. The distribution of these genotypes did not show any significant deviation from the Hardy–Weinberg equilibrium between patients and healthy controls. The association analysis between rs1734787 and rs1734791 SNPs and the risk of SLE in the Iranian population showed a significant association. Our data showed about 98 % linkage between rs1734787 and rs1734791 polymorphisms and 98 % linkage between rs1734792 and rs17435 polymorphisms (Table 2, Fig. 1). The rs1734787 C allele, and the rs1734791 T allele frequencies in the patients were significantly higher than the control group (Adj. P a = 0.028, OR = 1.26, 95 % CI = 1.04–1.52, and Adj. P a = 0.028, OR = 1.27, 95 % CI = 1.05–1.53 respectively). There were no significant association between the genotype frequencies and SLE (Table 3). In addition, a high CTAT haplotype frequency was observed in cases with SLE (P = 0.012, OR = 1.307, 95 % CI = 1.059–1.613). Furthermore, a high AAAT haplotype frequency was observed in the control group (P = 0.0003, OR = 0.653, 95 % CI = 0.518–0.823; Table 4). We next investigated whether these two SNPs (rs1734787 and rs1734791) were correlated with clinical manifestations of SLE, such as the malar rash, discoid rash, photosensitivity, aseptic necrosis, oral ulcer, arthritis, pleuritic, proteinuria, psychosis, pericarditis, vasculitis, hepatitis, leukopenia, anemia, hematuria, thrombocytopenia, anti-dsDNA, CH50 complement, and ANA (Table 5). After the Bonferroni correction, there was no significant association between the SNPs and clinical features (Data not shown). The result of this study suggests that the MCEP2 gene may play a role in the pathogenesis of Iranian SLE patients.
DISCUSSION
Systemic lupus erythematosus (SLE) is a chronic and systemic autoimmune disease that presents with various symptoms. The immune system is widely endangered in SLE patients, and malfunction of the regulatory system might lead to breakage of tolerance and cause many problems [16]. It has been shown that the incidence and prevalence rate and also the clinical and laboratory features are quite different in various populations. Furthermore, it has been reported that genetic and environmental factors can affect the onset, activity, damage, and mortality caused by the disease [17–19]. Epidemiological data suggests that the SLE concordance rate in monozygotic twins (24–69 %) is higher than the dizygotic twins (2–5 %). The data also confirms the role of genetics in SLE [20]. Several genes that have been determined as risk factors (odds ratios generally between 1.15 and 2.0) for SLE are include; MHC region genes, IRF5, ITGAM, STAT4, BLK, BANK1, PDCD1, PTPN22, TNFSF4, TNFAIP3, SPP1, IRAK1, MECP2, TREX1, C2, C4, C1q, and some of the Fcγ receptors [21]. As mentioned earlier, several studies have shown that the MECP2 is a susceptibility gene for SLE patients in different populations [11, 22]. MECP2 is an approximately small gene that is located on the X chromosome [5]. It is known the MECP2 gene has four exons which are translated into two isoforms, i.e., E1 and E2 [23]. The E1 isoform has 498 amino acids and encoded by exons 1, 3, and 4, whereas the E2 isoform has 486 amino acids and encoded by exons 2, 3, and 4 [24, 25]. MeCP2 is a protein that binds to the methylated DNA. It also binds to the unmethylated DNA with a weaker affinity. Furthermore, it has a high affinity and specificity for methylated CpG than other methyl-DNA-binding family members [26–29]. The consequence of the binding of MeCP2 to the methylated binding domain (MBD) is the recruitment of HDAC, which induces heterochromatin formation and finally gene suppression [30]. Another controversial role of MeCP2 is the recruitment of CREB1 as a co-activator and ultimately in gene expression. In a study by Webb et al. in European-derived lupus patients, it was shown that the expression level of CREB1 was higher in patients with the lupus-risk MECP2 haplotype than patients with the protective haplotypes. In the meantime, the reverse was true in the case of HDAC1. We selected the four mentioned SNPs to analyze whether these SNPs in the Iranian population are associated with SLE. In this case–control study, we showed that rs1734787 and rs1734791 polymorphisms of the MECP2 gene were significantly associated with SLE and could be risk factors of developing SLE. The result of these two SNPs (rs1734787 and rs1734791) was congruent with the results of other studies in other countries, but it was interesting that the result of two other SNPs (rs1734792 and rs17435) was different from other studies. The different results of different studies may be explained by differences in the sample size and genetic predisposition. Our findings suggest that the C allele of rs1734787 and the T allele of rs1734791 polymorphisms may increase the susceptibility to SLE. The risk allele of the rs1734787 polymorphism in studies by Webb et al. in European-derived lupus patients, Kaufman et al. in four different ancestries, Sawalha et al. in a population of Korean ancestry, and also in our study was the C allele. The risk allele of the rs1734791 polymorphism in our study and the study by Kaufman et al. was the T allele [5, 11, 22]. On the other hand, the AAAT haplotype may protect the individuals and the CTAT haplotype may predispose the individuals to SLE. The MECP2 gene was associated with susceptibility to SLE in the Iranian population, similar to other studies in different ethnic groups.
In conclusion, polymorphisms in MECP2 gene might change the gene expression and because of important role of MeCP2 in regulate the other gene expression with recruit the HDAC and the CREB1, as we mentioned earlier, might alter the expression of many genes [11] which may cause unexpected immune responses and act as a starting point for inflammatory reactions in the body. Hence, the MECP2 has a complex regulatory system that will require larger studies to assess the role of MECP2 genotype on lupus in concert with other genetic and environmental risk factors.
References
Levy, D.M., and S. Kamphuis. 2012. Systemic lupus erythematosus in children and adolescents. Pediatric Clinics of North America 59: 345–364.
Bagavant, H., and S.M. Fu. 2009. Pathogenesis of kidney disease in systemic lupus erythematosus. Current Opinion in Rheumatology 21: 489.
Lateef, A., and M. Petri. 2012. Unmet medical needs in systemic lupus erythematosus. Arthritis Research and Therapy 14: S4.
Rahman, A. 2004. Autoantibodies, lupus and the science of sabotage. Rheumatology 43: 1326–1336.
Sawalha, A.H., R. Webb, S. Han, J.A. Kelly, K.M. Kaufman, R.P. Kimberly, M.E. Alarcon-Riquelme, J.A. James, T.J. Vyse, and G.S. Gilkeson. 2008. Common variants within MECP2 confer risk of systemic lupus erythematosus. PloS One 3: e1727.
Martens, H., I. Nolte, G. Van der Steege, M. Schipper, C. Kallenberg, G. Te Meerman, and M. Bijl. 2009. An extensive screen of the HLA region reveals an independent association of HLA class I and class II with susceptibility for systemic lupus erythematosus. Scandinavian Journal of Rheumatology 38: 256–262.
Yang, W., P. Ng, M. Zhao, N. Hirankarn, C.S. Lau, C.C. Mok, T.-M. Chan, R. Wong, K.W. Lee, and M.Y. Mok. 2009. Population differences in SLE susceptibility genes: STAT4 and BLK, but not PXK, are associated with systemic lupus erythematosus in Hong Kong Chinese. Genes and Immunity 10: 219–226.
Rhodes, B., and T. Vyse. 2008. The genetics of SLE: an update in the light of genome-wide association studies. Rheumatology 47: 1603–1611.
Koelsch, K.A., R. Webb, M. Jeffries, M.G. Dozmorov, M.B. Frank, J.M. Guthridge, J.A. James, J.D. Wren, and A.H. Sawalha. 2013. Functional characterization of the MECP2/IRAK1 lupus risk haplotype in human T cells and a human MECP2 transgenic mouse. Journal of Autoimmunity 41: 168–174.
Alonso-Perez, E., M. Suarez-Gestal, M. Calaza, J. Ordi-Ros, E. Balada, M. Bijl, C. Papasteriades, P. Carreira, F.N. Skopouli, and T. Witte. 2012. Further evidence of subphenotype association with systemic lupus erythematosus susceptibility loci: a European cases only study. PloS One 7: e45356.
Webb, R., J.D. Wren, M. Jeffries, J.A. Kelly, K.M. Kaufman, Y. Tang, M.B. Frank, J. Merrill, R.P. Kimberly, and J.C. Edberg. 2009. Variants within MECP2, a key transcription regulator, are associated with increased susceptibility to lupus and differential gene expression in patients with systemic lupus erythematosus. Arthritis and Rheumatism 60: 1076–1084.
Mirkazemi, S., M. Akbarian, A.R. Jamshidi, R. Mansouri, S. Ghoroghi, Y. Salimi, Z. Tahmasebi, and M. Mahmoudi. 2013. Association of STAT4 rs7574865 with susceptibility to systemic lupus erythematosus in Iranian population. Inflammation 36: 1548–1552.
Roe, B. A., J. Crabtree, and A. Khan. 1995. Methods for DNA isolation. Part III. Protocols for recombinant DNA isolation, cloning, and sequencing [Internet edition]. Norman, OK: University of Oklahoma: 2488–2498.
Benjamini, Y., and Y. Hochberg. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B (Methodological): 289–300.
Yong, Y., and H. Lin. 2005. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Research 15: 97–98.
Crispín, J.C., S.-N.C. Liossis, K. Kis-Toth, L.A. Lieberman, V.C. Kyttaris, Y.-T. Juang, and G.C. Tsokos. 2010. Pathogenesis of human systemic lupus erythematosus: recent advances. Trends in Molecular Medicine 16: 47–57.
Jakes, R.W., S.C. Bae, W. Louthrenoo, C.C. Mok, S.V. Navarra, and N. Kwon. 2012. Systematic review of the epidemiology of systemic lupus erythematosus in the Asia Pacific region: prevalence, incidence, clinical features, and mortality. Arthritis Care & Research 64: 159–168.
Pons-Estel, G. J., G. S. Alarcón, L. Scofield, L. Reinlib, and G. S. Cooper. 2010. Understanding the epidemiology and progression of systemic lupus erythematosus. In Seminars in Arthritis and Rheumatism. Elsevier. 257–268
Connelly, K., E. Morand, and A. Hoi. 2013. Asian ethnicity in systemic lupus erythematosus: an Australian perspective. Internal Medicine Journal 43: 618–624.
Askanase, A., K. Shum, and H. Mitnick. 2012. Systemic lupus erythematosus: an overview. Social Work in Health Care 51: 576–586.
Moser, K.L., J.A. Kelly, C.J. Lessard, and J.B. Harley. 2009. Recent insights into the genetic basis of systemic lupus erythematosus. Genes and Immunity 10: 373–379.
Kaufman, K. M., J. Zhao, J. A. Kelly, T. Hughes, A. Adler, E. Sanchez, J. O. Ojwang, C. D. Langefeld, J. T. Ziegler, and A. H. Williams. 2012. Fine mapping of Xq28: both MECP2 and IRAK1 contribute to risk for systemic lupus erythematosus in multiple ancestral groups. Annals of the rheumatic diseases: annrheumdis-2012-201851.
Singh, J., A. Saxena, J. Christodoulou, and D. Ravine. 2008. MECP2 genomic structure and function: insights from ENCODE. Nucleic Acids Research 36: 6035–6047.
Mnatzakanian, G.N., H. Lohi, I. Munteanu, S.E. Alfred, T. Yamada, P.J. MacLeod, J.R. Jones, S.W. Scherer, N.C. Schanen, and M.J. Friez. 2004. A previously unidentified MECP2 open reading frame defines a new protein isoform relevant to Rett syndrome. Nature Genetics 36: 339–341.
Kriaucionis, S., and A. Bird. 2004. The major form of MeCP2 has a novel N-terminus generated by alternative splicing. Nucleic Acids Research 32: 1818–1823.
Lewis, J.D., R.R. Meehan, W.J. Henzel, I. Maurer-Fogy, P. Jeppesen, F. Klein, and A. Bird. 1992. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell 69: 905–914.
Meehan, R., J.D. Lewis, and A.P. Bird. 1992. Characterization of MeCP2, a vertebrate DNA binding protein with affinity for methylated DNA. Nucleic Acids Research 20: 5085–5092.
Nan, X., R.R. Meehan, and A. Bird. 1993. Dissection of the methyl-CpG binding domain from the chromosomal protein MeCP2. Nucleic Acids Research 21: 4886–4892.
Roloff, T.C., H.H. Ropers, and U.A. Nuber. 2003. Comparative study of methyl-CpG-binding domain proteins. BMC Genomics 4: 1.
Damen, D., and R. Heumann. 2013. MeCP2 phosphorylation in the brain: from transcription to behavior. Biological Chemistry 394: 1595–1605.
Acknowledgments
This work was supported by a research grant (grant number 93-01-30-24094) from the Deputy of Research, Tehran University of Medical Sciences, Tehran, Iran.
Declaration of conflict of interest
The author declares that they have no competing interests.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Alesaeidi, S., Karami, J., Mahmoudi, M. et al. Methyl-CpG-Binding Protein 2 (MECP2) Polymorphism in Iranian Patients with Systemic Lupus Erythematosus. Inflammation 38, 2185–2190 (2015). https://doi.org/10.1007/s10753-015-0201-6
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10753-015-0201-6