Abstract
Rice (Oryza sativa L.) is widely cultivated around the world and is known to be domesticated from its wild form, O. rufipogon. A loss of seed shattering is one of the most obvious phenotypic changes selected for during rice domestication. Previously, three seed-shattering loci, qSH1, sh4, and qSH3 were reported to be involved in non-shattering of seeds of Japonica-type cultivated rice, O. sativa cv. Nipponbare. In this study, we focused on non-shattering characteristics of O. sativa Indica cv. IR36 having functional allele at qSH1. We produced backcross recombinant inbred lines having chromosomal segments from IR36 in the genetic background of wild rice, O. rufipogon W630. Histological and quantitative trait loci analyses of abscission layer formation were conducted. In the analysis of quantitative trait loci, a strong peak was observed close to sh4. We, nevertheless, found that some lines showed complete abscission layer formation despite carrying the IR36 allele at sh4, implying that non-shattering of seeds of IR36 could be regulated by the combination of mutations at sh4 and other seed-shattering loci. We also genotyped qSH3, a recently identified seed-shattering locus. Lines that have the IR36 alleles at sh4 and qSH3 showed inhibition of abscission layer formation but the degree of seed shattering was different from that of IR36. On the basis of these results, we estimated that non-shattering of seeds in early rice domestication involved mutations in at least three loci, and these genetic materials produced in this study may help to identify novel seed-shattering loci.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Asian cultivated rice, Oryza sativa L., is one of the major crops supporting almost half of the world population and was domesticated from its wild ancestor, O. rufipogon (Oka 1988). During domestication, the loss of seed shattering was a major selected trait because it directly contributes to yield increase (Harlan 1975; Fuller and Allaby 2009). In general, Asian cultivated rice is classified into two types, Japonica and Indica. The former is mainly cultivated in northeast Asia, whereas the latter is widely cultivated in tropical Asian countries (Oka 1988). Most of the Japonica-type cultivated rice has strong non-seed shattering behaviour and few seeds drop from the panicles. But with Indica type, external forces such as hand gripping and hitting against stones will detach mature seeds. These differences in seed-shattering degree may be generated by spontaneous mutations and selected through various harvesting styles. Probably, natural variation of seed-shattering degree is quantitatively regulated by a combination of mutated alleles at shattering loci.
Seed shattering in rice is caused by the formation of an abscission layer that is developed in the basal part of grains. Several genes involved in abscission layer formation were identified through natural variation, mutagenesis and reverse genetic approaches (Dong and Wang 2015). A few major loci for loss of seed shattering selected for in rice domestication or modern breeding have been reported so far. First, sh4 was identified by QTL analysis using an F2 population between Indica-type cultivated rice and wild rice, O. nivara (an annual form of O. rufipogon) (Li et al. 2006a, b). Second, qSH1 was detected using segregating population between Japonica cultivar Nipponbare and Indica cultivar Kasalath (Konishi et al. 2006). For both loci, functional alleles are responsible for the formation and development of abscission layer at the basal part of seeds in the genetic background of cultivated rice. In addition, qSH3 was reported as a result of a QTL analysis between Japonica cultivated rice and wild rice although the effect of qSH3 on seed shattering was weaker than that of qSH1 or sh4 when evaluated with in the genetic background of cultivated rice (Onishi et al. 2007a, b). In our recent study, we also identified qSH3 with segregating population from introgression lines of cultivated O. sativa Japonica Nipponbare with a wild annual accession of O. rufipogon W630 (Htun et al. 2014). We further examined allele effects at three seed-shattering loci in the genetic background of wild rice, and found that the plants only having cultivated alleles at either locus (qSH1, sh4 or qSH3) shed all the seeds because they formed complete abscission layers between the pedicels and spikelets. Mutations at these three loci played roles in the establishment of non-shattering cultivated rice.
Apart from the genes identified from the natural variation of seed-shattering behaviour, some others were identified through mutagenesis and reverse genetic studies. OsCPL1, encoding a protein with a conserved carboxy-terminal domain (CTD) phosphatase domain, was shown to repress differentiation of the abscission layer (Ji et al. 2010). Suppressor mutagenesis analysis of an introgression line having the wild sh4 allele with easily shattering behaviour identified non-shattering mutants. SHAT1, a novel gene encoding an APETALA2 transcription factor, was found to specifying abscission zone development (Zhou et al. 2012). The BEL1-type homeobox gene SH5, a homolog of qSH1, was shown to inhibit lignin biosynthesis and enhance abscission layer development (Yoon et al. 2014). Although these genes are clearly involved in the control of seed shattering, their involvement in a loss of seed shattering during rice domestication is not known.
Among the three loci for the natural variation of seed shattering, a mutation at sh4 was shown to be conserved in all rice cultivars, whereas a loss-of-function mutation at qSH1 was mainly observed in Japonica-type cultivars (Konishi et al. 2006; Lin et al. 2007; Onishi et al. 2007b; Zhang et al. 2009). These findings imply that a non-functional allele at qSH1 was specifically selected in Japonica-type cultivars after the early stages of rice domestication. Regarding qSH3, the genomic region harbouring the locus was shown to have undergone artificial selection in both Indica and Japonica cultivars (Xu et al. 2012), which suggests that qSH3 may be involved in the loss of seed shattering in cultivated rice.
In the present study, we used an Indica cultivar of IR36 having a functional allele at qSH1. It was backcrossed twice with O. rufipogon W630, and backcross recombinant inbred lines (BRILs) at BC2F7 generation were generated. Their seed-shattering behaviour was examined based on the abscission layer formation and seed detachment force in order to estimate the loci involved in non-shattering of seeds in early rice domestication.
Materials and methods
Plant materials
An Indica-type cultivar, O. sativa IR36, and a wild accession, O. rufipogon W630, were used as parental lines for backcross recombinant inbred lines (BRILs). IR36 is a semi-dwarf cultivar with high resistance to major insect pests and diseases, and has contributed to the green revolution (Ballini et al. 2007; Spielmeyer et al. 2002). IR36 has a non-shattering behaviour but mature seeds can be detached using external force. As compared with Japonica cultivars, most Indica rice cultivars show such a characteristic, which arises as a consequence of a partial abscission layer at the basal part of the seeds (Konishi et al. 2006). O. rufipogon W630, an annual wild accession with complete seed-shattering behaviour, originates in Myanmar. Initially, IR36 was crossed twice with O. rufipogon W630, and 10 BC1F1 plants were obtained. They were further backcrossed, and 200 BC2F1 plants (20 plants each from 10 BC1F1 plants) were generated. From these, BRILs at the BC2F7 generation were developed using the single-seed-descendant method. The plants were not artificially selected during the selfing process, but more than 50 lines were eliminated because of weak growth and sterility. As a result, a total of 143 BRILs having wild genetic background were generated.
Evaluation of the seed-shattering degree
It was difficult to evaluate seed-shattering degree using BRILs in the field, because almost all plants of the BRILs shed seeds after maturing. They had long awns at the tip of the seeds and strong wind would cause seed shedding under natural conditions. Therefore, histological analysis was first carried out to observe abscission layer formation at the flowering stage when the first panicle appeared from the node as described by Htun et al. (2014), and the BRILs were classified into two groups; lines with complete and incomplete abscission layer formation. Since the lines with incomplete abscission layer formation had potential to keep mature seeds on the panicles, they were grown in the glasshouse and the seed-shattering degree was evaluated by measuring the breaking tensile strength (BTS) required to detach the seed from the pedicel using a digital force gauge (FGP 0.5, Nidec-Shimpo Co., Japan). Approximately 35 days after flowering, seed-shattering degree was measured with all the mature seeds from five panicles for each BRIL, and their average BTS values were calculated. The BTS value of seeds which had already shed before the day of measurement was scored as “zero gram-force (gf)”.
Marker genotyping and QTL analysis
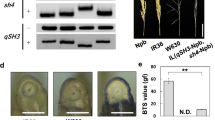
The genomic DNA of 143 BRILs was extracted, and 159 polymorphic simple sequence repeat (SSR) marker loci covering the whole rice genome were surveyed (Supplementary Fig. 1). PCR condition of SSR markers followed Ishikawa et al. (2010). The amplicons were electrophoresed in 4.0% polyacrylamide gels, and banding patterns were visualised by the silver staining method (Panaud et al. 1996). QTL analysis for seed shattering was performed with the abscission layer formation scores; “0” and “1” for partial and complete formation, respectively. A single marker analysis was applied to detect a putative QTL with LOD threshold value of 3.0 using WinQTL cartographer ver. 2.5 (Wang et al. 2012). Genotyping surveys of causative SNPs at sh4 and qSH1 were conducted using derived cleaved amplified polymorphic sequence (dCAPS) markers (Htun et al. 2011). Genotypes at qSH3 were classified into IR36 and W630 types using two flanking polymorphic SSR markers, RM3501 and RM3513 (Inoue et al. 2015).
Results
Seed-shattering characteristics in the parental lines
The wild accession of O. rufipogon W630 has strong seed-shattering behaviour, whereas the Indica cultivar IR36 keeps mature seeds on the panicles unless an external force acts on them. Their seed-shattering characteristics were evaluated by two methods. To begin with, BTS values were measured with mature seeds at 35 days after flowering (Table 1). As for IR36, a BTS value of 50.3 ± 5.9 gf was obtained with 50 randomly chosen seeds each from three panicles. However, the BTS value of W630 could not be determined because all the seeds shed on maturing. We further investigated abscission layer formation of two parental plants at the flowering stage when the first panicle appeared from the node (Fig. 1). Using ten longitudinal sections in the basal part of the pedicels stained with toluidine O solution, the ratio of abscission layer formation was calculated by the length of abscission layer divided by the entire outside length at one side of vascular bundle, which corresponds to the length from the external layer to white triangle (Fig. 1; Table 1). As a result, W630 and IR36 gave 100.0 and 72.5 ± 4.8% of formation ratios, respectively. These results showed that about 30% of non-layer tissues around the vascular bundle in IR36 connected the mature seeds on the pedicels.
Abscission layer formation of a wild rice accession of Oryza rufipogon W630 (a, b) and an Indica cultivar O. sativa IR36 (c, d). Magnified views of the abscission layers in dotted squares of a and c are shown in b and d, respectively. Black triangles indicate both edges of the abscission layer. White triangle indicates the position where the extended edge of abscission layer crossed with vascular bundle. VB vascular bundle. Bars 50 μm
Generation of the BRILs and their abscission layer formation
A total of 143 BRILs at BC2F7 generation between O. sativa IR36 and O. rufipogon W630 were produced. Theoretically, these BRILs are expected to have 12.5% of the IR36 genome in the genetic background of O. rufipogon W630. According to their genotypes at 159 SSR loci covering the whole genome, almost all marker loci showed homozygous alleles of either IR36 or W630. The frequency distribution of the proportion of IR36 genome introgressed into BRILs was calculated (Fig. 2). The introgression percentages ranged from 1.9 to 30.8%, with the mean of 15.3%.
Most of the BRILs showed strong seed-shattering behaviour under natural condition. Therefore we began by observing abscission layer formation of all BRILs using longitudinal sections in the basal part of the pedicels. Sixteen out of 143 BRILs showed partial inhibition of abscission layer formation. These were grown in the glasshouse, and the BTS values were examined. Although one line could not produce enough panicles, the other 15 gave BTS values from 2.2 to 26.9 gf (Supplementary Table 1). The highest value was observed in the A4 line showing BTS values corresponding to almost half of the IR36 value (50.3 gf).
QTL analysis of non-shattering of seeds of IR36
To identify the genomic loci involved in non-shattering of seeds of IR36, QTL analysis was carried out based on the presence or absence of abscission layer formation with 143 BRILs using 159 SSR marker genotypes. A single strong peak was detected near RM5506 on chromosome 4 with an LOD score of 23.9. It explained 53.7% of the total phenotypic variance with the IR36 allele enhancing the BTS value. No other loci were detected above LOD threshold of 3.0, suggesting the QTL on chromosome 4 had a large effect on abscission layer formation between O. sativa Indica IR36 and wild O. rufipogon W630.
Evaluation of the allele effects at the previously reported seed-shattering loci
In the genetic background of cultivars, a seed-shattering locus of sh4 was previously identified in the flanking region of the putative position of the QTL detected in this study (Li et al. 2006a; Onishi et al. 2007a; Lin et al. 2007). We examined the IR36 allele at sh4 using a dCAPS marker detecting the causative mutation (Htun et al. 2011), and found that IR36 had a non-functional allele at sh4 in the same way as the Japonica cultivar Nipponbare (Fig. 3). We carried out QTL analysis again by adding a sh4 dCAPS marker. As a result, a higher peak (LOD = 33.3) was detected with the new marker, indicating that the QTL was identical to sh4 (Table 2). As for qSH1, we could not detect any QTLs on chromosome 1, and IR36 showed the same banding pattern of the qSH1 dCAPS marker as a wild accession of W630 (Fig. 3). In order to examine whether IR36 has a functional allele at qSH1 or not, we further sequenced the coding region of the qSH1 (Os01g0848400) gene, and found that there are no differences in protein sequences between IR36 and W630 (Supplementary Fig. 2). Although the regulatory region, which has not been identified yet, could not be examined, qSH1 is not likely to be responsible for the difference in the seed-shattering characteristics of such degree between IR36 and W630.
We next investigated the genotypes at sh4 and abscission layer formation. Among 143 BRILs, we genotyped all the lines using the dCAPS marker. The results showed that 23 lines carried homozygous IR36 alleles at sh4, and 16 of them gave partial abscission layer formation (Table 3). This indicates that the loss-of-function mutation at sh4 was not sufficient to explain the inhibition of abscission layer formation in the genetic background of wild rice. Probably, other gene(s) may still have been required for non-shattering of seeds of IR36. An additional locus controlling seed shattering is qSH3, which was detected as the region involved in non-shattering of seeds between Japonica cultivar and O. rufipogon (Onishi et al. 2007a, b). Since we previously determined the putative location of qSH3 between RM3513 and RM3601 on chromosome 3 (Inoue et al. 2015), we examined genotypes at qSH3 using these two flanking markers among BRILs (Table 3). We found that all five lines having homozygous IR36 alleles at both sh4 and qSH3 showed the inhibition of abscission layer formation. Partial formation was also observed 11 out of 18 lines with IR36 alleles at sh4 and wild alleles at qSH3. All the lines with wild alleles at sh4 showed complete abscission layer formation. The comparison of genotypes at shattering loci with BTS values for 16 lines with partial abscission layer formation is shown in Supplementary Table 1.
Discussion
Generation of BRILs having IR36 chromosomal segments in the genetic background of wild rice
Most Japonica cultivars show strong non-seed shattering behaviour because they share a loss-of-function mutation at qSH1. Since Indica cultivars have functional alleles at qSH1, their genotypes at shattering loci may be closer to wild forms compared to Japonica cultivars. IR36 is a typical Indica cultivar with non-shattering behaviour. In the present study, we produced BRILs having the chromosomal segments of IR36 in the genetic background of wild rice, O. rufipogon W630. Theoretically, these BRILs have 12.5% of the IR36 chromosome segments in the genome, however, allele survey at 159 SSR loci gave an average of 15.3% of IR36 segments (Fig. 2). Probably, cultivated alleles might have some advantages for growth and fertility in glasshouse condition. Since the BRILs produced in this study have various combinations of cultivated chromosomal blocks in the genetic background of wild rice, they are good materials to analyse interactions between alleles at different loci, especially for domestication-related traits.
Loci involved in non-shattering of seeds in early rice domestication
During rice domestication, a loss of seed shattering has played the most vital role in harvesting efficiency, and has been acquired by selection of natural mutations at seed-shattering loci (Fuller 2007). Among them, sh4 was identified as a major locus for seed shattering in the genetic background of cultivated rice, and its loss-of-function mutation was observed in all cultivars (Li et al. 2006b; Lin et al. 2007; Onishi et al. 2007b). However, in the wild genetic background, we previously found that a near isogenic line carrying the cultivated alleles at sh4 showed strong seed-shattering behaviour with complete abscission layer formation, just as wild rice does (Ishii et al. 2013; Htun et al. 2014; Inoue et al. 2015). This finding suggested that the mutation at sh4 was not sufficient to confer the phenotypic change on the abscission layer. In support of this, some strains of weedy rice were reported to show complete seed shattering despite carrying the non-functional allele at sh4 (Thurber et al. 2010). Moreover, a recent study showed that some accessions of wild rice have a non-functional allele at sh4 with complete seed shattering (Zhu et al. 2012). Therefore, some other gene(s) may have been required to cause non-shattering of seeds in early rice domestication. In the present study, we only detected a strong QTL for abscission layer formation in the same region of sh4. Since IR36 has non-functional allele at sh4, the QTL must be identical to sh4. Using BRILs, chromosomal blocks were surveyed to detect interaction with sh4 for abscission layer formation. As a result, a combination of IR36 alleles at sh4 and qSH3 was found to cause inhibition of abscission layer formation (Table 3). The BRILs having homozygous IR36 alleles at both sh4 and qSH3 gave BTS values ranging from 10.4 to 26.9 gf (Supplementary Table 1). These values are not sufficient to keep mature seeds on the pedicels because of the long awns, but we could see a few non-dispersed seeds on the panicles of A4 (26.9 gf) and H6 (17.5 gf) lines under natural condition. Interestingly, a recent study reported that the genomic region around qSH3 had undergone strong artificial selection in both Indica and Japonica cultivated rice (Xu et al. 2012). This suggests that cultivated alleles at qSH3 might have an important role for non-seed shattering. As for BRILs having IR36 alleles at sh4 and wild alleles at qSH3, 11 out of 18 lines showed partial abscission layer formation, indicating the existence of additional loci involved in non-shattering of seeds. Considering the above, primitive rice farmers may have selected non-shattering plants through a combination of mutations at major locus of sh4 and at some minor loci including qSH3 in early rice domestication. Therefore, we estimated that non-shattering of seeds in early rice domestication involved mutations in at least three seed-shattering loci.
Utilization of minor loci in the control of the seed-shattering degree of cultivated rice
Generally Indica rice shows partial abscission layer formation that results in a loss of the yield in tropical Asian countries. In addition, seeds dispersed onto or around paddy fields may germinate in the next cropping season. If different cultivars are grown in the same paddy field, a quality or a value of harvested rice will decrease because of the variety mixture. Therefore, control of the degree of seed-shattering is still important in tropical Asian countries to maximise the yield potential and seed quality. To optimise the degree of seed-shattering, a combination of the alleles at seed-shattering loci may be useful for breeding purposes in the future. Because the non-functional allele at sh4 is fixed in all cultivated rice, a combination of non-functional alleles at qSH1 and/or minor loci may help to fine-tune the degree of seed-shattering.
References
Ballini E, Berruyer R, Morel JB, Lebrun MH, Nottéghem JL, Tharreau D (2007) Modern elite rice varieties of the ‘Green Revolution’ have retained a large introgression from wild rice around the Pi33 rice blast resistance locus. New Phytol 175:340–350
Dong Y, Wang YZ (2015) Seed shattering: from models to crops. Front Plant Sci 6:476
Fuller DQ (2007) Contrasting patterns in crop domestication and domestication rates, recent archaeobotanical insights from the Old World. Ann Bot 100:903–924
Fuller DQ, Allaby R (2009) Seed dispersal and crop domestication: shattering, germination and seasonality in evolution under cultivation. In: Ostergaard L (ed) Annual plant reviews, fruit development and seed dispersal, vol 38. Wiley-Blackwell, Oxford, pp 238–295
Harlan JR (1975) Crops and man. American Society of Agronomy, Madison
Htun TM, Ishii T, Ishikawa R (2011) Temporal changes of seed-shattering degree of substitution lines having non-shattering alleles from cultivated rice (Oryza sativa) in the genetic background of wild rice (O. rufipogon). J Crop Res 56:39–44
Htun TM, Inoue C, Chhourn O, Ishii T, Ishikawa R (2014) Effect of quantitative trait loci for seed shattering on abscission layer formation in Asian wild rice Oryza rufipogon. Breed Sci 64:199–205
Inoue C, Htun TM, Inoue K, Ikeda K, Ishii T, Ishikawa R (2015) Inhibition of abscission layer formation by an interaction of seed-shattering loci, sh4 and qSH3, in rice. Genes Genet Syst 90:1–9
Ishii T, Numaguchi K, Miura K, Yoshida K, Thanh PT, Htun TM, Yamasaki M, Komeda N, Matsumoto T, Terauchi R, Ishikawa R, Ashikari M (2013) OsLG1 regulates a closed panicle trait in domesticated rice. Nat Genet 45:462–465
Ishikawa R, Thanh PT, Nimura N, Htun TM, Yamasaki M, Ishii T (2010) Allelic interaction at seed-shattering loci in the genetic backgrounds of wild and cultivated rice species. Genes Genet Syst 85:265–271
Ji H, Kim SR, Kim YH, Kim H, Eun MY, Jin ID, Cha YS, Yun DW, Ahn BO, Lee MC, Lee GS, Yoon UH, Lee JS, Lee YH, Suh SC, Jiang W, Yang JI, Jin P, McCouch SR, An G, Koh HJ (2010) Inactivation of the CTD phosphatase-like gene OsCPL1 enhances the development of the abscission layer and seed shattering in rice. Plant J 61:96–106
Konishi S, Izawa T, Lin SY, Ebana K, Fukuta Y, Sasaki T, Yano M (2006) An SNP caused loss of seed shattering during rice domestication. Science 312:1392–1396
Li C, Zhou A, Sang T (2006a) Genetic analysis of rice domestication syndrome with the wild annual species, Oryza nivara. New Phytol 170:185–193
Li C, Zhou A, Sang T (2006b) Rice domestication by reducing shattering. Science 311:1936–1939
Lin Z, Griffith ME, Li X, Zhu Z, Tan L, Fu Y, Zhang W, Wang X, Xie D, Sun C (2007) Origin of seed shattering in rice (Oryza sativa L.). Planta 226:11–20
Oka HI (1988) Origin of cultivated rice. Elsevier, Amsterdam
Onishi K, Horiuchi Y, Ishigoh-Oka N, Takagi K, Ichikawa N, Maruoka M, Sano Y (2007a) A QTL cluster for plant architecture and its ecological significance in Asian wild rice. Breed Sci 57:7–16
Onishi K, Takagi K, Kontani M, Tanaka T, Sano Y (2007b) Different patterns of genealogical relationships found in the two major QTLs causing reduction of seed shattering during rice domestication. Genome 50:757–766
Panaud O, Chen X, McCouch SR (1996) Development of microsatellite markers and characterization of simple sequence length polymorphism (SSLP) in rice (Oryza sativa L.). Mol Gen Genet 252:597–607
Spielmeyer W, Ellis MH, Chandler PM (2002) Semidwarf (sd-1), “green revolution” rice, contains a defective gibberellin 20-oxidase gene. Proc Natl Acad Sci USA 99:9043–9048
Thurber CS, Reagon M, Gross BL, Olsen KM, Jia Y, Caicedo AL (2010) Molecular evolution of shattering loci in U.S. weedy rice. Mol Ecol 19:3271–3284
Wang S, Basten CJ, Zeng ZB (2012) Windows QTL cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh, NC. http://statgen.ncsu.edu/qtlcart/WQTLCart.htm
Xu X, Liu X, Ge S, Jensen JD, Hu F, Li X, Dong Y, Gutenkunst RN, Fang L, Huang L, Li J, He W, Zhang G, Zheng X, Zhang F, Li Y, Yu C, Kristiansen K, Zhang X, Wang J, Wright M, McCouch S, Nielsen R, Wang J, Wang W (2012) Resequencing 50 accessions of cultivated and wild rice yields markers for identifying agronomically important genes. Nat Biotechnol 30:105–111
Yoon J, Cho LH, Kim SL, Choi H, Koh HJ, An G (2014) The BEL1-type homeobox gene SH5 induces seed shattering by enhancing abscission-zone development and inhibiting lignin biosynthesis. Plant J 79:717–728
Zhang LB, Zhu Q, Wu ZQ, Ross-Ibarra J, Gaut BS, Ge S, Sang T (2009) Selection on grain shattering genes and rates of rice domestication. New Phytol 184:708–720
Zhou Y, Lu D, Li C, Luo J, Zhu BF, Zhu J, Shangguan Y, Wang Z, Sang T, Zhou B, Han B (2012) Genetic control of seed shattering in rice by the APETALA2 transcription factor SHATTERING ABORTION1. Plant Cell 24:1034–1048
Zhu Y, Ellstrand NC, Lu BR (2012) Sequence polymorphisms in wild, weedy, and cultivated rice suggest seed-shattering locus sh4 played a minor role in Asian rice domestication. Ecol Evol 2:2106–2113
Acknowledgements
We thank Dr. Cristina Castillo for her critical reading and editing of our manuscript. The wild rice accession, O. rufipogon W630, used in this study was provided by the National Institute of Genetics supported by the National Bioresource Project, MEXT, Japan. This study was supported in part by Grants-in-Aid from the Japan Society for the Promotion of Science (JSPS) to T.I. (No. 26292004) and R.I. (No. 26450003) and by the JSPS Bilateral Open Partnership Joint Research Project to R.I.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ishikawa, R., Nishimura, A., Htun, T.M. et al. Estimation of loci involved in non-shattering of seeds in early rice domestication. Genetica 145, 201–207 (2017). https://doi.org/10.1007/s10709-017-9958-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10709-017-9958-x