Abstract
Genetic diversity among 115 coffee accessions from the Coffea Germplasm Collection of IAC was assessed using SSR markers. The germplasm represents 73 accessions of Coffea arabica derived from spontaneous and subspontaneous plants in Ethiopia and Eritrea, species center of origin and diversity, 13 commercial cultivars of C. arabica developed by the Breeding Program of IAC, 1 accession of C. arabica cv. ‘Geisha’, 13 accessions of C. arabica from Yemen, 5 accessions of C. eugenioides, 4 accessions of C. racemosa and 6 accessions of C. canephora. Genetic analysis was performed using average number of alleles per locus (A), proportion of polymorphic loci (P), Shannon’s genetic index (H′ and G′ST) and clustering analysis. All evaluated species were distinguished by a cluster analysis based on Jaccard’s coefficient. Differentiation between the cultivated plants of C. arabica and accessions derived from spontaneous and subspontaneous plants was observed. Spontaneous and subspontaneous accessions from Ethiopia were separated according to the geographical origin: east and west of the Great Rift Valley. Cultivated plants showed a low genetic diversity with a division in two groups: accessions from Yemen (H′=0,028) and Brazilian commercial cultivars (H′=0,030). The results agreed with previously reported narrow genetic basis of cultivated plants of C. arabica and supported the hypotheses about domestication of the species. This study also showed a significant genetic diversity among accessions from Ethiopia and Eritrea present in the Germplasm Collection of IAC. This diversity is specially observed in accessions from Sidamo (H′=0,143), Kaffa (H′=0,142) and Illubabor (H′=0,147) indicating their importance as source of genetic variability for coffee breeding programs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Although the genus Coffea contains approximately 80 taxa (Bridson and Verdcourt, 1988), only two species are commercially exploited, Coffea arabica L. and Coffea canephora Pierre ex Froehner. C. arabica is an autogamous species and the only allotetraploid (2n = 4x = 44) of the genus while the other species are diploids (2n = 2x = 22) and generally self-incompatible (Krug and Carvalho, 1951). C. arabica is native of the highlands of Ethiopia (Sylvain, 1955), but there are also records of wild arabica coffee in the Boma Plateau of Sudan (Thomas, 1942) and Mount Marsabit of Kenya (Berthaud and Charrier, 1988).
The arabica coffee cultivation started in Arabia, specifically in Yemen, five centuries ago. In the early 18th century, progenies from a single Indonesia plant cultivated in Europe were spread out to South America and turned out to be the genetic basis of main cultivars of Brazil and other countries (Chevalier and Dagron, 1928; Carvalho, 1945).
In view of this narrow genetic basis and the necessity of genetic resources conservation, FAO organized in 1964–65 a mission to collect spontaneous and subspontaneous coffee germplasm in probable native regions of the species (FAO, 1968). This initiative resulted in 621 samples of seeds from various collecting sites in Ethiopia and the Republic of Eritrea which were carefully documented and sent to six institutions in India, Tanzania, Ethiopia, Costa Rica, Peru and Portugal for further research (FAO, 1968).
The accessions represented in the Coffee Germplasm Collection of Instituto Agronômico de Campinas (IAC) include plant material originated from 308 of those seed sample, obtained from plants of the Centro Agronómico Tropical de Investigación y Enseñanza (CATIE) at Turrialba, Costa Rica, collected in the years 1973 and 1987. There is also a substantial number of cultivated coffee germplasm in the IAC’s collection. Besides this, several samples of plants from Yemen were introduced by A.B. Eskes, a researcher from IRCC, Montpellier, after a detailed morphologic and agronomic characterization (Eskes and Mukred, 1990).
Since their introduction at IAC, all those materials are under continuous evaluation regarding botanic, agronomic, and technologic aspects (Carvalho et al., 1983; Mazzafera et al., 1990; Silvarolla et al., 2000). However, despite all these studies, the degree of genetic diversity and structure of the IAC collection is poorly understood. In order to fulfill this analysis, molecular markers could help reveal the structure of genetic diversity, which is an important information for setting up a core collection and to improve the conservation, accessibility and use of genetic resources of this collection (Hamon et al., 1999).
Previous studies have already evaluated the genetic diversity of some Ethiopian and FAO accessions through morphologic and agronomic characteristics (Montagnon and Bouharmont, 1996), RAPD markers (Lashermes et al., 1996; Anthony et al., 2001; Chaparro et al., 2004; Aga et al., 2003) and ISSR markers (Aga et al., 2005). However, in these studies only few accessions from Yemen were analyzed (Montagnon and Bouharmont, 1996; Lashermes et al., 1996; Anthony et al., 2002) and were reported different results regarding grouping and genetic structure of those accessions collected in different regions from Ethiopia and Yemen. Chaparro et al. (2004) did not found an association between grouping and the site of origin in Ethiopia. Montagnon and Bouharmont (1996) showed that there is a separation between coffee trees growing east and west of the Great Rift Valley, with accessions from south–eastern and southern of Ethiopia being grouped together with cultivated plants from Yemen. Anthony et al. (2001) and Aga et al. (2003, 2005) also distinguished eastern and western groups in accessions from Ethiopia, but with a low genetic differentiation between them and cultivated plants arising as a group apart (Anthony et al., 2001).
Microsatellites markers (SSR) have shown good efficiency to assess genetic diversity and relationships among coffee trees (Moncada and McCouch 2004; Maluf et al., 2005). Thus, we thought SSR could be an alternative tool to elucidate the questions about genetic structure of C. arabica as well to characterize diversity levels of genetic resources included in the Coffea Germplasm Collection of IAC. Furthermore, this is the first study analyzing a representative sample of both materials collected from the center of the origin and diversity of the C. arabica species and cultivated plants from Yemen and Brazil through microsatellites markers.
Material and methods
Plant material
The evaluated plant material consisted of 115 coffee accessions from the Coffea Germplasm Collection of IAC. These included 73 accessions of C. arabica L. derived from spontaneous and subspontaneous trees from various regions of Ethiopia and Eritrea (Fig. 1), 13 accessions of C. arabica from Yemen, 1 accession of C. arabica cv. ‘Geisha’ (Table 1), and 13 commercial cultivars of C. arabica developed by the IAC Breeding Program (Table 2). Also, 5 accessions of C. eugenioides S. Moore, 4 accessions of C. racemosa Lour. and 6 accessions of Coffea canephora Pierre ex Froehner were included as outgroup species due to their importance in the evolutionary history of coffee and as source of target genes for breeding programs. Sampling of spontaneous and subspontaneous accessions from Ethiopia and Eritrea was representative of total accession number at the Germplasm Collection of IAC.
Collecting sites of Coffea arabica accessions evaluated. Ethiopia political organization is according to FAO (1968), except by separation of Eritrea
Genomic DNA extraction
Total genomic DNA was extracted from frozen young leaves according to Stewart Jr. and Via (1993), using CTAB as detergent. All DNA samples were diluted to a final concentration of 20 ng/μl.
SSR amplification
Primer sequences to amplify SSR locus were obtained from Combes et al. (2000) and Rovelli et al. (2000) (Table 3). Final reaction conditions were 40 ng of genomic DNA, 1.5× reaction buffer, 0.1 mmol l−1 of dNTP, 2 mmol l−1 of MgCl2, 5 ρmol of each primer and 1.5 U of Taq DNA polymerase. The complete thermal cycle program was 5 min at 95°C, followed by 30 cycles of 1 min at 95°C, 1 min at 60°C and 1 min at 72°C, and a final 5 min of elongation time at 72°C.
Primers were fluorescently labelled and amplified products were separated on a 5% acrylamide gel using an ABI 377 automated sequencer (Applied Biosystems). Products were detected by the applicative Gene Scan (Applied Biosystems) using internal molecular size markers (GENESCAN-500 ROX), and analyzed by the applicative Genotyper v 2.0 (Applied Biosystems).
Data analysis
Gels were scored by presence or absence of bands or alleles. Due to the tetraploid condition of C. arabica, it is impossible to distinguish between diallelic duplex and simplex and among different types of triallelic combinations of SSR loci. Therefore, for each individual plant, fragment frequencies were analyzed as multilocus fingerprints, in which each allele was either scored present or absent.
Genetic diversity within groups of C. arabica and within diploids species was evaluated by average number of alleles per locus (A), proportion of polymorphic loci (P) and the Shannon’s genetic index (H′) (Bussel, 1999). Groups of C. arabica consisted of accessions of the same collecting region (Table 1) and commercial cultivars developed by IAC. Cultivar ‘Geisha’ was excluded of the genetic diversity analysis because it was represented by only one accession/group. Estimation of A was an exception to the multilocus fingerprints approach once amplified products of each pair of primers were considered as alleles of the same locus. P was calculated dividing the number of polymorphic bands by total number of amplified bands in each group. Shannon’s genetic index for each marker was calculated for each group as:
H′ = - ∑ p i *log2 p i
where p i is the frequency of the presence or absence of a band in that group.
Following the method of Bussel (1999), the partitioning of genetic variation within and between groups of C. arabica was estimated. The average diversity over all populations for each locus (H′pop) and the total diversity in the 99 accessions of C. arabica for each locus (H′sp) were calculated (for more details see Bussel, 1999). Then the component of diversity within populations (H′pop/H′sp) and the component between populations (G′ST = (H′sp–H′pop)/H′sp) were estimated.
H′, H′pop, H′sp and G′ST were the average values per locus calculated over all loci, including monomorphic ones, according to Bussel (1999). We categorized the partitioning of genetic diversity analysis according to the groups of accessions analyzed. These groups included all accessions (Analysis 1), all accessions without Eritrea group (Analysis 2), spontaneous and subspontaneous accessions (Analysis 3) and spontaneous and subspontaneous accessions without Eritrea group (Analysis 4). Eritrea group was excluded from Analysis 2 and 4 to improve understanding of species genetic structure. This was carried out because Eritrea group contains just one individual and this could lead to bias G′ST values by decreasing H′pop.
Genetic distance among all accessions (including C. arabica cv. ‘Geisha’) was estimated as the complement of Jaccard’s (1908) coefficient (Link et al., 1995). Genetic distances were also estimated using Dice coefficient (Dice, 1945), which is equivalent to Nei and Li (1979), in order to compare values with other studies. Cluster analysis was performed using the matrix distance based on the complement of Jaccard’s coefficient employing the UPGMA method. Bootstrap analysis (Felsenstein, 1985) was performed to evaluate the tree topology reliability for 1,000 simulations using the software Treecon (Van de Peer and Watcher, 1994).
Results
The multilocus fingerprints approach was used to analyze the genetic diversity and structure of coffee accessions. In spite of this, specific results for each primers pairs were pointed out for characterization of individual SSR locus.
SSR locus characterization
The E12-3CTG (SSR9) locus showed a profile with 3 and 4 peaks even in diploid species such as C. canephora and C. eugenioides. The 4-1CTG (SSR5) locus was monomorphic (band of 97 bp) in C. arabica and C. eugenioides and did not amplify any fragment in C. racemosa and C. canephora species. Primers E6-3CTG, E8-3CTG and M3 also amplified monomorphic bands within C. arabica accessions (a total of 12 bands), and those were polymorphic among evaluated species.
Genetic diversity
Sixteen SSR primers pairs produced a total of 121 bands or alleles. All tested markers detected polymorphisms among the accessions evaluated, being 54 bands among C. arabica accessions and 15 bands among cultivated plants (Yemen, ‘Geisha’ and Brazilian cultivars). Also, 7 out of 54 polymorphic bands of C. arabica, were present at high frequencies (0.8 or more), 31 as rare alleles in low frequency (0.2 or less) and 16 with frequencies between 0.2 and 0.8.
Genetic diversity analysis showed the highest values of H′ in diploid species (Table 4). Values for A in diploid species were similar or even lower than those observed in C. arabica groups, probably due to the allotetraploid nature of C. arabica which results in duplicate A values per plant. H′ values were relatively high in accessions from Kaffa, Illubabor and Sidamo provinces, moderate in Gojjam and Harar, and very low in cultivated plants. The P index was higher in diploid species and within Kaffa and Illubabor groups. The discrepancy of H′ relative to A and P values in Kaffa and Illubabor groups occurred due to the presence of rare alleles (Table 4).
Genetic distances ranged from 0 to 0.88 between all possible pairs of genotypes, from 0 to 0.37 among C. arabica accessions, from 0 to 0.30 among spontaneous and subspontaneous accessions from Ethiopia and Eritrea, and from 0 to 0.19 among cultivated accessions (Fig. 1). Genetic distances among C. arabica accessions were also calculated using only the polymorphic bands exclusive to C. arabica (44.6% of total fragments). Results showed that this distance ranged from 0 to 0.65 using Jaccard’s coefficient or its complement and from 0 to 0.49 using Dice’s coefficient.
Genetic structure
G′ST values showed a strong genetic structure in all accessions of C. arabica (Table 5). A strong genetic structure was also observed in spontaneous and subspontaneous accessions from Ethiopian. Exclusion of group Eritrea, which contains just one individual, increased H′pop values and decreased G′ST values but yet results showed the strong genetic structure in the evaluated accessions (Table 5).
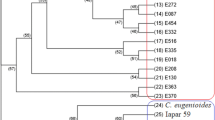
The hierarchical clustering analysis presented in Fig. 2 showed four major clusters comprising grouped accessions of each species. The analysis of species relationships showed C. canephora closer to C. arabica, followed by C. eugenioides. Also, C. racemosa was distantly related to C. arabica.
Dendrogram of the 115 Coffea accessions listed in Tables 1 and 2 based on Jaccard genetic distance obtained from SSR markers using the UPGMA method. Numbers (%) on the branches correspond to bootstrap values above 50% (1,000 replications). Letters indicate the geographical origin of accessions: W (West of the Great Rift Valley) and E (East of the Great Rift Valley)
Grouping of C. arabica accessions (Fig. 2) revealed two main clusters. The first contains only cultivated plants, including the accessions of Yemen, cultivar ‘Geisha’ and commercial cultivars from Brazil. An exception was the accession of Sidamo (SI4) also included in this group with materials from Yemen. Yemen accessions of type Tessawi (Table 1) were distinguished from the others in a separated cluster. The second group encompassed basically all spontaneous and subspontaneous accessions from Ethiopia and Eritrea. Despite the low boostrap values (Fig. 2), there was a clear separation of these accessions in two large subgroups: the first included mainly accessions from Sidamo and the second subgroup included all other accessions from the west side of the Great Rift Valley.
Discussion
SSR locus characterization
In general, the amplification patterns for each SSR locus evaluated corresponded to those previously reported. The profile of E12-3CTG (SSR9) locus with 3 and 4 peaks in the diploid species C. canephora and C. eugenioides confirmed that this pair of primers amplified a number of independent loci (Rovelli et al., 2000). Considering this, E12-3CTG locus has not been used to estimate number of alleles per locus (A).
The locus 4-1CTG (SSR5) was monomorphic in C. arabica and C. eugenioides and did not amplify any fragment in C. racemosa and C. canephora species. According Rovelli et al. (2000) this locus showed diploid type segregation in C. arabica. Thus, it is probable that the amplification has just occurred in putative genome provided by C. eugenioides (Lashermes et al., 1999). However, in other analysis Poncet et al. (2004) using different primer sequences to amplify this same SSR locus identified a monomorphic band of 239 bp also in C. canephora and other diploid species, including C. eugenioides. This new primer pair amplified the total SSR sequence present at GENBANK, while Rovelli et al. (2000)’s primers amplified only part of the SSR sequence.
These results suggested that the 4-1CTG locus is present in C. arabica, C. eugenioides and C. canephora and other Coffea species (see Poncet et al., 2004). However, there is an interspecific polymorphism in the flanking regions of SSR that prevent amplification in C. canephora, C. racemosa and in the putative C. canephora genome of C. arabica (Lashermes et al., 1999). Interestingly, Rovelli et al. (2000) identified heterozygotes in the accessions of C. arabica var. Caturra showing a polymorphism in repeat motifs within cultivated plants of C. arabica that was not observed in our study.
Genetic diversity
The high variability detected in spontaneous and subspontaneous accessions of C. arabica was observed mainly in coffee trees from Sidamo, Kaffa and Illubabor provinces although it also must be noted that 50% of accessions were sampled from Kaffa and Illubabor regions. Significant levels of genetic diversity in coffee plants from Kaffa and Illubabor were also reported by Anthony et al. (2001) and Chaparro et al. (2004). Indeed, the high variability among accessions from these regions in the collections is a consequence of the great effort in collecting samples with as much visual, botanical and agronomical diversity as possible (FAO, 1968). The high genetic diversity detected in Sidamo accessions could be visualized by the proportion of shared alleles among genotypes within both divergent groups of C. arabica, cultivated plants and spontaneous and subspontaneous accessions from Ethiopia.
Comparing the genetic distance values among C. arabica accessions, we found values similar to those reported by Orozco-Castillo et al. (1994), Anthony et al. (2001, 2002), but lower values than Moncada and McCouch (2004). Genetic diversity evaluated by allele distribution (29.6% of alleles with frequencies between 0.2 and 0.8) revealed a diversity degree intermediate between that identified by Anthony et al. (2001) and that found by Chaparro et al. (2004), with 17 and 39.6% of alleles with frequencies between 0.2 and 0.8, respectively.
Proportion of polymorphic loci and Shannon’s index values estimated in this study can be considered in the same range of a similar analysis of arabica coffee in natural populations from Ethiopia (Aga et al., 2003). These authors determined P and H′ values ranging from 37% to 73% and 0.2 to 0.4, respectively, in 9 plants per population. However, P and H′ were calculated based on C. arabica amplified bands while in the present study all amplified bands, including those specific of diploid species and monomorphic to C. arabica were included in the index calculation. On the other hand, if only C. arabica amplified bands were considered for calculating P and H′ values, these would be for Kaffa group, for example, P= 55.6% and H′= 0.328, identical to that observed by Aga et al. (2003). Besides this, observed values of genetic distances among accessions were similar in both studies.
Therefore, these results suggest that there is a significant genetic diversity in the coffee collection of IAC. Furthermore, the genetic diversity estimated for Ethiopian accessions was higher than that of cultivated plants. Thus there is a large variation that can be source for introgression of desirable characteristics in commercial cultivars that has already been used by the coffee breeding program of IAC (Bettencourt and Carvalho, 1968; Moraes et al., 1974; Fazuoli, 1981; Silvarolla et al., 2004).
Genetic structure
Results of amplification of 4-1CTG locus in C. arabica, C. eugenioides and C. canephora and analysis of species relationship observed in this work are in agreement with the hypothesis about the botanic origin of C. arabica species as a natural hybrid between C. canephora and C. eugenioides (Lashermes et al., 1999). Despite contrasting results concerning which of both species is closer to C. arabica, several molecular markers as well as phylogenetics studies have recognized C. canephora and C. eugenioides closer related to C. arabica than other Coffea species (Lashermes et al., 1993, 1997; Cros et al., 1998; Ruas et al., 2000, 2003). These diversity analyses also identified C. racemosa as the most genetically distant species of C. arabica.
According to Bussel (1999) calculation of G′ST using the Shannon’s index are in accordance with GST estimated by other methods, such as AMOVA and modified F-statistics. Also, G′ST is a good estimate to evaluate genetic structure of tetraploid, autogamous collections such as this of C. arabica plants, where heterozygosity cannot be assessed and Hardy–Weinberg equilibrium assumptions cannot be considered. G′ST values of C. arabica observed in this work fitted into expected values based on the breeding system. In that case, according to Bussel (1999), G′ST values for autogamous species from natural populations is around 60% and around 15% for allogamous species.
Results of G′ST for C. arabica accessions associated with cluster analysis indicated a strong genetic structure in the species. Therefore, the genetic diversity was observed among groups rather than within groups. Cluster analysis clearly showed the separation of cultivated plants from Ethiopian and Eritrean spontaneous and subspontaneous accessions. Besides, the analysis allowed distinguishing a morphologic type of Yemen (Tessawi) and Yemen accessions from Brazilian cultivars, although both groups exhibited a very low genetic diversity. These results are in agreement with the well-described narrow genetic basis of cultivated plants of C. arabica (Lashermes et al., 1996; Anthony et al., 2001; Moncada and McCouch 2004; Maluf et al., 2005) and historical data. Brazilian coffee originated from a few plants introduced in the early 18th century and these plants were originated from the first cultivated plants in Yemen (Chevalier and Dagron, 1928; Carvalho, 1945).
Partitioning of genetic diversity of the spontaneous and subspontaneous accessions showed a lower value of G′ST (0.464) than that observed when all accessions were analyzed. However, the G′ST value still indicated a strong genetic structure in these accessions. Hence, accessions of Gojjam were grouped altogether and there was a separation between Sidamo accessions and the others from the opposite side of Great Rift Valley: Kaffa, Gojjam, and Illubabor. However, these grouping associations were not supported by high bootstrap values indicating that the cluster was established based on a few distinct markers.
A genetic structure and low differentiation between southern/south–eastern and south–western coffee trees of Ethiopia were also verified by Anthony et al. (2001) and Aga et al. (2003, 2005). All together, these analyses may support the hypothesis that southern and south–eastern coffee trees of Ethiopia were introduced from the South West (Montagnon and Bouharmont, 1996; Anthony et al., 2001).
Based on morphological and agronomical traits, Montagnon and Bouharmont (1996) were the first to point out a genetic structure in C. arabica accessions, with a division between accessions from west and east side of the Great Rift Valley. However, accessions from east side of Ethiopia were grouped with cultivated plants of Yemen. According to Montagnon and Bouharmont (1996), most of the evaluated characteristics were affected by domestication and the similarity found could be the result from the fact that plants collected from Sidamo, Harar and other provinces of the East are cultivated rather than wild-type once primary forests were eradicated in this region. The authors suggested then two hypotheses to explain the origin of the cultivated plants of C. arabica: eastern plants could have been introduced from the West or could have been selected from wild-type plants situated to the East of the Rift. Nevertheless, the authors claimed unlikely the existence of a single center of wild coffee trees in Ethiopia located west of the Great Rift Valley. They also suggested that arabica plants transferred to Yemen could be originated from south–eastern of Ethiopia.
In the present study, cluster analysis showed accessions from Sidamo closer related to the cultivated plants, with one accession from Sidamo (SI4) being grouped within Yemen group. Similar results were achieved by Moncada and McCouch (2004). It is interesting that the accession from Eritrea and two accessions from Shoa were clustered with Sidamo’s accessions (Fig. 2). According to FAO (1968), E579 (ER) was introduced in Eritrea probably from Yemen, E37 (SH3) was a cultivated plant and E16 (SH1) was also a cultivated plant that is originated from Harar seeds. This observed similarity between accessions from Yemen and from eastern Ethiopia agrees with results from Montagnon and Bouharmont (1996). In the same way, these relationships corroborate the putative separation between Ethiopian accessions from east and west of the Great Rift Valley, but with a low differentiation between both groups (Anthony et al., 2001).
The cultivar ‘Geisha’, derived of plants from Kaffa province (Jones, 1956; FAO, 1968), was grouped with accessions from Yemen and was very distant from Kaffa accessions. This suggests that plants of the same origin were genetically separated by the domestication process and that the West is the primary origin of plants from the East.
Hence, our results suggest that there was a straight evolutionary pathway in domestication of C. arabica trees. The species origin is the Southwest highlands of Ethiopia, and thereafter it was introduced into the South and South East (natural colonization or/and by humans). Later on, some plants were transferred by Arabs to Yemen. After a long period in Yemen, these cultivated plants were spread out around the world. Also, possible occurrences of wild plants in south and south–east of Ethiopia cannot be ruled out.
SSR markers confirmed previously reported phylogenetic relationships among Coffea species as well as they demonstrated to be an efficient method to analyze genetic diversity and structure within C. arabica species. Our results agreed with the well-described narrow genetic basis of cultivated plants of C. arabica and showed a significant genetic diversity of accessions from Ethiopia and Eritrea included in the Coffea Germplasm Collection of IAC. These results indicate the importance of non-cultivated accessions as source of genetic variability for coffee improvement.
References
Aga E, Bryngelsson T, Bekele E, Salomon B (2003) Genetic diversity of forest arabica coffee (Coffea arabica L.) in Ethiopia as revealed by random amplified polymorphic DNA (RAPD) analysis. Hereditas 138:36–46
Aga E, Bekele E, Bryngelsson T (2005) Inter-simple sequence repeat (ISSR) variation in forest coffee trees (Coffea arabica L.) populations from Ethiopia. Genetica 124:213–221
Anthony F, Bertrand B, Quiros O, Wilches A, Lashermes P, Berthaud J, Charrier A (2001) Genetic diversity of wild coffee (Coffea arabica L.) using molecular markers. Euphytica 118:53–65
Anthony F, Combes MC, Astorga C, Bertrand B, Graziosi G, Lashermes P (2002) The origin of cultivated Coffea arabica L. varieties revealed by AFLP and SSR markers. Theor Appl Genet 104:894–900
Berthaud J, Charrier A (1988) Genetics resources of Coffea. In: Clarke RJ, Macrae R (eds) Coffee, vol 4. Agronomy. Elsevier Applied Science, London, pp 1–42
Bettencourt AJ, Carvalho A (1968) Melhoramento do cafeeiro visando resistência à ferrugem. Bragantia 27:35–68
Bridson DM, Verdcourt B (1988) Rubiaceae (Part 2). In: Polhill RM (eds) Flora of Tropical East Africa. Balkema, Rotterdam/Brookfield, pp 703–723
Bussel JD (1999) The distribution of random amplified polymorphic DNA (RAPD) diversity amongst populations of Isotoma petraea (Lobeliaceae). Mol Ecol 8:775–789
Carvalho A (1945) Distribuição geográfica e classificação botânica do gênero Coffea com referência especial à espécie Arabica. Boletim da Superintendência dos Serviços do Café, 21:174–180
Carvalho A, Fazuoli LC, Levy FA, Guerreiro Filho O, Mazzafera P (1983) Observações sobre características dos frutos de introduções de Coffea arabica da Etiópia. In: Resumos. 10o Congresso Brasileiro de Pesquisas Cafeeiras. Poços de Caldas, MG, pp 90–92
Chaparro AP, Cristancho MA, Cortina HA, Gaitán AL (2004) Genetic variability of Coffea arabica L. accessions from Ethiopia evaluated with RAPDs. Genet Resour Crop Evol 51:291–297
Chevalier A, Dagron M (1928) Recherches historiques sur les débuts de la culture du caféier en Amérique. Communications et Actes de l’Académie des Sciences Coloniales, Paris
Combes MC, Andrzejewski S, Anthony F, Bertrand B, Rovelli P, Graziosi G, Lashermes P (2000) Characterization of microsatellites loci in Coffea arabica and related coffee species. Mol Ecol 9:1171–1193
Cros J, Combes MC, Trouslot P, Anthony F, Hamon S, Charrier A, Lashermes P (1998) Phylogenetic analysis of chloroplast DNA variation in Coffea L. Mol Phyl Evol 9:109–117
Dice LR (1945) Measures of the amount of ecologic association between species. Ecology 26:297–302
Eskes AB, Mukred AWO (1990) Coffee survey in PDR Yemen. Colloque Scientifique International sur le Café, 13. ASIC, Paipa, Colombia, p 582
FAO (1968) FAO Coffee mission to Ethiopia 1964–1965. FAO, Rome. p 200
Fazuoli LC (1981) Resistance of coffee to the root-knot nematode species Meloidogyne exigua and M. incognita. Colloque International sur la protection des cultures tropicales, Lyon, France, p 57
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Jones PA (1956) Notes on the varieties of Coffea arabica in Kenya. Coffee Board of Kenya Monthly Bull 21:305–309
Krug CA, Carvalho A (1951) The genetics of Coffea. In: Demerec M (eds) Advances in genetics, vol. 4. Academic Press, New York, pp 127–158
Hamon P, Seguin M, Perrier X, Glaszmann C (eds) (1999) Diversité génétique des plantes tropicales cultivées. CIRAD, Paris, France
Jaccard P (1908) Nouvelle recherches sur la distribution florale. Bull Soc Vaud Sci Nat 44:223–270
Lashermes P, Cros J, Marmey P, Charrier A (1993) Use of random amplified DNA markers to analyze genetic variability and relationships of Coffea species. Genet Resour Crop Evol 40:91–93
Lashermes P, Trouslot P, Anthony F, Combes MC, Charrier A (1996) Genetic diversity for RAPD markers between cultivated and wild accessions of Coffea arabica. Euphytica 87:59–64
Lashermes P, Combes MC, Trouslot P, Charrier A (1997) Phylogenetic relationships of coffee-tree species (Coffea L.) as inferred from ITS sequences of nuclear ribosomal DNA. Theor Appl Genet 94:947–955
Lashermes P, Combes MC, Robert J, Trouslot P, D’Hont A, Anthony F, Charrier A (1999) Molecular characterization and origin of the Coffea arabica L. genome. Mol Gen Genet 261:259–266
Link W, Dixkens C, Singh M, Schwall M, Melchinger AE (1995) Genetic diversity in European and Mediterranean faba bean germ plasm revealed by RAPD markers. Theor Appl Genet 90:27–32
Maluf MP, Silvestrini M, Ruggiero LMC, Guerreiro Filho O, Colombo C (2005) Genetic diversity of cultivated Coffea arabica inbred lines assessed by RAPD, AFLP and SSR marker systems. Sci Agric 62(4):366–373
Mazzafera P, Eskes AB, Parvais J, Carvalho A (1990) Stérilité mâle détectée chez Coffea arabica et C. canephora au Brésil. In: XIII Colloque Scientifique International du Café, vol. 1. Paipa, Colombia, pp 466–473
Moncada P, McCouch S (2004) Simple sequence repeat diversity in diploid and tetraploid Coffea species. Genome 47:501–509
Montagnon C, Bouharmont P (1996) Multivariate analysis of phenotypic diversity of Coffea arabica. Genet Res Crop Evol 43:221–227
Moraes SA, Sugimori MH, Thomazielo FM, Carvalho PTC (1974) Resistência de cafeeiros a Pseudomonas garceae. In: Anais do 2° Congresso Brasileiro de Pesquisas Cafeeiras, Poços de Caldas, MG. Rio de Janeiro, IBC, p 183
Nei M, Li WH (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA 76:5269–5273
Orozco-Castillo C, Chalmers KJ, Waugh R, Powel W (1994) Detection of genetic diversity and selective gene introgression in coffee using RAPD markers. Theor Appl Genet 87:934–940
Poncet V, Hamon P, Minier J, Carasco C, Hamon S, Noirot M (2004) SSR cross-amplification and variation within coffee trees (Coffea spp.). Genome 47:1071–1081
Rovelli P, Mettulio R, Anthony F, Anzueto F, Lashermes P, Graziosi G (2000) Microsatellites in Coffea arabica L. In: Sera T, Soccol CR, Pandey A, Roussos S (eds) Coffee biotechnology and quality. Proceedings of the 3rd International Seminar on Biotechnology in the Coffee Agroindustry, Londrina, Brazil. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 123–133
Ruas PM, Diniz LEC, Ruas CF, Sera T (2000) Genetic polymorphism in species and hybrids of Coffea revealed by RAPD. In: Sera T, Soccol CR, Pandey A, Roussos S (eds) Coffee biotechnology and quality. Proceedings of the 3rd International Seminar on Biotechnology in the Coffee Agroindustry, Londrina, Brazil. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 187–195
Ruas PM, Ruas CF, Rampim L, Carvalho VP, Ruas EA, Sera T (2003) Genetic relationship in Coffea species and parentage determination of interspecific hybrids using ISSR (Inter-Simple Sequence Repeat) markers. Genet Mol Biol 26(3):319–327
Silvarolla MB, Mazzafera P, Lima MMA (2000) Caffeine content of Ehiopian Coffea arabica beans. Genet Mol Biol 23(1):213–215
Silvarolla MB, Mazzafera P, Fazuoli LC (2004) Plant biochemistry: a naturally decaffeinated arabica coffee. Nature 429(24):826
Stewart Jr CN, Via LE (1993) A rapid CTAB DNA isolation technique useful for RAPD fingerprinting and other PCR applications. BioTechniques 14(5):748–750
Sylvain PG (1955) Some observations on Coffea arabica L. in Ethiopia. Turrialba 5:37–53
Thomas AS (1942) The wild arabica coffee on the Boma Plateau, Anglo–Egyptean Sudan. Emp J Exp Agric 10:207–212
Van de Peer Y, Wachter R (1994) TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comp Appl Biosci 10:569–570
Acknowledgements
Special thanks to Luiz Henrique Mazzotini Gomes for drawing the Ehiopia map. This work was supported by The State of São Paulo Research Foundation (FAPESP). Milene Silvestrini was supported by the Brazilian Consortium for Coffee Research and Development (CBPandD). Michele G. Junqueira and Andréa C. Favarin were supported by FAPESP.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Silvestrini, M., Junqueira, M.G., Favarin, A.C. et al. Genetic diversity and structure of Ethiopian, Yemen and Brazilian Coffea arabica L. accessions using microsatellites markers. Genet Resour Crop Evol 54, 1367–1379 (2007). https://doi.org/10.1007/s10722-006-9122-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-006-9122-4