Abstract
Rice seed storability is an important characteristic of seed quality so that the cultivars with strong seed storability are expected in the production of hybrid seeds. Presently, little is known about the genetic and physiological mechanisms controlling rice seed storability. In this study, a double haploid population derived from the cross between a japonica cultivar CJ06 and an indica cultivar TN1 was used to identify the quantitative trait loci (QTLs) for seed germination percentage (GP) and fatty acid content (FA) during natural storage or artificial aging. A total of 19 QTLs, including ten QTLs for GP and nine QTLs for FA, were identified on nine chromosomes with the phenotypic variations ranged from 2.1 to 22.7%. Besides, six and three pairs of epistatic interactions were identified for GP and FA, respectively. Moreover, qGP-9, a QTL for germination percentage, was delimited in an interval of 92.8 kb between two STS markers P6 and P8, which contains 15 putative open reading frames. These results provide important information for understanding the genetic mechanisms on rice seed storability, and will be useful for breeding new rice varieties with high seed storability.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hybrid rice has been cultivated commercially since 1976 and has covered more than 50% of the total rice planting areas in China since 1988 (Lu et al. 2013). However, if an air-conditioned room is unavailable, it will be problematic to maintain hybrid rice seeds vigor and viability. As a result, rice seeds become more sensitive to the stresses during germination, and will ultimately die (Nguyen et al. 2012). Owing to the great importance of hybrid rice in grain production, rapid development of hybrid rice planting areas, complexity of hybrid rice seeds and unpredictability of seed market, a substantial amount of hybrid rice seeds should be controlled and regulated as a whole among different years and regions. Therefore, it often happens that a great quantity of hybrid rice seeds is discarded due to their poor storability or improper seed storage measures, which thus seriously affect rice production. Nevertheless, the influencing factor of seed storability is more complex than us imagines. It is urgent to breed the hybrid rice genotypes with their seeds tolerating long time storage or having better storability.
Seed storability is known to be affected by many factors, e.g., genetic background, the environment during seed development and storage stages, leading to the great difficulty in genetic analysis for seed storability (Wang et al. 2010). During rice seed storage, the lipids and lipids-degrading enzymes in bran and germ will cause lipid degradations (Suzuki 2011). Especial to high temperatures condition, a rapid lipid hydrolysis and following an oxidative rancidity will be induced (Goffman and Bergman 2003; Maraschin et al. 2008). Lipids degradations of the grains during storage are usually initiated by hydrolysis of lipids by lipases and free fatty acids are released. The tracing FAs accumulation is able to assess the degradative reactions of lipid hydrolysis. Thus, the low FA is more desirable than high FA to seed storage.
With the technical developments on DNA molecular marker and genome mapping, the quantitative trait loci (QTLs) for seed vigor, seed longevity and other seed quality-related characteristics have been reported. Miura et al. (Miura et al. 2002) discover three putative QTLs for seed longevity, qLG2, qLG4 and qLG9, using 98 backcross inbred lines. Sasaki et al. (2005) identify 12 QTLs for seed germination and three QTLs for seed longevity in a recombinant inbred line (RIL) population derived from the combination of Milyang 23 and Akihikari. Zeng et al. (2006) detect three QTLs for seed storability using a set of 127 double haploid (DH) lines derived from the anther culture of a typical indica/japonica hybrid, ‘ZYQ8’/‘JX17’. Xue et al. (2008) map three QTLs for seed storability based on relative GP using RILs derived from a cross between japonica cultivar Asominori (Oryza sativa L.) and indica cultivar IR24 (Oryza sativa L.). Wang et al. (2010) discover three QTLs for rice seed vigor in the germination stage, and 7.5–68.5% variations can be explained by an individual QTL. Li et al. (2012) identify six QTLs for seed storability under natural aging. Among them, qSS9 is most stable and detected in seeds from all environments and storage periods.
Although there are a few reports related to the QTLs for rice seed storability, the physiological or molecular mechanisms underlying rice seed storability are still unclear. In the present study, the QTLs for germination percentage (GP) and fatty acid content (FA) under natural storage and artificial aging were detected using the DH population derived from a cross between the indica variety TN1 and the japonica variety CJ06. The qGP-9 was located in an interval of 92.8 kb on chromosome 9. Our results provide new insights into the genetic mechanism controlling seed storability in rice. In addition, the QTLs reported here will be useful for marker-assisted breeding to achieve high seed storability.
Materials and Methods
Plant Materials and Construction of Chromosome Segment Substitution Lines (CSSLs)
A DH population consisting of 120 lines was developed via anther culture of an F1 hybrid between the japonica cultivar CJ06 and the indica cultivar TN1. A uniform amount of seeds in all DH lines and their parents were soaked in distilled water in dark at 30 °C for 2 days, followed by germination in distilled water at 35 °C for 12 h. The germinated seeds were sown in the paddy field in Hangzhou. The seedlings at the fifth leaf stage were transplanted to field. Each DH line and their parents were planted in four rows with six plants in each row (planting density, 20 cm × 20 cm).
To obtain a relatively simple genetic background and to weaken or remove the impacts from other minor QTLs on target traits, we constructed the CSSLs related to seed germination percentage (GP). DH population was screened by markers at both sides of GP-related QTL peak values. Individual plant with TN1 genotype was continuously backcrossed to CJ06. After each backcross, target plants were screened using molecular markers. After the continuous backcross for four generations and selfing, plant genetic background would become relatively simple, beneficial for further fine mapping. After the continuous backcross for four generations and selfing, we used 71 microsatellite markers (SSRs) to initially detect and select the target genetic backgrounds in BC4F2 lines (Table 1). When PG-related QTLs were detected, two STS markers P1 and P2 at both sides of peak values were used to identify TN1 genotype in backcross generations.
Generation of Seeds for Storability Test
The period with 90–100% of grains turning yellow and hard was defined as the fully mature stage. All seeds were harvested at fully mature stage, kept in oven at 50 °C for seven days to break seed dormancy (Zeng et al. 2006), and then dried at 30 °C until seed water content dropped to 11–13%. Subsequently, some seeds were stored at room temperature for 6 months for testing natural aging. The others were stored under dry conditions at 4 °C for artificial aging.
Aging Treatment
Artificial aging treatment was based on an improved method originally proposed by Zeng et al. (2002). The sample seeds were treated at high temperature (40 °C) and high relative humidity (95%) (high T and RH) for 10 days in a KCH-1000 thermostatic moisture regulator (Tokyo Rikakikai Co., Ltd., Tokyo, Japan).
Seed Germination Test
Some seeds for natural aging and artificial aging tests were germinated on paper toweling according to Zeng et al. (2006). The germination was conducted at 30°C, 100% RH under 8 h of light per day with the light intensity of 15,000 l× in an FLI-301 N incubator (Japan). The number of germinated seeds was counted after 10 days. A total of 50 seeds from each DH line were used in each of the three experimental replicates.
Fatty Acids Detection
Fatty acids were detected by titration (Xu et al. 2007). The brown rice under natural aging and artificial aging were grounded into rice flour. Ten gram rice flour was added in 50 mL petroleum benzin in a conical flask with cover. Flasks were oscillated on oscillator for 10 min and then allowed to stand for 7 min. The suspending liquid was collected by filtering. A total of 25 mL filtrate was pipetted into a triangular flask, followed by the addition of 75 mL 50% ethanol and 2–3 drips of 1% phenolphthalein indicator, and then immediately titrated with 0.01 M KOH + 95% ethanol until it became reddish and the color did not fade. Fatty acids content (FA) was represented by the amount of KOH needed for neutralizing the fatty acid in 100 g flour sample.
QTL Mapping and Data Analysis
Population distribution and correlation analysis were performed using the SPSS statistical software package (p < 0.05). A genetic linkage map was constructed using a total of 227 SSR and STS markers distributed evenly on all 12 rice chromosomes, as previously described (Rao et al. 2010). The QTLs for GP and FA were analyzed. The QTLs and epistatic effects in DH population were mapped by the software QTL Network version 2.0 (Yang and Zhu 2005) based on the mixed linear model (Wang et al. 1999). Target QTLs were identified when p < 0.005. The genetic parameters, additive effects and the accounted variations of each QTL were estimated. The relative contribution of a genetic component was calculated by the proportion of phenotypic variance explained by that component in the selected model (McCouch et al. 1997).
For fine mapping of qGP-9, the STS and CAPS makers were developed based on the sequence differences between indica var. 93-11 and japonica var. Nipponbare (http://www.ncbi.nlm.nih.gov). These developed markers were further used to distinguish the polymorphism between CJ06 and CSSL4-2. The polymorphic markers were used for the fine mapping of qGP-9. Primers were designed using the software Primer Premier 5.0. The PCR procedure for mapping was as follows: denaturation at 94 °C for 4 min, followed by 35 cycles of 98 °C for 10 s, annealing for 30 s (annealing temperature determined by primer pair sequence), 72 °C for 30 s, and a final extension step at 72 °C for 10 min. The PCR products were analyzed on 4% agarose gels. The developed makers in this study were shown in Table 2.
Results
The Performances of GP and FA in DH Population and Their Parents
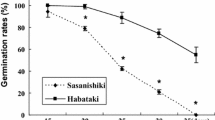
The GPs and FAs of parents and all DH lines during natural storage and artificial aging are shown (Fig. 1; Table 3). After the natural storage at room temperature for 12 months, the FA in TN1 was drastically lower than those of CJ06, whereas the GP in TN1 was higher than that of CJ06. It indicates that the stronger lipids degradations in CJ06 than that in TN1. Moreover, there were considerable differences in GPs among DH lines which were 64.6% on average and ranged from 15.7 to 92.0%. The FAs in DH lines were 264 mg kg−1 on average and ranged from 20 to 646 mg kg−1.
After artificial aging under high temperature and high relative humidity (high T and RH), GP and FA showed considerable differences in DH population and their parents. The GP of CJ06 decreased by 61.2% (from 91.7% at room temperature to 30.5% at high T and RH), while the GP of TN1 only decreased by 12.4% (from 94.9% at room temperature to 82.5% at high T and RH). These results imply that the seeds of TN1 have significantly better storability than those of CJ06. The FA was increased by 32.3 mg KOH/100 g in CJ06 and 19.7 mg KOH/100 g in TN1 after artificial aging treatment. Correspondingly, the changes of GP and FA in DH population also showed considerable differences. The average GP in DH population was reduced by 40.5% (from 64.6% at room temperature to 24.1% at high T and RH), while FA was increased by 4.5 mg KOH/100 g (from 26.4 mg KOH/100 g at room temperature to 30.9 mg KOH/100 g at high T and RH). The two parents showed significant differences in GP and FA between natural aging (room temperature) and artificial aging (high T and RH) treatment. The seed properties of DH population displayed normal distribution under both storage environments.
QTL Mapping for GP and FA
A total of 19 QTLs, including ten QTLs for GP and nine QTLs for FA, were identified on nine chromosomes under natural storage and artificial aging (Fig. 2, Table 4). Among the main-effect QTLs, additive heritability ranged from 2.1% to 22.7%, with the additive effects of 4.6–19.86% on GP and 4.0–15.1%, with the additive effects of 23.4–58.3 mg KOH/100 g on FA. 6 QTLs for GP were detected on chromosome 1, 2, 3, 9, 11 and 12 at room temperature, while 4 QTLs were identified on chromosome 1, 3, 8 and 9 at high T and RH. Among them, qGP-1, qGP-3 and qGP-9 were identified consistently either at room temperature or at high T and RH. The qGP-1 allele from CJ06 advanced seed germination under both storage conditions, while the qGP-9 allele from TN1 increased GP by approximate 12.64% at room temperature and 19.86% at high T and RH. The qGP-3 allele from TN1 also improved seed germination under both storage conditions. Unfortunately, the major QTL for GP, qGP-12 on chromosome 12, was only detected at room temperature, which explained approximate 22.7% variations with the additive effects of 15.28%. In contrast, qGP-8 was only found at high T and RH. These results showed that qGP-12 and qGP-8 were possibly expression in special environments.
Four and five QTLs for seed FA were located to chromosomes 1, 4, 6, 8, and 9 under natural storage and artificial aging respectively. These QTLs explained 4.0–15.1% phenotypic variations with seed FA ranging from 23.4 to 58.3 mg KOH/100 g. Among them, the QTL on chromosome 8 was only identified at room temperature, which contributed to 4.0% of seed FA. It is suggested that seed storage conditions affected fatty acids content in rice seed. qFA-1, qFA-4, qFA-6 and qFA-9 located on chromosome 1, 4, 6, and 9 were all detected under both storage conditions, suggesting that these four QTLs were constitutively expressed and associated with seed FA. The flanking between RM7364 and RM3700 on chromosome 9 was identified two main-effect QTLs for FA under room temperature and high T and RH, which explained 10.7 and 15.0% of the phenotypic variance under two different condictions, respectively. It indicates that ‘CJ06’ allele at the locus of qFA-9 also could increase the phenotypic values of fatty acid content whether at room temperature or high T and RH. Interestingly, the location between RM7364 and RM3700 on chromosome 9 significantly affected GP as well. Nevertheless, the allele from CJ06 at the similar region reduced rice seed germination.
Epistatic Interaction Loci for GP and FA
Four and two pairs of epistatic loci were identified for GP and FA at natural storage, whereas two and one pairs of epistatic loci were detected for GP and FA at high T and RH. These epistatic loci explained 2.39–10.81% of the phenotypic variance, with the additive effects of 4.26–10.58% on GP and 25.7–37.5 mg KOH/100 g on FA (Fig. 2; Table 5). Four pairs of epistatic loci were found for GP at room temperature with about 23% total phenotypic variation. Among them, the digenic interaction between chromosome 7 and chromosome 12 was estimated to explain 10.88% of the total phenotypic variation under natural storage. Two pairs of epistatic loci were detected for GP with 5.7% phenotypic variation with high T and RH.
Development of the Chromosome Segment Substitution Lines for qGP-9
Based on the QTL analysis for qPG9, the flanking between RM7364 and RM3700 on chromosome 9 was similarly identified under two different conditions. The allele from TN1 could increase GP about 19.8%, and explain 22.3% of the GP variations with the LOD score of 4.83 at artificial aging (Fig. 3). One line from the DH population was selected to cross with CJ06. Synchronously, the genotype screening with RM7364 and RM3700 were performed, and the plant carried the TN1 genotype in the flanking between RM7364 and RM3700 was selected to backcross with CJ06.
Followed by three successive backcrosses to CJ06, the BC3F2 generation was scanned with a set of SSR markers, which were uniformly distributed on the previous linkage map. One plant was selected, CSSL4-2, which carried a homozygous introgression from TN1 across the entire qGP-9 region in the CJ06 genetic background and was devoid of other QTLs in the region (Fig. 4a, b). In detail, we discovered that CSSL4-2 carried TN1 fragments at the end of the chromosome 3 and 10, besides the qGP-9 fragment derived from TN1. Base on the results of QTL analysis, there was no GP-related QTL at the end of chromosome 3 and 10. A comparison was conducted on GP during artificial aging. The GP of CSSL4-2 was 60.2%, much higher than and about twice as the 30.5% GP in its recurrent parent CJ06 (Fig. 4c). However, the GP of CSSL4-2 was still lower than the 82.5% GP in its donor parent TN1, which might be due to the existence of other QTLs affecting seed storability in TN1.
Fine Mapping of qGP-9
The segregation population derived from a residual heterozygous line in the BC3F2 generation with heterozygosity only in the qGP-9 region was used for fine mapping. The SSR marker RM7364 on one side of the qGP-9 target region and RM3700 on the other side were used to identify recombination break points. About 1800 F2 plants were used to screen for recombinants and 264 individuals were selected and selfed to generate a homozygous genotype. These seeds were harvested to examine seed storability. After artificial aging treatment, the GPs of these individuals displayed a clear bimodal distribution with the separation ratio close to 3:1 and were classified into low and high germination percentage groups. Moreover, the high GP plants were obviously more than the low GP plants (Fig. 5). The 55 plants with the minimum GP (≤35%) were selected for further gene mapping. Among them, 21 individuals with chromosome recombination were identified by the marker RM7364 on one side of qGP-9, and 34 individuals with chromosome recombination were identified by the marker RM3700 on the other side of the qGP-9.
The 15 developed Indel (insertion/ deletion) markers were selected to scan for polymorphisms between CJ06 and CSSL4-2, and ten polymorphic markers were available to narrow down the region of the qGP-9 locus (Table 2). P1 detected 5 recombinants, whereas Indel marker P7 co-segregated with qGP-9. The markers P4, P5, and P6 revealed 3, 1, and 1 recombinants, respectively, on one side. While markers P8, P9, P10, and P2 indicated 1, 1, 3, and 3 recombinants, respectively, on the other side (Fig. 6). Therefore, the qGP-9 locus was finally delimited to an approximately 92.8 kb DNA region between the two Indel markers.
This 92.8 kb region of the Nipponbare rice genome retrieved from the Rice Genome Annotation Project database (http://rice.plantbiology.msu.edu/annotation_pseudo_current.shtml) encoded 15 genes in its annotation (gene IDs from LOC_Os09g17690 to LOC_Os09g17830). Five candidate genes, LOC_Os09g17730, LOC_Os09g17740, LOC_Os09g17750, LOC_Os09g17810 and LOC_Os09g17830, were predicted to be proton pump interactor, chlorophyll A–B binding protein, oxidoreductase, leucine zipper protein-like and protein transport protein Sec61 alpha subunit, respectively (Table S1). One candidate gene encodes a rice transposon-related protein and four encode rice retrotransposon-related proteins; the others are unknown proteins. These results can provide important information for better understanding the genetic mechanisms controlling rice seed storability, and will be useful for breeding new rice varieties with high seed storability.
Discussion
By comparing GP of rice seeds with and without artificial aging, 6 and 4 QTLs were respectively identified under the two storage conditions. Among them, qGP-1, qGP-3 and qGP-9 were identified both at room temperature and high T and HR. Three QTLs on chromosomes 2, 11 and 12 were detected only at room temperature while one QTL was only identified at high T and HR. Li et al. (2012) detect 6 QTLs for seed storability under room temperature conditions after storing for 32 and 48 months. Among them, qSS9 is the most stable QTL for seed storability at all environments and storage periods and located between R10783S and R1751 on chromosome 9. We also found that qGP-9 was a stable QTL for rice germination whether at natural condition or under high temperature & relative humidity. The delimited 92.8 kb region of qGP-9 was located in the flank of qSS9 by Li et al. (2012). Li et al. (2012) also map two QTLs for seed storability on chromosomes 2 and 11, which are close to qGP-2 and qGP-1, respectively. These overlapping and adjacent regions for seed storability suggest the universal regulatory mechanism of seed storability in rice. Additionally, the main QTL qGP-12 on chromosome 12 has an additive effect of 15.28%, which is detected for the first time. The QTL qGP-9 on chromosome 9 can be considered as a stable QTL related to seed storability. The location of qGP-9 coincides with that reported by Miura et al. (2002) and Zeng et al. (2006), suggesting that this region may accommodate an anti-stress QTL. In our report, qGP-9 was first fine-mapped in 92.8 kb region between P6 and P8, and then 19 candidate open reading frames were predicted. Other studies have reported that there is a main QTL for seed germination capacity on chromosome 2 (Wang et al. 2010), and it shows high consistency with the region holding qGP-2. Seed germination capacity, fat content and storage proteins are all the important factors affecting rice seed vigor (Ren et al. 2014). Isolating and cloning these stable QTLs can guide the breeding for rice seeds with high storability.
GP is the most important and easiest ponderable indicator for rice seed storability. The first report on rice seed longevity was published in 2002 by Miura. It locates three QTLs controlling rice seed longevity on chromosome 2, 4 and 9, respectively (Miura et al. 2002). Thereafter, a succession of experiments has been conducted on identifying the QTLs related to seed longevity, seed storability and seed dormancy (Shen et al. 2005; Zeng et al. 2006; Whitehouse et al. 2015; Wang et al. 2011; Ellis and Hong 2006; Sasaki et al. 2013). In fact, the physiological and agronomical mechanisms controlling seed storability, seed longevity and seed dormancy have already been reported in the 1990 s (Ellis and Hong 1994; Zhang et al. 1998; Kameswara and Jackson 1997; Powell and Matthews 1984). However, the physiological and agronomical mechanisms underlying rice seed storability remain unclarified. Previous reports use relative GP at high T and RH/GP at normal temperature to measure seed storability, while the present study takes the absolute GP at high T and RH or normal temperature as an independent indicator for seed storability. This approach allows us to distinguish the contribution of internal effect of seeds and the comprehensive effects of seeds and environmental factors during storage. At room temperature, 6 QTLs involved in seed germination were respectively identified on six chromosomes, which could be considered as the internal effects from seeds, such as the constituents of rice seeds controlled by seed developmental processes. However, at high T and RH, only 4 QTLs were found on chromosome 1, 3, 8 and 9, respectively, which were considered as the impacts from storage. Among all the identified QTLs under both conditions, the QTLs located in the flanking of RM6349–RM6849 on chromosome 3 and RM7364–RM3700 on chromosome 9 were stable whether at room condition or high T and RH.
FA is an important component of rice seed. It is reported to be associated with seed storability and the rice grain quality as food. However, there is no genetic evidence supporting the direct relationship between FA and seed storability. In the present study, 4 and 5 QTLs were respectively identified at room temperature and high T and RH, including the 4 QTLs detected under both storage conditions. It is interesting that the QTL located between RM7364 and RM3700 on chromosome 9 was closely related to both GP and FA irrespective of storage conditions. It clearly shows that seed FA is a direct factor affecting seed germination, and this QTL may be very useful for breeding new rice varieties with high seed storability. Furthermore, the japonica rice CJ06 shows higher FA than that in indica rice TN1 after artificial ageing, while TN1 presents higher germination percentage than CJ06. It implies that seed FA maybe poses a negative effect on seed germination. Kim et al (2007) also reported that rice seed germination rate and cooked rice palatability were lower with long storage, while fat acidity was higher after long storage. This negative effect is consistent with the relationship found between seed FA and GP in the RIL population. As is known, plant fatty acid components are mainly composed of unsaturated fatty acids that can be more easily oxidized by the oxygen from air than saturated fatty acids.
References
Ellis RH, Hong TD (1994) Desiccation tolerance and potential longevity of developing seeds of rice (Oryza sativa L.). Ann Bot 73(5):501–506
Ellis RH, Hong TD (2006) Temperature sensitivity of the low-moisture-content limit to negative seed longevity-moisture content relationships in hermetic storage. Ann Bot 97:785–791
Goffman FD, Bergman C (2003) Hydrolytic degradation of triacylglycerols and changes in fatty acid composition in rice bran during storage. Cereal Chem 80:459–461
Kameswara RN, Jackson MT (1997) Variation in seed longevity of rice cultivars belonging to different isozyme groups. Genet Resour Crop Evol 44(2):159–164
Kim KY, Lee GM, Noh KI, Ha KY, Son JY, Kim BK, Ko JK, Kim CK (2007) Varietal difference of germination, fat acidity, and lipoxygenase activity of rice grain stored at high temperature. Kor J Crop Sci 52(1):29–35
Li L, Lin Q, Liu S, Liu X, Wang W, Hang N, Liu F, Zhao Z, Jiang L, Wan J (2012) Identification of quantitative trait loci for seed storability in rice (Oryza sativa L.). Plant Breed 131:739–743
Lu C, Liu J, Jiang J, Breria CB, Tan H, Ichii M, Hong D (2013) Development and application of SCAR markers for discriminating cytoplasmic male sterile lines from their cognate maintainer lines in indica rice. Rice Sci 20:191–199
Maraschin C, Robert H, Boussard A, Potus J, Baret J-L, Nicolas J (2008) Effect of storage temperature and flour water content on lipids, lipoxygenase activity, and oxygen uptake during dough mixing. Cereal Chem 85:372–378
McCouch S, Cho Y, Yano M, Paul E, Blinstrub M, Morishima H, Kinoshita T (1997) Report on QTL nomenclature. Rice Genet Newsl 14:1
Miura K, Lin Y, Yano M, Nagamine T (2002) Mapping quantitative trait loci controlling seed longevity in rice (Oryza sativa L.). Theor Appl Genet 104(6–7):981–986
Nguyen T, Keizer P, Eeuwijk F, Smeekens S, Bentsink L (2012) Natural variation for seed longevity and seed dormancy are negatively correlated in Arabidopsis. Plant Physiol 160:2083–2092
Powell A, Matthews S (1984) Application of the controlled deterioration vigour test to detect seed lots of Brussels sprouts with low potential for storage under commercial conditions. Seed Sci Technol 12:649–657
Rao Y, Dong G, Zeng D, Hu J, Zeng L, Gao Z, Zhang G, Guo L, Qian Q (2010) Genetic analysis of leaffolder resistance in rice. J Genet Genom 37:325–331
Ren Y, Wang Y, Liu F, Zhou K, Ding Y, Zhou F, Wang Y, Liu K, Gan L, Ma W, Han X, Zhang X, Guo X, Wu F, Cheng Z, Wang J, Lei C, Lin Q, Jiang L, Wu C, Bao Y, Wan J (2014) GLUTELIN PRECURSOR ACCUMULATION3 encodes a regulator of post-golgi vesicular traffic essential for vacuolar protein sorting in rice endosperm. Plant Cell 26(1):410–425
Sasaki K, Fukuta Y, Sato T (2005) Mapping of quantitative trait loci controlling seed longevity of rice (Oryza sativa L.) after various periods of seed storage. Plant Breed 124:361–366
Sasaki K, Kazama Y, Chae Y, Sato T (2013) Confirmation of novel quantitative trait loci for seed dormancy at different ripening stages in rice. Rice Sci 20:207–212
Shen B, Zhuang JY, Fan YY, Ying JZ, Yang J, Sun LX, Fei JH, Zheng KL (2005) Genetic analysis of starch branching enzyme activity in rice grain. Zhi Wu Sheng Li Yu Fen Zi Sheng Wu Xue Xue Bao=J Plant Physiol. Mol Biol 31(6):631–636 (in Chinese)
Suzuki Y (2011) Isolation and characterization of a rice (Oryza sativa L.) mutant deficient in seed phospholipase D, an enzyme involved in the degradation of oil-body membranes. Crop Sci 51:567–573
Wang D, Zhu J, Li Z, Paterson AH (1999) Mapping QTLs with epistatic effects and QTL environment interact ions by mixed linear model approaches. Theor Appl Genet 99:1255–1264
Wang Z, Wang J, Bao Y, Wang F, Zhang H (2010) Quantitative trait loci analysis for rice seed vigor during the germination stage. J Zhejiang Univ Sci B 11:958–964
Wang Z, Wang J, Bao Y, Wu Y, Zhang H (2011) Quantitative trait loci controlling rice seed germination under salt stress. Euphytica 178(3):297–307
Whitehouse KJ, Hay FR, Ellis RH (2015) Increases in the longevity of desiccation-phase developing rice seeds: response to high-temperature drying depends on harvest moisture content. Ann Bot 116(2):247–259
Xu X, Ying X, Duan B (2007) Study on determination of fatty acid value of rice. J Chin Cereals Oils Assoc 22:105–107
Xue Y, Zhang S, Yao Q, Peng R, Xiong A, Li X, Zhu Y, Zha D (2008) Identification of quantitative trait loci for seed storability in rice (Oryza sativa L.). Euphytica 164:739–744
Yang J, Zhu J (2005) Predicting superior genotypes in multiple environments based on QTL effects. Theor Appl Genet 110:1268
Zeng D, Qian Q, Yasukumi K, Teng S, Hiroshi F (2002) Study on storability and morphological index in rice (Oryza sativa L.) under artificial ageing. Acta Agron Sin 28:551–554
Zeng D, Guo L, Xu Y, Yasukumi K, Zhu L, Qian Q (2006) QTL analysis of seed storability in rice. Plant Breed 125:57–60
Zhang WM, Ni AL, Wang CC (1998) Study on seed vigor of hybrid rice. Seed 2:7–10
Acknowledgements
We thank Dr. D. L. Zeng, G. H. Zhang for the useful advice on designed the experiment, and thank for Dr. L. Zhu at the China National Rice Research Institute for the critical reading of the manuscript; this work was supported by grants from National Science and Technology Support Project of China (Grant No. 2011BAD35B09).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, C.S., Shao, G.S., Wang, L. et al. QTL Identification and Fine Mapping for Seed Storability in Rice (Oryza sativa L.). Euphytica 213, 127 (2017). https://doi.org/10.1007/s10681-017-1913-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-017-1913-5