Abstract
Potassium (K) is one of the essential macronutrients of rice growth and eventually affects yields. To investigate the genetics of low K tolerance and related ions concentrations at the seedling stage in rice, quantitative trait loci (QTLs) were detected using a doubled haploid population derived from a cross between a japonica cultivar CJ06 and an indica accession TN1. A total of 96 QTLs were identified with phenotypic variation 5–29 %, including 30 QTLs found to be associated with shoot height, root length, shoot dry weight, root dry weight and total dry weight under the control (40 mg L−1 K+) and low K stress conditions (4 mg L−1 K+), 14 putative QTLs associated with the K tolerance coefficient of all examined traits and synthetic appraisal index, and 52 QTLs controlling four ions (Na+, K+, Ca2+ and Mg2+) concentrations of root and shoot under two treatments. The results indicated that low K tolerance and related ions concentrations were quantitatively inherited, and the detected major QTLs may be useful for marker-assistant selection.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Potassium (K) is one of the essential mineral elements for plant growth and development (White and Karley 2010), which account for 10 % dry weight in a plant (Watanabe et al. 2007). The deficiency of K leads to growth arrest, impaired nitrogen and sugar balance, increased susceptibility to pathogens (Armengaud et al. 2004) and tremendously affects crop yield (Laegreid et al. 1999). Rice (Oryza sativa L.) is one of the most important food crops in the world which consumes 13 % world K fertilizer supplies (Heffer and Prud’homme 2010). The growth and development of rice requires large amounts of available K, and the K deficiency ultimately limits rice yields (Dobermann et al. 1998). To solve this problem, one effective way is to develop new varieties with higher tolerance to K deficient. Some studies have reported on morphological, physiological, biochemical analysis and K use efficiency under low K stress, and found there is a significant difference among rice genotypes in K deficient tolerance (Liu and Liu 2002; Liu et al. 2003; Tai et al. 2004; Yang et al. 2004). However, the genetic mechanisms of controlling low K tolerance in rice are not completely clear.
Quantitative trait locus (QTL) analysis is a powerful approach to understand the genetic mechanisms of complicated traits which controlled by multiple genes and wildly used in identifying genetic loci of tolerance to low N and low P growing conditions in rice (Cho et al. 2007; Lian et al. 2005; Wissuwa et al. 1998; Hu et al. 2001). However, genetic analysis associated with low K tolerance has little reported in rice. Wu et al. (1998) conducted QTL analysis for relative changes of the parameters of plant height, shoot and root dry matter, K concentration in plant tissue, K uptake, and K use efficiency using a DH population, and a total of 21 QTLs were detected which explained 8.6–15.3 % phenotypic variety. Recently, Miyamotoa et al. (2012) identified two QTLs associated with shoot Na+ concentration with 8 and 74 % phenotypic variation and one QTL with shoot K+ concentration explaining 12 % phenotypic variation under low K condition.
The genotypic differences of low K tolerance in different rice varieties indicate it may be possible to develop the cultivars with high tolerance and high K use efficiency. In this study, we conducted the genetics analysis to identify QTLs for low K tolerance and related ions concentrations (Na+, K+, Mg2+ and Ca2+) at seedling stage with a doubled haploid (DH) lines derived from a cross of japonica and indica, which will facilitate to understand the genetics mechanisms of low K tolerance and gain major QTLs for breeder to carry out molecular marker assisted breeding.
Materials and methods
Plant materials and growth condition
Due to indica rice cultivar TN1 was more tolerant to low K stress than japonica cultivar CJ06 (Wang et al. 2015), a total of 116 DH lines were used in the present study which was developed from a cross of CJ06 and TN1 (Sogava et al. 2005; Yang et al. 2004 ).
The seeds of DH lines and two parents were surface sterilized in 3 % H2O2 solution for 5 min, and rinsed three times with deionized water. Then the seeds were soaked in deionized water at 30 °C for 2 days in the dark, germinated on a plastic net floating on deionized water at 30 °C for 1 day, and sown into sand in a controlled chamber with photoperiod of 14 h light/10 h dark. The light/dark temperatures were set at 31 °C/28 °C, and relative humidity was kept at 80 %. After 10 days, rice seedlings with similar size were selected and transplanted into 20 L plastic containers containing nutrient solution and cultivated in the hydroponic form. The nutrient solution was prepared according to Yoshida et al. (1976) and the pH value was adjusted to 5.2 every 2 days. One week after transplanting to the basic solution, two treatments were established: control (40 mg L−1 K+) and low K stress (4 mg L−1 K+). The experiment was laid out as a split-plot design with three separate repeats.
Measurements
After 3 weeks, plants were carefully taken out of the containers, and washed with distilled water three times, then emerged into 1.0 mM EDTA for 2 h to remove the metals from plant surface. Shoot height (SH) was measured from the coleoptilar node to the tip of the longest leaf and root length (RL) from the coleoptile node to the tip of the longest root. The mean values were calculated from data of 4–6 plants. The sampled plants were separated into roots and shoots, dried at 105 °C for 1 h and then at 70 °C for 48 h in an oven. The samples of shoot dry weight (SDW) and root dry weight (RDW) were measured and 0.5 g sample was put into a digestion tube with 0.5 mL of HF (GR, ≥30 %), 1 mL of H2O2 (GR, ≥40 %) and 5 mL of HNO3 (GR, ≥65 %). The solution was digested using a microwave digester (CEM, MARS). The concentrations of Na+, K+, Mg2+ and Ca2+ in the digestion solution were measured by graphite furnace atomic absorption spectrophotometry (PE Company, A800). Total dry weight (TDW) was calculated according to SDW+RDW, and K tolerance coefficient (KTC) of SH, RL, SDW, RDW and TDW was calculated using the following formula: KTC = the value in K stress treatment (SH, RL, SDW, RDW and TDW)/the corresponding value in the control. SAI (Synthetic appraisal index) = (KTC of SH + KTC of RL + KTC of SDW + KTC of RDW)/4.
Data and QTL analysis
Population distribution analysis was performed using SPSS 16.0 statistical software and correlation analyses were performed using SAS8.0 statistical software. The genetic linkage map was constructed using a total of 227 SSR and STS markers distributed evenly on all 12 rice chromosomes. The 1674.8 cM linkage map covering nearly the whole rice genome was used for QTL analysis (Zeng et al. 2009). The QTLNetwork version 2.0 (Yang et al. 2008) was used in QTL detection and the nomenclature of QTL followed the system of McCouch et al. (1997).
Results
Phenotypic variation
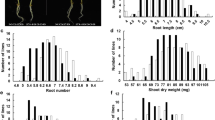
According to the measured data, the values of growth traits in two parents were all reduced in the low K stress compared to control except for RDW and RL in TN1. As shown in Table 1, the KTC of all traits were greater in TN1 than in CJ06 and there were significant differences between two parents under low K stress condition which indicating TN1 has higher tolerance to low K than CJ06. In control condition, the CJ06 had higher SNC (shoot Na+ concentration) and RNC values than TN1, whereas opposite results were observed under K stress condition. In addition, the Na+, Mg2+ and Ca2+ concentrations of two parents in roots and shoots were all increased in low K stress condition than control (Table 1), but opposite results were found in K+ concentration.
For DH population, the SDW, TDW, SH, RCC, RKC, SCC and SKC values under K stress condition were all lower than control, whereas opposite results were observed in RDW, RL, RNC, RMC, SNC and SMC (Table 1). There was a continuous frequency distribution and transgressive segregation in these parameters among DH population under both conditions, indicating that the traits appeared to be quantitative and matched the request of QTL mapping.
Correlation analysis
There were positive correlations between SAI and KTC of all measured growth traits (Supplementary Table 1) and it were also found among the KTC of SH, SDW, RDW and TDW, but the correlarion was not found in KTC of RL with other four measured growth traits (Supplementary Table 1). Among them, the correlation coefficient of KTC of RDW and SDW reached to 0.989 which indicated the significantly positive correlations of biomass between aboveground and underground part. In addition, the significantly positive correlations were also found in KTC of SH with other three traits of KTC of RDW, SDW, and TDW which suggested shoot height is an important index in plant aboveground and underground development. Supplementary Table 2 showed that there were significantly positive correlations among Na+, K+, Mg2+ and Ca2+ concentrations in roots and shoots respectively under both the control condition and low K stress condition, which suggested there may be some common QTLs play role in regulating the ion absorption.
QTL analysis
A total of 96 QTLs were identified and distributed across all 12 chromosomes clustering in 17 chromosomal regions (Table 2 and Supplementary Fig. 1). The phenotypic variation explained by each QTL ranged from 5 to 29 %.
QTL analysis for growth traits
Quantitative trait loci (QTLs) for five traits (RDW, SDW, TDW, SH and RL) under the control and K stress were mapped to rice chromosomes 1, 2, 3, 4, 5, 6, 7, 8, 10 and 11, and a total of 30 putative QTLs were identified including thirteen QTLs under control conditions and seventeen QTLs under K stress conditions. The detected QTLs accounted for 5–22 % of the phenotypic variation, and the main effects and graphical locations were shown in Table 2 and Supplementary Fig. 1.
Under control condition, two QTLs associated with cRDW were identified on chromosomes 1 and 5 explaining phenotypic variance 14 and 16 % respectively. Another two QTLs were also detected for tRDW on chromosomes 1 and 5 under the low K stress, but not located in the same interval with qcRDW1 and qcRDW5 (Supplementary Fig. 1). For the traits of cSDW and tSDW, six QTLs were detected on chromosomes 3 (qtSDW3), 4 (qcSDW4), 5 (qcSDW5 and qtSDW5), 6 (qcSDW6) and 7 (qtSDW7) which accounted for 5–19 % phenotypic variation (Table 2). Among them, the alleles of qtSDW3 and qtSDW7 from CJ06 posed an increase in character values and the alleles of other four QTLs from TN1 tended to increase the value. Moreover, a total of eight QTLs for cTDW and tTDW were detected in control and K treatments. Three QTLs were mapped on chromosomes 4 (qcTDW4), 5 (qcTDW5) and 6 (qcTDW6) under the control, explaining 11, 11 and 17 % of total RDW variation respectively. Under K stress, five QTLs were mapped on chromosome 4 (qtTDW4), 5 (qtTDW5), 7 (qtTDW7), 10 (qtTDW10) and 11 (qtTDW11), and totally explained 33 % phenotypic variation. Two alleles of qtTDW7 and qtTDW10 from CJ06 tended to increase the value under K stress, whereas other five QTLs from TN1 caused an increase in the value.
Two locus (qcSH8 and qcSH10) influencing cSH were identified on chromosome 8 and 10 under control condition. The qcSH8 explained 9 % phenotypic variation and increased 4.46 cm shoot height with positive effect from CJ06, whereas qcSH10 allele in TN1 had additive effect of 5.43 cm for increasing SH. Under K stress, four QTLs associated with tSH were detected on chromosomes 2, 4, 6 and 8 which totally explained 36 % phenotypic variation. In addition, three QTLs (qcRL4, qcRL7 and qcRL11) related to root length were identified on chromosome 4, 7 and 11 under the control, whereas the other three QTLs (qtRL4, qtRL7 and qtRL8) were found in K stress which explained 7–15 % of the total phenotypic variation.
QTL analysis for KTC of growth traits
A total of 14 QTLs were found to be associated with the KTC of all examined traits and SAI on chromosomes 1, 4, 7, 8, 9 and 10 (Table 2 and Supplementary Fig. 1) which explained phenotypic variation ranged from 5 to 23 % (Table 2). Among them, three QTLs (qRDW1, qRDW4 and qRDW9) for the KTC of RDW were detected on chromosome 8 and 9 which accounted for 25 % of the total phenotypic variation. Two QTLs (qSDW4 and qSDW10) for KTC of SDW were mapped within the regions of RM1359-RM1155 and RM228-RM4771 on chromosome 4 and 10 with total 19 % phenotypic variance, and the positive additive effect indicated the alleles from CJ06 increased KTC of SDW. Two QTLs (qTDW4 and qTDW9) affecting KTC of TDW were identified on chromosome 4 and 9. The QTL qTDW4 had a relatively large effect which explained 23 % of the total phenotypic variance and had the positive effect from CJ06. There were three QTLs for KTC of SH on chromosome 1, 7 and 8, and explained 18 % of the total phenotypic variation. Two QTLs related to the KTC of RL (qRL7 and qRL8) were mapped on chromosome 7 and 8 which accounted for 12 % of the total phenotypic variance, and the allele from CJ06 had an additive effect for the increase of KTC of RL. For SAI, two QTLs were identified on chromosomes 1 (qSAI4) and 10 (qSAI10) with 10 and 9 % of total SAI variation respectively.
QTL analysis for ion concentration
A total of five QTLs (qcRNC2, qtRNC4, qtRNC7, qcRNC8 and qcRNC11) associated with Na+ concentration of roots were detected (Table 2 and Supplementary Fig. 1). Under control condition, qcRNC2, qcRNC8 and qcRNC11 were mapped to chromosome 2, 8 and 9 in the regions of RM208-RM207, RM6976-ISA and RM286-RM1812 with 8, 9 and 20 % of phenotypic variance respectively. The other two QTLs qtRNC4 and qtRNC7 under K stress condition were located on chromosome 11 in the marker interval of RM16874-RM1359 and RM418-RM11 which explained 14 and 9 % of phenotypic variance respectively. For shoot Na+ concentration, seven QTLs were mapped on the chromosome 1, 3, 4, 6 and 10. Among them, qcSNC4, qcSNC6 and qcSNC10 were detected under control conditions and totally explained 36 % of the phenotypic variation. Under K stress condition, the qtSNC4 was located on chromosomes 4 with 15 % phenotypic variance and the additive effects showed that the trait-enhancing allele was derived from the parent TN1 (Table 1).
For K+ concentration, seven QTLs (qtRKC3, qcRKC7, qcRKC8, qtRKC9, qtRKC11, qcRKC12 and qtRKC12) were identified under control and K stress conditions in root. The QTLs qcRKC7, qcRKC8 and qcRKC12 were detected with the phenotypic variance of 8–29 % under control condition, and qtRKC3, qtRKC9, qtRKC11 and qtRKC12 were mapped to chromosomes 3, 9, 11 and 12 with 7–11 % of phenotypic variance under K stress condition. Among them, qcRKC12 and qtRKC12 were mapped on the same region of RM3226-RM12 on the chromosome 12 and the positive alleles for both of them were contributed by TN1. Six QTLs associated with shoot K+ concentrations were detected under control and K stress conditions. Three QTLs (qcSKC2, qcSKC4 and qcSKC7) were mapped under control condition on chromosomes 2, 4 and 7 in the marker intervals of RM5472-RM240, RM6997-RM252 and RM505-RM234 which explained 6–9 % of phenotypic variance. There are also three QTLs for K+ in shoots (qtSKC1, qtSKC5 and qtSKC6) under K stress condition which were mapped to chromosomes 1, 5 and 6 in the marker interval of RM246-RM1297, RM548-RM17954 and SBE1-RM494 with 5–20 % phenotypic variance respectively. Among them, qtSKC5 was detected with the additive effect value of 0.27 mg/g which explained 20 % phenotypic variation and the positive allele derived from CJ06.
Under the control condition, the Ca2+ concentration of roots and shoots were measured and a total of eight QTLs were identified. Among them, four QTLs (qcRCC2, qcRCC4, qcRCC8 and qcRCC10) for RCC were detected on chromosomes 2, 4, 8, and 10 explaining 10, 9, 7 and 12 % phenotypic variation respectively and qcSCC1, qcSCC2, qcSCC3 and qcSCC9 for SCC were mapped on chromosomes 1, 2, 3 and 9 with the 5–8 % phenotypic variance respectively. Under K stress condition, three QTLs (qtRCC1, qtRCC6 and qtRCC10) for root Ca2+ concentrations were located on chromosomes 1, 6 and 10 which explained 8–14 % phenotypic variance, and the three QTLs qtSCC1, qtSCC4 and qtSCC6 for shoots Ca2+ concentrations were detected on chromosomes 1, 4 and 6 explaining 7, 15 and 11 % phenotypic variance respectively.
For Mg2+ concentration, two QTLs (qcRMC4 and qcRMC8) in roots under control condition were identified on chromosomes 4 and 8 which explained 9 and 8 % phenotypic variation respectively, and four QTLs (qcSMC4, qcSMC8, qcSMC9 and qcSMC10) in shoots were detected with the phenotypic variations 5–12 %. Under K stress condition, four and three QTLs for roots and shoots Mg2+ concentrations were detected respectively which explained 6–12 % phenotypic variance (Table 2, Supplementary Fig. 1). Among them, qtRMC1 and qtSMC1 were detected at the same locus between RM8094 and RM6716 on the chromosome 1 and the CJ06 alleles of this locus increased the root and shoot Mg2+ concentrations.
Discussion
Rice is the staple food for more than 50 % of the world population and consumes large K fertilizer supplies (Dobermann et al. 1998). Therefore, understanding the genetic mechanism of low K tolerance is important for selecting and breeding rice cultivars with low K tolerance. In this study, rice low K tolerance and the concentrations of four ions in roots and shoots of DH population were determined using a hydroponic experiment. The transgressive segregation of all measured traits suggested that low K tolerance and ion concentration were quantitatively inherited traits and were controlled by multiple genes (Table 1). In addition, the different QTLs for the same trait were detected under the control and low K stress which indicated that the expression of QTLs associated with low K tolerance is readily affected by K stress level. Similarly, the different QTLs for K tolerance coefficient (KTC) were identified under different K concentration indicated the suitable K level should be selected to detect the QTL for K tolerance.
In previous study, the 123 double haploid (DH) lines from the parents IR64 and Azucena have been used to detect QTLs under low K stress condition and five QTLs for plant height have been detected on the chromosomes 1, 2, 3, 7 and 8 (Wu et al. 1997, 1998). In the current study, four QTLs associated with tSH were identified on chromosomes 2, 4, 6 and 8 under K stress. Among them, the qtSH8 located on the region of RM5493 and RM447 was found in the same interval identified by Wu et al. (1997) which suggested the QTL for shoot height under low K stress had a stable inheritance and not affected by the mapping populations and environments. In addition, the QTL qtSDW3 located on chromosome 3 between the markers STS146.1 and RM143 was also similar with the QTL reported by Wu et al. (1997) under low K stress. Although some QTLs (qcSDW4, qtRDW5, qtSDW5 and qcSDW6) for dry shoot and root weight under control and low K stress were located on the same chromosomes with previous report (Wu et al. 1997), but not showed the same loci which indicated that the genetic basis of dry shoot and root weight variation is difference with different parents. We also carried out the genetics analysis of KTC of growth traits and twelve putative QTLs were detected on chromosomes 1, 4, 7, 8, 9 and 10. Moreover, we firstly detected the QTLs of synthetic appraisal index of low K tolerance and two QTLs qSAI1 and qSA10 were located on chromosome 4 and 10 within the RM1359-RM1155 and RM228-RM4771 region. As expected, the two QTLs were located on the same region as the qSDW4 and qSDW10 indicated the significantly positive correlation between the two traits.
We examined the related ion concentrations of Na+, K+, Mg2+ and Ca2+ in roots and shoots under the control and low K stress condition. The correlation analysis showed there were significantly positive correlations among Na+, K+, Mg2+ and Ca2+ concentrations in roots and shoots. In fact, a lot of QTLs for Na+ or K+ concentrations have been identified in different populations under salt stress conditions (Ahmadi et al. 2011; Cheng et al. 2012; Koyama et al. 2001; Lin et al. 2004; Wang et al. 2012). However, there were little studies directly for QTL mapping association with Na+, K+, Mg2+ and Ca2+concentrations under low K stress condition. In this study, a total of 52 QTLs associated with the four ion concentrations in roots and shoots were detected in rice seedlings (Table 2). Lin et al. (2004) identified a major QTL qSKC-1 for shoot K+ concentration on the chromosome 1 under salt stress and the locus has been cloned (Ren et al. 2005). But we not detected QTLs for shoot K+ concentration under low K stress on chromosome 1 which suggested the genetic mechanisms of regulating shoot K+ concentration under salt and K stress may be different in rice. It has been reported that several QTLs for K concentration of rice plant have been detected on chromosomes 1 and 8 by Wu et al. (1997) under both control and low K stress, and one significant QTL for shoot K+ concentration has been identified near the marker C25 on the chromosome 3 under low K condition (Miyamotoa et al. 2012). However, no one K+ concentration QTL correspond to these QTLs was detected under control and low K condition in this study. One possible explanation is the different genetic background between parent cultivars or different culture conditions. In addition, the QTL qcSNC6 for shoot Na+ concentrations was located in the same region with previous report on chromosome 6 under culture solution (Miyamotoa et al. 2012), which suggested that the QTL for shoot Na+ concentration could be stably detected by different mapping populations and environments. For Mg2+ and Ca2+, the concentrations of them in roots and shoots of both parents under low K stress condition were higher than control, but Ca2+ concentrations in roots and shoots of DH lines were lower than control. A total of 27 QTLs for Mg2+ and Ca2+ concentrations were detected under control and low K condition in rice seedling. The QTLs for SCC and RCC under both conditions were not mapped to the same locations and the QTLs associated with SMC and RMC were also different. These QTLs are helpful to understand the interaction among the ions under low K stress.
There was a major locus which associated with cSDW, tTDW, cTDW, RDW and tSCC, and all of these QTLs were mapped on the same region of chromosome 4. The region flanked by the markers RM1112 and RM280 contained the qcSDW4, qtTDW4, qcTDW4, qRDW4 and qtSCC4, and the positive alleles of these QTLs except for qRDW4 were all from TN1. The results indicated that the QTLs in the same region controlling different traits could be closely linked or play a pleiotropic role. Similarly, the region flanked by the markers RM6972 and RM3321 on chromosome 5 contained the qcSDW5, qtSDW5, qcTDW5 and qtTDW5 and all their positive alleles also came from TN1 which suggested that the QTL is a stable locus controlling shoot dry weight and total dry weight under control and low K stress conditions. A pleiotropic region containing qtSDW7, qtTDW7 and qtRL7 between the markers RM3555 and RM1306 on chromosome 7 were detected and the positive alleles of the three QTLs were from CJ06 under low K stress condition. The results indicated that this region of chromosome 7 is important for low K tolerance in rice seedlings and the region may be used as an important target for improving rice low K tolerance. The other similar regions were also found on the rest of the chromosomes including one on chromosome 1 (qtRDW1, qcSCC1 and qtSKC1), one on chromosome 4 (qcRCC4, qtRNC4 and qtSNC4), one on chromosome 5 for qtRDW5, qtRMC5 and qtSKC5 and one on chromosome 10 for qcRCC10, qcSH10 and qSAI10 which indicated these QTLs loci controlled more than one trait.
There was little research for QTL analysis of low K tolerance and related ions concentrations of shoots and roots in rice seedlings. The present results showed that some QTLs under low K stress might exhibit the ability of low K tolerance, and they might be useful in marker-assisted selection (MAS) for developing the low K tolerance rice cultivars. In addition, the results obtained in the current study may be helpful for further understanding of genetic mechanisms of ion absorption on the tolerance for low K tolerance in rice.
Conclusion
In this study, the genetics of low K tolerance and related ions concentrations were investigated in a DH population at the seedling stage and a total of 96 QTLs were identified with 5–29 % phenotypic variation. Among them, there are six major QTLs more than 20 % effects were detected. The qcRNC11 and qtRKC11 were located in the same region between the markers RM286 and RM1812 explaining 20 and 11 % phenotypic variation. In the segment of RM6997-RM252, qcSKC4, qcSMC4 and qTDW4 were identified with the phenotypic variation of 9, 10 and 23 % respectively, and another qcSNC4 was located in the adjacent region with the phenotypic variation of 20 %. The qtSKC5, qtRMC5 and qtRDW5 explaining 20, 9 and 22 % phenotypic variation respectively were detected in the interval of RM6972-RM3321. The QTLs of qcRKC12 and qtRKC12 with 29 and 11 % phenotypic variation were also identified in the same region of RM3226-RM12. These results indicated that these major QTLs have pleiotropic effects in K tolerance and related ions absorption, and the major QTLs of qcSNC4, qcRNC11 and qcRKC12 may be useful for marker-assistant selection in regulating the absorption of Na+ and K+ under the control condition. In addition, the QTL qtSKC5 also could be used in rice molecular breeding in regulating K+ absorption under low K stress condition.
Abbreviations
- DH:
-
Doubled haploid
- QTLs:
-
Quantitative trait loci
- K:
-
Potassium
- RDW:
-
Root dry weight
- SDW:
-
Shoot dry weight
- TDW:
-
Total dry weight
- SH:
-
Shoot height
- RL:
-
Root length
- RNC:
-
Root Na+ concentration
- RCC:
-
Root Ca2+ concentration
- RKC:
-
Root K+ concentration
- RMC:
-
Root Mg2+ concentration
- SNC:
-
Shoot Na+ concentration
- SCC:
-
Shoot Ca2+ concentration
- SKC:
-
Shoot K+ concentration
- SMC:
-
Shoot Mg2+ concentration
- KTC:
-
K tolerance coefficient
- SAI:
-
Synthetic appraisal index
References
Ahmadi N, Negrão S, Katsantonis D, Frouin J, Ploux J, Letourmy P, Droc G, Babo P, Trindade H, Bruschi G, Greco R, Oliveira M, Piffanelli P, Courtois B (2011) Targeted association analysis identified japonica rice varieties achieving Na+/K+ homeostasis without the allelic make-up of the salt tolerant indica variety Nona Bokra. Theor Appl Genet 123:881–895
Armengaud P, Breitling R, Amtmann A (2004) The potassium-dependent transcriptome of Arabidopsis reveals a prominent role of jasmonic acid in nutrient signaling. Plant Physiol 136(1):2556–2576
Cheng L, Wang Y, Meng L, Hu X, Cui Y, Sun Y, Zhu L, Ali J, Xu J, Li Z (2012) Identification of salt tolerant QTLs with strong genetic background effect using two sets of reciprocal introgression lines in rice. Genome 55:45–55
Cho YI, Jiang WZ, Chin JH, Piao Z, Cho YG, McCouch S, Koh HJ (2007) Identification of QTLs associated with physiological nitrogen use efficiency in rice. Mol Cells 23(1):72–79
Dobermann A, Cassman KG, Mamaril CP, Sheehy JE (1998) Management of phosphorus, potassium and sulfur in intensive, irrigated lowland rice. Field Crops Res 56:113–138
Heffer P, Prud’homme M (2010) Fertilizer outlook 2010–2014. In: 78th IFA Annual Conference, Paris
Hu B, Wu P, Liao C, Zhang W, Ni J (2001) QTLs and epistasis underlying activity of acid phosphatase under phosphorus sufficient and deficient condition in rice (Oryza sativa L.). Plant Soil 230:99–105
Koyama ML, Levesley A, Koebner RM, Flowers TJ, Yeo AR (2001) Quantitative trait loci for component physiological traits determining salt tolerance in rice. Plant Physiol 125(1):406–422
Laegreid M, Bockman OC, Kaarstad O (1999) Agriculture, fertilizers and the environment. CABI, Oxon
Lian XM, Xing YZ, Yan H, Xu C, Li X, Zhang Q (2005) QTLs for low nitrogen tolerance at seedling stage identified using a recombinant inbred line population derived from an elite rice hybrid. Theor Appl Genet 112:85–96
Lin HX, Zhu MZ, Yano MJ, Gao P, Liang ZW, Su WA, Hu XH, Ren ZH, Chao DY (2004) QTLs for Na+ and K+ uptake of the shoots and roots controlling rice salt tolerance. Theor Appl Genet 108:253–260
Liu GD, Liu GL (2002) Screening indica rice for K-efficient genotypes. Acta Agron Sin 28(2):161–166
Liu J, Yang X, Yang Y, Wu L (2003) Some agronomic and nutritional characteristics for potassium efficient rice genotypes under low potassium stress. Plant Nutr Fert Sci 9(2):190–195
McCouch SR, Cho YG, Yano M, Paul E, Blinstrub M (1997) Report on QTL nomenclature. Rice Genet Newsl 14:11–13
Miyamotoa T, Ochiaia K, Takeshitaa S, Matoha T (2012) Identification of quantitative trait loci associated with shoot sodium accumulation under low potassium conditions in rice plants. Soil Sci Plant Nutr 58:728–736
Ren ZH, Gao JP, Li LG, Cai XL, Huang W, Chao DY, Zhu MZ, Wang ZY, Luan S, Lin HX (2005) A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat Genet 37:1141–1146
Sogava K, Qian Q, Zeng DL, Hu J, Zeng LJ (2005) Differential expression of whitebacked planthopper resistance in the japonica/indica doubled haploid rice population under field evaluation and seedbox screening test. Rice Sci 12:63–67
Tai D, Zhang X, Su Z, Wang Y, Luo Y, Xia J (2004) Screening for low-kalium tolerance varieties at seedling stage from the core germplasm of integrated international rice molecular breeding program. J Plant Genet Resour 5(4):356–359
Wang Z, Chen Z, Cheng J, Lai Y, Wang J, Bao Y, Huang J, Zhang H (2012) QTL analysis of Na+ and K+ concentrations in roots and shoots under different levels of NaCl stress in rice (Oryza sativa L.). PLoS One 7(12):e51202
Wang G, Lu W, Chen H, Zhang X, Xue D (2015) Seedling screening of rice germplasm resources with low potassium tolerance. J Hangzhou Normal Univ (Nat Sci Ed) 14(1):46–50
Watanabe T, Broadley MR, Jansen S, White PJ, Takada J, Satake K, Takamatsu T, Tuah SJ, Osaki M (2007) Evolutionary control of leaf element composition in plants. New Phytol 174:516–523
White PJ, Karley AJ (2010) Potassium. In: Hell R, Mendel R (eds) Cell biology of metals and nutrients in plants. Springer, Dordrecht, pp 119–224
Wissuwa M, Yano M, Ae N (1998) Mapping of QTLs for phosphorus-deficiency tolerance in rice (Oryza sativa L.). Theor Appl Genet 97:777–783
Wu P, Ni J, Luo A, Jin G, Tao Q (1997) Investigation of QTLs underlying rice tolerance for potassium deficiency via molecular markers. Plant Nutr Fert Sci 3(3):209–217
Wu P, Ni JJ, Luo AC (1998) QTLs underlying rice tolerance to low-potassium stress in rice seedlings. Crop Sci 38:1458–1462
Yang X, Liu J, Wang W, Ye Z, Luo A (2004) Potassium internal use efficiency relative to growth vigor, potassium distribution, and carbohydrate allocation in rice genotypes. J Plant Nutr 27:837–852
Yang J, Hu CC, Hu H, Yu RD, Xia Z, Ye XZ, Zhu J (2008) QTLNetwork: mapping and visualizing genetic architecture of complex traits in experimental populations. Bioinformatics 24:721–723
Yoshida S, Forna DA, Cock JH, Gomez KA (1976) Laboratory manual for physiological studies of rice. Intemational Rice Research Institute, Los Banos, pp 62–63
Zeng DL, Hu J, Dong GJ, Liu J, Qian Q (2009) Quantitative trait loci mapping of flag–leaf ligule length in rice and alignment with gene. J Integr Plant Biol 51(4):360–366
Acknowledgments
This work was supported by Zhejiang Provincial Science and Technology Bureau (2012C22039); National Natural Science Foundation of China (31171535; 31101135); Hangzhou Scientific and Technological Program (20130432B04). The authors are grateful to the editors and the anonymous reviewers for their valuable comments and help.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Yunxia Fang, Weiming Wu and Xiaoqin Zhang have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Fang, Y., Wu, W., Zhang, X. et al. Identification of quantitative trait loci associated with tolerance to low potassium and related ions concentrations at seedling stage in rice (Oryza sativa L.). Plant Growth Regul 77, 157–166 (2015). https://doi.org/10.1007/s10725-015-0047-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10725-015-0047-9