Abstract
The southeastern United States (U.S.) has experienced dynamic climatic changes over the past several million years that have impacted species distributions. In many cases, contiguous ranges were fragmented and a lack of gene flow between allopatric populations led to genetic divergence and speciation. The Southern Red-backed Salamander, Plethodon serratus, inhabits four widely disjunct regions of the southeastern U.S.: the southern Appalachian Mountains, the Ozark Plateau, the Ouachita Mountains, and the Southern Tertiary Uplands of central Louisiana. We integrated phylogenetic analysis of mitochondrial DNA sequences (1399 base pairs) with ecological niche modeling to test the hypothesis that climate fluctuations during the Pleistocene drove the isolation and divergence of disjunct populations of P. serratus. Appalachian, Ozark, and Louisiana populations each formed well-supported clades in our phylogeny. Ouachita Mountain populations sorted into two geographically distinct clades; one Ouachita clade was sister to the Louisiana clade whereas the other Ouachita clade grouped with the Appalachian and Ozark clades but relationships were unresolved. Plethodon serratus diverged from its sister taxon, P. sherando, ~5.4 million years ago (Ma), and lineage diversification within P. serratus occurred ~1.9–0.6 Ma (Pleistocene). Ecological niche models showed that the four geographic isolates of P. serratus are currently separated by unsuitable habitat, but the species was likely more continuously distributed during the colder climates of the Pleistocene. Our results support the hypothesis that climate-induced environmental changes during the Pleistocene played a dominant role in driving isolation and divergence of disjunct populations of P. serratus.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The geographic distribution of a species is determined by interactions among multiple biotic, abiotic, and historical factors (MacArthur 1984; Brown et al. 1996; Peterson et al. 2011). Biotic factors include interspecific ecological interactions such as competition, predation, and parasitism, whereas historical factors are related to dispersal opportunities and range dynamics of the species and its ancestors over evolutionary time (Chase and Leibold 2003; Gaston 2003; Morrone 2009). Abiotic factors include physical barriers to dispersal such as rivers and mountain ranges as well as climatic variables such as temperature, precipitation, and seasonality (Brown et al. 1996; Gaston 2003; Peterson et al. 2011). Many abiotic and biotic factors vary spatially and temporally, especially climate, which has changed considerably over the past 65 million years (Myr), greatly impacting the distributions and evolution of species (Webb and Bartlein 1992; Zachos et al. 2001; Jansson and Dynesius 2002).
The biological effects of climate fluctuations associated with Pleistocene glacial cycles [~2.58 million years ago (Ma)–11.7 thousand years ago (ka)] are well documented and are often evident at the level of populations and closely related species because of their recency (Bennett 1990; Hewitt 2000, 2004). These effects are pronounced in species in areas formerly covered by glaciers and recolonized after glacial retreat, but species in many unglaciated areas were also affected (Hewitt 1996, 2000, 2004). For example, the southeastern United States (U.S.) has experienced dynamic environmental changes over the past several million years that impacted species distributions, population connectivity, and speciation (Watts 1980; Soltis et al. 2006). In some cases, climatic changes facilitated range expansion and dispersal to new regions whereas in other cases range contraction and fragmentation resulted (Hewitt 1996; Soltis et al. 2006). In fact, the cyclical nature of Pleistocene climatic fluctuations means that many species experienced alternating episodes of range expansion and contraction (Bennett 1990; Hewitt 2004). In species whose ranges were fragmented, a lack of gene flow between allopatric populations sometimes led to genetic divergence and speciation (Hewitt 1996). Many responses to climatic fluctuations in southeastern U.S. species are related to the expansion and contraction of deciduous forest (Watts 1980; Davis 1983; Jackson et al. 2000). In addition, this region has physical barriers to dispersal such as the Mississippi and Apalachicola rivers as well as montane regions such as the Appalachian Mountains and Interior Highlands (Ozark Plateau and Ouachita Mountains), further complicating phylogeographic history of species in the region (Swenson and Howard 2005; Soltis et al. 2006; Rissler and Smith 2010).
One of the most prominent biotic components of southeastern U.S. forests is plethodontid salamanders, which often dominate vertebrate biomass (Petranka and Murray 2001; Crawford and Peterman 2013; Semlitsch et al. 2014). Salamanders of the family Plethodontidae are the largest and most diverse group of salamanders with >450 currently recognized species worldwide (Frost 2015). The genus Plethodon includes 55 recognized species, many of which can only be distinguished using molecular techniques due to conserved morphology (Highton 1995; Wake 2009; Frost 2015). These terrestrial salamanders require mesic environments for survival and reproduction so their distributions are strongly influenced by moisture and temperature (Spotila 1972; Feder 1983). Many closely related and similar-looking species of Plethodon are allopatric or parapatric, and many have restricted distributions (Highton 1995; Kozak and Wiens 2006). Pleistocene climatic fluctuations have been implicated in fragmenting distributions and driving lineage diversification in several species (Highton 1995; Weisrock and Larson 2006; Shepard and Burbrink 2008, 2009).
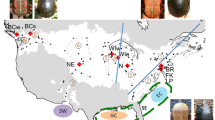
Among species of Plethodon, the Southern Red-backed Salamander, Plethodon serratus Grobman 1944, has one of the most intriguing distributions, consisting of four widely disjunct regions: (1) the southern Appalachian Mountains of western North Carolina, southeastern Tennessee, northwestern Georgia, and northeastern Alabama, (2) the Ozark Plateau of southeastern Missouri, (3) the Ouachita Mountains of southeastern Oklahoma and west-central Arkansas, and (4) the Southern Tertiary Uplands of central Louisiana (Fig. 1; Highton and Webster 1976; Petranka 1998). This small-bodied (<10 cm total length) woodland salamander was recognized as a subspecies of the widespread P. cinereus until 1976, when it was elevated to species based on allozyme data (Highton and Webster 1976). In addition to showing high genetic differentiation between P. serratus and P. cinereus, allele frequencies indicated “modest” differentiation between geographic isolates of P. serratus (Nei’s D = 0.01–0.19; Highton and Webster 1976). Genus-level studies including samples of P. serratus from Oklahoma and Georgia also documented substantial mitochondrial DNA (mtDNA) divergence within the species (Sites et al. 2004). The disjunct distribution and documented genetic variation among geographic isolates suggests that Pleistocene climatic fluctuations may have impacted the distribution and lineage diversification of P. serratus.
Here we test the hypothesis that the disjunct distribution of P. serratus is the result of fragmentation driven by climatic changes during the Pleistocene. To evaluate this, we use mtDNA sequences from multiple populations within each of the four geographic isolates to test if populations within each isolate are genetically differentiated and form monophyletic groups, which would be indicative of long-term isolation. We also use divergence-dating methods to estimate the times that populations from the four isolates became isolated. We then use ecological niche modeling to predict the distribution of P. serratus to test if the availability of suitable environmental conditions limits the species’ distribution and if the four isolates are separated by unsuitable areas. Lastly, we project the inferred ecological niche of P. serratus onto climate layers for the Last Glacial Maximum (LGM; ~21 ka) to test if the species was likely more widely distributed in the past with suitable environmental conditions connecting the four current isolates.
Materials and methods
Sampling and DNA sequencing
Between September 2010 and April 2012, we collected tissue samples of P. serratus Grobman 1944 throughout the species’ distribution, including multiple locations within each of the four geographic isolates. We also obtained tissue samples of P. serratus through loans from the Louisiana Museum of Natural History at Louisiana State University (LSUMZ) and Bell Museum of Natural History at the University of Minnesota (JFBM). In total, we obtained 43 samples of P. serratus: 25 from the Ouachita Mountains, five from the Ozark Plateau, seven from the southern Appalachian Mountains, and six from central Louisiana (see Appendix S1 in Supporting Information for specific locality data). Specimens and tissues from our fieldwork were deposited at LSUMZ (Appendix S1).
We extracted genomic DNA from tissues using commercial DNA extraction kits (Qiagen DNeasy; Viogene Blood & Tissue Miniprep) and used polymerase chain reaction (PCR) to amplify portions of the mitochondrial cytochrome b (cytb) and NADH dehydrogenase subunit 4 (ND4) genes. For cytb, we used the primers Pcytb419F and PThrR1 from Wiens et al. (2006) and we used the primers ND4(F) and Ephist from Arévalo et al. (1994) and Wiens et al. (2006) for ND4. Each PCR contained 10 µl ddH20, 6 µl GoTaq® Green Master Mix (Promega), 2.0 µl DNA template (10 ng/µl), and 1 µl of each primer (10 µM). For both loci, PCRs consisted of an initial denaturing stage of 94 °C for 2 min, followed by 34 cycles of 94 °C for 1 min, 55 °C for 45 s, and 72 °C for 2 min, with a final extension of 72 °C for 5 min. We verified PCR products by gel electrophoresis and cleaned PCR reactions using an exonuclease/shrimp alkaline phosphatase protocol (Werle et al. 1994). We prepared sequencing reactions using 2 µl of purified PCR product and 5 pmol of either the same forward or reverse primer used in PCR; PCR products were sequenced in both directions. Sequencing reactions and automated sequencing were performed by the University of Missouri DNA Core Facility using an ABI3730xl 96-capillary sequencer. We edited chromatograms by eye to verify base calls and assembled contigs from forward and reverse sequences using Sequencher v.5.0 (Gene Codes Corp.). We aligned sequences for each gene using the MUSCLE algorithm in MEGA v.5 (Tamura et al. 2011) and visually inspected alignments to verify an open reading frame for these protein-coding genes.
In total, we sequenced 1399 base pairs (bp) of mtDNA for 43 P. serratus. The cytb alignment comprised 698 bp of which 112 sites were variable and 96 were parsimony informative within P. serratus. The ND4 alignment was 701 bp with 106 variable sites of which 91 were parsimony informative. We deposited sequences generated in this study on GenBank (see Appendix S1 in Supporting Information for GenBank accession numbers). We obtained cytb and ND4 sequences for P. cinereus (NC_006343) from GenBank to use as an outgroup (Mueller et al. 2004; Kozak et al. 2009) and concatenated alignments for phylogenetic inference.
Phylogenetic and divergence dating analyses
We assessed phylogeographic relationships and estimated divergence times within P. serratus using BEAST v.1.8.0 (Drummond and Rambaut 2007), which employs a Bayesian approach to simultaneously infer topology and divergence times, thus accounting for phylogenetic uncertainty. To do this, we first estimated the phylogeny and divergence times of Plethodontidae using the taxa and sequences from Kozak et al. (2009). This aligned data set comprises 5514 bp of DNA sequence from three mitochondrial genes (cytb, ND2, ND4) and three nuclear genes (BDNF, POMC, RAG1) for 184 plethodontid taxa and the outgroup, Amphiuma tridactylum. We used PartitionFinder (Lanfear et al. 2012) to evaluate different data-partitioning schemes and choose the best substitution model for each partition. Based on the Bayesian Information Criterion (BIC), the best partitioning scheme consisted of eight partitions with the following substitution models: (1) cytb first codon positions: SYM + Γ + I, (2) cytb and ND4 second codon positions: HKY + Γ + I, (3) cytb and ND4 third codon positions: GTR + Γ + I, (4) ND2 and ND4 first codon positions: GTR + Γ + I, (5) ND2 second codon positions: GTR + Γ + I, (6) ND2 third codon positions: TrN + Γ, (7) POMC and RAG1: SYM + Γ + I, and (8) BDNF: K80 + Γ + I. We unlinked substitution model parameters among partitions, linked clock models of mitochondrial DNA partitions (1–6 above), and linked clock models of nuclear DNA partitions (7–8 above). We used uncorrelated lognormal relaxed-clock models and a birth–death tree model. For this analysis, we inferred a single tree from the concatenated data set and provided six fossil-based calibration points (see Appendix S2 in Supporting Information for details). We estimated tree topology and divergence times using three independent searches of 30 million generations each, sampling every 1000th iteration with 10 % of the initial samples discarded as burn-in. We viewed log files in Tracer v.1.6 (Rambaut and Drummond 2009) to verify that parameter estimates had reached stationarity and that ESS values in the combined post-burn-in samples were >200.
To infer phylogeographic relationships and estimate divergence times within P. serratus, we performed a second analysis in BEAST in which we used the mean age estimate (12.7 Myr) and 95 % Highest Posterior Density (HPD; 9.3–16.8 Myr) for the MRCA of P. serratus and P. cinereus from the Plethodontidae analysis above to place a lognormally distributed prior (Mean = 12.7, SD = 0.15) on the root age of a tree containing all samples of P. serratus and the outgroup, P. cinereus. Additionally, we included a cytb sequence from GenBank (JF731320) for P. sherando, a recently described, narrowly distributed species from the Appalachian Mountains that was not included in the Kozak et al. (2009) study but was shown to be the sister taxon of P. serratus in recent analyses (Bayer et al. 2012; Highton et al. 2012). For our analysis of P. serratus, we used PartitionFinder (Lanfear et al. 2012) to evaluate different data-partitioning schemes and choose the best substitution model for each partition. Based on BIC, the best scheme consisted of three partitions that combined like codon positions of the two genes. The best substitution models were: HKY + Γ for first codon positions, HKY + I for second codon positions, and TrN + Γ for third codon positions. We unlinked substitution models among partitions, linked the clock models and tree among partitions, and used a constant population size tree prior. We ran analyses under a strict-clock and a relaxed-clock model (Drummond et al. 2006) and then compared them using path sampling and stepping-stone sampling in BEAST following the methods of Baele et al. (2012, 2013). In both cases, we simultaneously estimated topology and divergence times in a single search of 20 million generations sampling every 1000th iteration with 10 % of the initial samples discarded as burn-in. We viewed log files in Tracer v.1.6 to verify that parameter estimates had reached stationarity and that all ESS values were >200. For both path sampling and stepping-stone sampling, the difference in marginal likelihoods between strict-clock and relaxed-clock analyses, termed the Bayes factor (lnBF), was <1. Kass and Raftery (1995) state that 2lnBF values between 2 and 6 are positive evidence favoring one model over another, thus for our data set the more parameterized relaxed-clock model was not positively favored over the simpler strict-clock model (2lnBF = 0.9 for path sampling and −0.25 for stepping-stone sampling). We therefore used TreeAnnotator (Drummond and Rambaut 2007) to summarize the post-burn-in sample of trees from the strict-clock analysis and generate a maximum clade credibility tree with mean node heights. Nodes were considered to have high support when the Bayesian posterior probability (Bpp) was ≥0.95. We used the chronogram and estimates of node ages (means and 95 % HPDs) from this analysis to determine the approximate time that P. serratus diverged from its sister taxon, P. sherando, and also the timing of divergences within P. serratus. We calculated mean uncorrected pairwise sequence divergence between clades using MEGA v.5 (Tamura et al. 2011).
Ecological niche modeling
To test if the availability of suitable environmental conditions limits the distribution of P. serratus to the four current isolates and whether these populations were likely connected under past climatic conditions, we used geographic information system (GIS)-based ecological niche modeling (Peterson 2001; Kozak et al. 2008; Elith and Leathwick 2009). We obtained 678 unique georeferenced localities for P. serratus from several sources including the Global Biodiversity Information Facility (www.gbif.org), HerpNET (www.herpnet2.org), the Arkansas Natural Heritage Commission, and from our own collections and fieldwork (see Appendix S3 in Supporting Information for locality data). Next, we acquired GIS layers for 19 environmental variables from the Worldclim project (www.worldclim.org). These data comprise 19 bioclimatic variables (BIO1–BIO19) related to temperature, precipitation, and seasonality (Hijmans et al. 2005). We downloaded layers for both current conditions and the Last Glacial Maximum (LGM; ~21 ka) at 2.5 arc-min (~5 km2) resolution. For building the model, we drew a polygon around all locality points and used the geometry of that polygon to clip layers for current conditions to a spatial extent that provided adequate information to train the model while also restricting analysis to a geographic area in which the species could possibly occur (Barve et al. 2011). For projecting the model, we clipped layers for current conditions and the LGM to the southeastern U.S.
We constructed an ecological niche model (ENM) for P. serratus using a maximum entropy method implemented in Maxent v.3.3.3 (Phillips et al. 2006; Phillips and Dudík 2008; Elith et al. 2011). The output of Maxent consists of a threshold-independent measure of the overall performance of the model (Area Under the Receiver Operating Curve or AUC) and a grid map with each cell having an index of suitability between 0 and 1 (logistic output); higher values indicate higher predicted suitability. An AUC value of 0.5 indicates the predictive model is no better than random, whereas higher AUC values indicate better predictive ability with a value of 1 indicating perfect prediction. We used auto-features and the default regularization multiplier parameter (1.0). Additionally, we removed duplicate presence points (i.e., points mapping to the same grid cell) and increased the number of iterations to 5000.
To gauge the sensitivity of the ENM to the samples used in training and test the predictive ability of the model, we performed 10 replicate runs using a different random seed and subsampling with 70 % of samples allotted for training and 30 % for testing. We examined the mean AUC for test data across the 10 replicates and considered a mean AUC ≥ 0.9 as very good support for the model (Swets 1988). To construct the final ENM for P. serratus, we used all locality points. We projected this ENM onto layers for current conditions and the LGM for the southeastern U.S., and used the Minimum Training Presence threshold to classify grid cells as unsuitable if they fell below this value (Liu et al. 2005). We retained values above this threshold to group cells into classes representing different levels of predicted suitability.
Results
Phylogeny and divergence times
The Bayesian phylogeny from BEAST revealed five geographically distinct lineages within P. serratus distributed across the four geographic isolates (Fig. 2). The Appalachian, Ozark, and Louisiana populations each formed well-supported clades (Bpp = 0.91–1.0). In contrast, Ouachita Mountain populations sorted into two clades, an eastern (Bpp = 0.82) and a western (Bpp = 1.0), that do not form a monophyletic group. The western Ouachita clade was sister to the Louisiana clade (Bpp = 1.0) whereas the eastern Ouachita clade was sister to the Ozark clade (Bpp = 0.50); however, support for the monophyly of the eastern Ouachita clade and its relationship to the Ozark clade was too low to have sufficient confidence about this relationship. The Appalachian clade grouped with Ozark and eastern Ouachita populations (Bpp = 1.0), but relationships among these three clades were unresolved. The greatest sequence divergence (6.5 %) was between the eastern and western Ouachita clades whereas the lowest (2.1 %) was between the Louisiana and western Ouachita clades. Sequence divergence between other clades ranged from 4.4 to 6.4 % (Table 1). Divergence times indicated that P. serratus diverged from its sister taxon, P. sherando, ~5.4 Ma (95 % HPD: 3.1–8.1 Ma; Fig. 2). The MRCA of all P. serratus was ~1.9 Ma (95 % HPD: 1.1–2.8 Ma). Divergences among the Appalachian, Ozark, and eastern Ouachita mtDNA lineages dated to ~1.3–1.4 Ma (95 % HPD: 0.8–2.1 Ma) whereas the divergence between the Louisiana and western Ouachita mtDNA lineages was more recent, dating to ~0.6 Ma (95 % HPD: 0.4–0.9 Ma).
Bayesian maximum clade credibility chronogram for Plethodon serratus based on concatenated analysis of cytb and ND4 mitochondrial DNA sequences in BEAST. Nodes are labeled with Bayesian posterior probabilities (%); asterisks indicates 100 %. Node ages are presented as means with bars representing 95 % Highest Posterior Densities around age estimates. Phylogeographic lineages are labeled and bars next to labels are colored to match distributions in Fig. 1. (Color figure online)
Ecological niche modeling
Across the ten subsampling replicates, the ENM for P. serratus had a mean test AUC of 0.949 (SD = 0.009), indicating a very good ability to discriminate between presence and absence locations (Swets 1988). The Minimum Training Presence threshold was 0.02; thus, areas with values lower than this were classified as unsuitable. The predicted distribution of P. serratus fits well with its actual present-day distribution restricted to four geographic isolates (Fig. 3a). All four isolates of P. serratus are separated from each other by large expanses of unsuitable habitat. A few areas within the circumscribed range of P. serratus as well as a few areas outside the current range are predicted to be suitable, but are not currently occupied by the species. The ENM also predicted suitable conditions along the Gulf Coast and the Kansas/Missouri border, but predictions in these regions should be treated with caution because climatic variables were outside the range of training data.
Predicted distribution of Plethodon serratus under a current climate conditions and b at the Last Glacial Maximum (~21,000 years ago). Models were constructed in Maxent using 19 bioclimatic variables at 2.5 arc-min resolution (~5 km2). Locality points for P. serratus (n = 678) used in modeling are shown as black points and the dashed polygon is the spatial extent used to train the model. Suitability values range from 0 to 1 with higher values indicating higher predicted suitability. (Color figure online)
When we project the ENM onto climate layers for the LGM, the distribution of P. serratus is predicted to have been considerably expanded in the past compared to present-day (Fig. 3b). The model predicts suitable environmental conditions existed at the LGM everywhere within the current circumscribed range of P. serratus, although some areas along the Mississippi River in northeastern Louisiana and west-central Mississippi are marginal. Thus, this ENM predicts that the four current isolates of P. serratus were connected by suitable habitat during the LGM.
Discussion
Significant levels of genetic differentiation among the four isolates of P. serratus was not surprising given the geographic distances among them and results of previous work (Highton and Webster 1976; Sites et al. 2004). The Appalachian, Ozark, and Louisiana populations each formed monophyletic groups, indicating long-term isolation. What was surprising is that the Ouachita Mountains contain two non-sister parapatric lineages that show the highest level of sequence divergence within the species (6.5 %). In their allozyme study, Highton and Webster (1976) sampled only two populations of P. serratus from the Ouachitas, but it appears that they sampled both lineages. Our study included samples from the same two localities, FG and BB, which belong to the eastern and western lineages, respectively. Furthermore, their BB population was more similar genetically to their Louisiana population than it was to the FG population, which is consistent with our results showing a sister-taxon relationship between the western Ouachita and Louisiana lineages. Highton and Webster (1976) could not resolve relationships among Appalachian, Ozark, and Ouachita populations of P. serratus with 20 nuclearly encoded allozyme loci. Similarly, we could not resolve relationships among Appalachian, Ozark, and eastern Ouachita clades. Data from multiple nuclear DNA sequence loci combined with more extensive population sampling might provide better resolution. We also note that our results are based on mtDNA, which provides only a single genealogical estimate of evolutionary history that may be different from the true species history (Edwards 2009).
The eastern and western Ouachita clades abut along a roughly north–south line that is coincident with where the ranges of the Ouachita endemics P. ouachitae and P. fourchensis meet. These sister taxa were estimated to have diverged at approximately the same time as we inferred for the MRCA of eastern and western Ouachita lineages of P. serratus (~2.2 vs. ~1.9 Ma, respectively; Shepard and Burbrink 2008, 2009). There are no physical barriers to dispersal that would explain the distinct genetic break between eastern and western lineages of P. serratus; however, the contact zone appears to correspond to an extended divide separating major eastward- and westward-draining river systems (Arkansas and Ouachita rivers from the Red River). Studies on plethodontid species with aquatic larval phases have found a relationship between phylogeographic structure and river drainages (Jones et al. 2006; Kozak et al. 2006a); however, Plethodon are terrestrial, direct-developers and thus such a relationship would not be predicted. The congener, P. caddoensis, is sympatric with P. serratus in one region of the Ouachita Mountains, and also shows genetic structure associated with stream drainages, which was posited to be due to the unique geology of the region and flash flood events transporting individuals downstream (Shepard and Burbrink 2011). The process by which river drainage patterns might contribute to genetic differentiation of populations of P. serratus is unknown, but more detailed sampling of the contact zone and estimating gene flow between eastern and western lineages would be informative.
Although the biogeographic history of P. serratus would be better addressed within a broader taxonomic context including all species of Plethodon or all North American plethodontids, we can still make some inferences about the roles of dispersal and vicariance in shaping the species’ distribution. Given that most species of Plethodon occur in the eastern U.S. (Kozak et al. 2009; Kozak and Wiens 2012), including P. serratus and its sister taxon, P. sherando, we can infer at least one dispersal from the Appalachians westward across the Mississippi River to the Interior Highlands. Dowling (1956) hypothesized that plethodontid salamanders colonized the Interior Highlands multiple times from different sources, including the Appalachians and southern Coastal Plain. The Appalachians are also considered the source of other Interior Highlands fauna, including some darters (Nothonotus; Near and Keck 2005), crayfishes (Orconectes; Crandall and Templeton 1999), and insects (Allen 1990). The lower Mississippi River appears to have been a major barrier to east–west dispersal between the Appalachians and Interior Highlands since well before the Pleistocene (>2.5 Ma), but these two regions are thought to have been connected via a large upland north of the present-day Ohio River until the onset of Pleistocene glaciations and subsequent vicariance (Mayden 1985, 1988; Soltis et al. 2006; Pyron and Burbrink 2010). Studies on eastern North American fishes and crayfishes have found that both dispersal and vicariance have been important in shaping species distributions and biogeographic patterns, but species vary in how they responded to historical events (Strange and Burr 1997; Crandall and Templeton 1999; Near et al. 2001).
During the Miocene (23–5.3 Ma), central North America experienced a transition from warm and humid climates to cool and dry climates with increased seasonality (Zachos et al. 2001; Kohn and Fremd 2008; Kürschner et al. 2008). This change in climate led to a contraction of forest and an expansion of grasslands accompanied by major faunal changes (Axelrod 1985; Kohn and Fremd 2008; Kürschner et al. 2008). Since the Late Miocene, a steep longitudinal moisture gradient has characterized this region with mesic conditions in the east and xeric conditions in the west (Axelrod 1985; Costa et al. 2008). The Ouachitas and Ozarks occur at the eastern edge of this gradient and are thought to have served as refugia for mesic-adapted organisms (e.g., salamanders) whenever xeric conditions expanded eastward in the past (Dowling 1956; Costa et al. 2008). The edges of this gradient fluctuated along an east–west axis during the climatic oscillations of the Pleistocene, which would have provided multiple opportunities for dispersal among regions when mesic conditions expanded westward and also multiple instances of vicariance when they contracted eastward (Axelrod 1985; Webb and Bartlein 1992; Grundstein 2009).
The divergence of P. serratus and P. sherando near the Miocene/Pliocene boundary (~5.4 Ma) corresponds to a period of rapid speciation in eastern North American Plethodon that has been attributed to widespread aridification (Highton 1995; Kozak et al. 2006b; Wiens et al. 2006; Highton et al. 2012). Using different methods, Bayer et al. (2012) estimated the MRCA of P. serratus and P. sherando to be 6.0 Ma (95 % HPD: 4.4–7.6 Ma), which is similar to our estimate (~5.4 Ma; 95 % HPD: 3.1–8.1 Ma). The divergence of mtDNA lineages within P. serratus ~1.9–0.6 Ma is consistent with the hypothesis that Pleistocene climatic fluctuations facilitated multiple instances of dispersal and vicariance. We note that our divergence time estimates are based gene tree inference and that gene coalescence will predate species/population divergence (Edwards and Beerli 2000; Edwards 2009). Thus, divergences among geographically isolated populations of P. serratus would likely be more recent than our mtDNA-based estimates, but still well within the Pleistocene.
Ecological niche models indicated that abiotic factors primarily limit the distribution of P. serratus. Our present-day ENM for P. serratus predicted that suitable habitat is largely restricted to the four geographic isolates and that intervening areas are unsuitable. In contrast, the ENM for the LGM predicted that P. serratus was widely distributed during colder climates of the Pleistocene and the four current isolates were connected by suitable habitat. Although divergences among the four isolates of P. serratus are older than the time periods of available climate layers, the current climate is similar to earlier interglacial climates of the Pleistocene and the LGM climate is similar to climates of earlier Pleistocene glacial periods (Petit et al. 1999). Therefore, ENMs based on these climate data likely capture the extent of broad-scale impacts of climatic fluctuations throughout the Pleistocene. A few areas within the current circumscribed range of P. serratus as well as some areas outside the species’ current range were predicted to be suitable but are not currently occupied by the species. These areas are, however, occupied by other species of small-bodied Plethodon (Petranka 1998), which may preclude P. serratus from occurring in those areas. Species of Plethodon of similar body size tend not to coexist due to competitive interactions (Hairston 1951; Jaeger 1971; Adams 2007; Kozak et al. 2009). Thus, although abiotic factors appear to have been the major driver of the disjunct distribution of P. serratus, biotic interactions have also likely had an influence on where the species occurs.
Niche conservatism is considered to be the primary mechanism by which speciation in temperate-zone plethodontids has occurred (Kozak and Wiens 2006, 2010). Under this model, the distribution of a wide-ranging species is fragmented into multiple isolated populations as individuals track their ecological niche when climate changes. Over time, these isolated populations diverge and become new species (Wiens 2004; Wiens and Graham 2005). The five lineages within P. serratus are geographically distinct and reciprocally monophyletic for mtDNA. Nuclear loci also indicate significant genetic differentiation (Highton and Webster 1976). Furthermore, the four geographic isolates of P. serratus are separated by both physical barriers (e.g., Mississippi River, Arkansas River) and environmental barriers (e.g., unsuitable habitat, closely related species). Given these factors and current climate trends, it is unlikely that these disjunct populations will become connected again in the near future and they will likely continue to diverge genetically. The amount of sequence divergence found between most lineages of P. serratus (4.4–6.5 %) is comparable to the divergence between some currently recognized species of Plethodon. For example, comparing a similar region of cytochrome b, P. virginia and P. hoffmani had an average divergence of 4.1 % and P. nettingi and P. hubrichti were on average 5.4 % divergent (Sites et al. 2004). Given our hypothesis of phylogenetic relationships, whether eastern and western Ouachita lineages interbreed where they come into contact is central to determining whether lineages should be recognized as species. Because of a longitudinal climatic gradient in the Ouachita Mountains, the eastern and western Ouachita lineages likely occupy different climatic niches (Shepard and Burbrink 2009). Whether this niche variation is of an extent that would impact ecological exchangeability is unknown, but would be important for delimiting species (Rader et al. 2005; Stockman and Bond 2007; Bond and Stockman 2008). Furthermore, such information would be important for defining and managing conservation units to preserve diversity within what is currently considered a single, broadly distributed species (Moritz 1994; Crandall et al. 2000; Fraser and Bernatchez 2001).
References
Adams DC (2007) Organization of Plethodon salamander communities: guild-based community assembly. Ecology 88:1292–1299
Allen RT (1990) Insect endemism in the Interior Highlands of North America. Fla Entomol 73:539–569
Arévalo E, Davis SK, Sites JW Jr (1994) Mitochondrial DNA sequence divergence and phylogenetic relationships among eight chromosome races of the Sceloporus grammicus complex (Phrynosomatidae) in Central Mexico. Syst Biol 43:387–418
Axelrod DI (1985) Rise of the grassland biome, central North America. Bot Rev 51:163–201
Baele G, Lemey P, Bedford T, Rambaut A, Suchard MA, Alekseyenko AV (2012) Improving the accuracy of demographic and molecular clock model comparison while accommodating phylogenetic uncertainty. Mol Biol Evol 29:2157–2167
Baele G, Lok W, Li S, Drummond AJ, Suchard MA, Lemey P (2013) Accurate model selection of relaxed molecular clocks in Bayesian phylogenetics. Mol Biol Evol 30:239–243
Barve N, Barve V, Jiménez-Valverde A, Lira-Noriega A, Maher SP, Peterson AT, Soberón J, Villalobos F (2011) The crucial role of the accessible area in ecological niche modeling and species distribution modeling. Ecol Model 222:1810–1819
Bayer CSO, Sackman AM, Bezold K, Cabe PR, Marsh DM (2012) Conservation genetics of an endemic mountaintop salamander with an extremely limited range. Conserv Genet 13:443–454
Bennett KD (1990) Milankovitch cycles and their effects on species in ecological and evolutionary time. Paleobiology 16:11–21
Bond JE, Stockman AK (2008) An integrative method for delimiting cohesion species: finding the population-species interface in a group of Californian trapdoor spiders with extreme genetic divergence and geographic structuring. Syst Biol 57:628–646
Brown JH, Stevens GC, Kaufman DM (1996) The geographic range: size, shape, boundaries, and internal structure. Annu Rev Ecol Syst 27:597–623
Chase JM, Leibold MA (2003) Ecological niches: linking classical and contemporary approaches. University of Chicago Press, Chicago
Costa GC, Wolfe CA, Shepard DB, Caldwell JP, Vitt LJ (2008) Detecting the influence of climatic variables on species distributions: a test using GIS niche-based models along a steep longitudinal environmental gradient. J Biogeogr 35:637–646
Crandall KA, Templeton AR (1999) The zoogeography and centers of origin of the crayfish subgenus Procericambarus (Decapoda: Cambaridae). Evolution 53:123–134
Crandall KA, Bininda-Emonds ORP, Mace GM, Wayne RK (2000) Considering evolutionary processes in conservation biology. Trends Ecol Evol 15:290–295
Crawford JA, Peterman WE (2013) Biomass and habitat partitioning of Desmognathus on wet rock faces in southern Appalachian Mountains. J Herpetol 47:580–584
Davis MB (1983) Quaternary history of deciduous forests of eastern North America and Europe. Ann Mo Bot Gard 70:550–563
Dowling H (1956) Geographic relations of Ozarkian amphibians and reptiles. Southwest Nat 1:174–189
Drummond AJ, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7:214
Drummond AJ, Ho SYW, Phillips MJ, Rambaut A (2006) Relaxed phylogenetics and dating with confidence. PLoS Biol 4:699–710
Edwards SV (2009) Is a new and general theory of molecular systematics emerging? Evolution 63:1–19
Edwards SV, Beerli P (2000) Perspective: gene divergence, population divergence, and the variance in coalescence time in phylogeographic studies. Evolution 54:1839–1854
Elith J, Leathwick JR (2009) Species distribution models: ecological explanation and prediction across space and time. Annu Rev Ecol Evol Syst 40:677–697
Elith J, Phillips SJ, Hastie T, Dudík M, Chee YE, Yates CJ (2011) A statistical explanation of MaxEnt for ecologists. Divers Distrib 17:43–57
Feder ME (1983) Integrating the ecology and physiology of plethodontid salamanders. Herpetologica 39:291–310
Fraser DJ, Bernatchez L (2001) Adaptive evolutionary conservation: towards a unified concept for defining conservation units. Mol Ecol 10:2741–2752
Frost DR (2015) Amphibian Species of the World: an online reference. Version 6.0. American Museum of Natural History, New York. http://research.amnh.org/vz/herpetology/amphibia/. Accessed 8 July 2015
Gaston KJ (2003) The structure and dynamics of geographic ranges. Oxford University Press, Oxford
Grundstein A (2009) Evaluation of climate change over the continental United States using a moisture index. Clim Change 93:103–115
Hairston NG (1951) Interspecies competition and its probable influence upon the vertical distribution of Appalachian salamanders in the genus Plethodon. Ecology 32:266–274
Hewitt GM (1996) Some genetic consequences of ice ages, and their role in divergence and speciation. Biol J Linn Soc 58:247–276
Hewitt GM (2000) The genetic legacy of the Quaternary ice ages. Nature 405:907–913
Hewitt GM (2004) Genetic consequences of climatic oscillations in the Quaternary. Philos Trans R Soc B 359:183–195
Highton R (1995) Speciation in eastern North-American salamanders of the genus Plethodon. Annu Rev Ecol Syst 26:579–600
Highton R, Webster TP (1976) Geographic protein variation and divergence in populations of the salamander Plethodon cinereus. Evolution 30:33–45
Highton R, Hastings AP, Palmer C, Watts R, Hass CA, Culver M, Arnold SJ (2012) Concurrent speciation in the eastern woodland salamanders (Genus Plethodon): DNA sequences of the complete albumin nuclear and partial mitochondrial 12s genes. Mol Phylogenet Evol 63:278–290
Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A (2005) Very high resolution interpolated climate surfaces for global land areas. Int J Climatol 25:1965–1978
Jackson ST, Webb RS, Anderson KH, Overpeck JT, Webb T III, Williams JH, Hansen BCS (2000) Vegetation and environment in eastern North America during the last glacial maximum. Quat Sci Rev 19:489–508
Jaeger RG (1971) Competitive exclusion as a factor influencing the distributions of two species of terrestrial salamanders. Ecology 52:632–637
Jansson R, Dynesius M (2002) The fate of clades in a world of recurrent climatic change: Milankovitch oscillations and evolution. Annu Rev Ecol Syst 33:741–777
Jones MT, Voss SR, Ptacek MB, Weisrock DW, Tonkyn DW (2006) River drainages and phylogeography: an evolutionary significant lineage of shovel-nosed salamander (Desmognathus marmoratus) in the southern Appalachians. Mol Phylogenet Evol 38:280–287
Kass RE, Raftery AE (1995) Bayes factors. J Am Stat Assoc 90:773–795
Kohn MJ, Fremd TJ (2008) Miocene tectonics and climate forcing of biodiversity, western United States. Geology 36:783–786
Kozak KH, Wiens JJ (2006) Does niche conservatism promote speciation? A case study in North American salamanders. Evolution 60:2604–2621
Kozak KH, Wiens JJ (2010) Niche conservatism drives elevational diversity patterns in Appalachian salamanders. Am Nat 176:40–54
Kozak KH, Wiens JJ (2012) Phylogeny, ecology, and the origins of climate-richness relationships. Ecology 93:S167–S181
Kozak KH, Blaine RA, Larson A (2006a) Gene lineages and eastern North American paleodrainage basins: phylogeography and speciation in salamanders of the Eurycea bislineata species complex. Mol Ecol 15:191–207
Kozak KH, Weisrock DW, Larson A (2006b) Rapid lineage accumulation in a non-adaptive radiation: phylogenetic analysis of diversification rates in eastern North American woodland salamanders (Plethodontidae: Plethodon). Proc R Soc B 273:539–546
Kozak KH, Graham CH, Wiens JJ (2008) Integrating GIS-based environmental data into evolutionary biology. Trends Ecol Evol 23:141–148
Kozak KH, Mendyk RW, Wiens JJ (2009) Can parallel diversification occur in sympatry? Repeated patterns of body-size evolution in coexisting clades of North American salamanders. Evolution 63:1769–1784
Kürschner WM, Kvacek Z, Dilcher DL (2008) The impact of Miocene atmospheric carbon dioxide fluctuations on climate and the evolution of terrestrial ecosystems. Proc Natl Acad Sci USA 105:449–453
Lanfear R, Calcott B, Ho SYW, Guindon S (2012) PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analysis. Mol Biol Evol 29:1695–1701
Liu C, Berry PM, Dawson TP, Pearson RG (2005) Selecting thresholds of occurrence in the prediction of species distributions. Ecography 28:385–393
MacArthur RH (1984) Geographical ecology: patterns in the distribution of species. Princeton University Press, Princeton
Mayden RL (1985) Biogeography of Ouachita Highland fishes. Southwest Nat 30:195–211
Mayden RL (1988) Vicariance biogeography, parsimony, and evolution in North American freshwater fishes. Syst Zool 37:329–355
Moritz C (1994) Defining ‘evolutionarily significant units’ for conservation. Trends Ecol Evol 9:373–375
Morrone JJ (2009) Evolutionary biogeography: an integrative approach with case studies. Columbia University Press, New York
Mueller RL, Macey JR, Jaekel M, Wake DB, Boore JL (2004) Morphological homoplasy, life history evolution, and historical biogeography of plethodontid salamanders inferred from complete mitochondrial genomes. Proc Natl Acad Sci USA 101:13820–13825
Near TJ, Keck BP (2005) Dispersal, vicariance, and timing of diversification in Nothonotus darters. Mol Ecol 14:3485–3496
Near TJ, Page LM, Mayden RL (2001) Intraspecific phylogeography of Percina evides (Percidae: Etheostomatinae): an additional test of the Central Highlands pre-Pleistocene vicariance hypothesis. Mol Ecol 10:2235–2240
Peterson AT (2001) Predicting species’ geographic distributions based on ecological niche modeling. Condor 103:599–605
Peterson AT, Soberón J, Pearson RG, Anderson RP, Martínez-Meyer E, Nakamura M, Araújo MB (2011) Ecological niches and geographic distributions. Princeton University Press, Princeton
Petit JR, Jouzel J, Raynaud D, Barkov NI, Barnola JM, Basile I, Bender M, Chappellaz J, Davis M, Delaygue G, Delmotte M, Kotlyakov VM, Legrand M, Lipenkov VY, Lorius C, Pépin L, Ritz C, Saltzman E, Stievenard M (1999) Climate and atmospheric history of the past 420,000 years from Vostok ice core, Antarctica. Nature 399:429–436
Petranka JW (1998) Salamanders of the United States and Canada. Smithsonian Institution Press, Washington, DC
Petranka JW, Murray SS (2001) Effectiveness of removal sampling for determining salamander density and biomass: a case study in an Appalachian streamside community. J Herpetol 35:36–44
Phillips SJ, Dudík M (2008) Modeling of species distributions with Maxent: new extensions and a comprehensive evaluation. Ecography 31:161–175
Phillips SJ, Anderson RP, Schapire RE (2006) Maximum entropy modeling of species geographic distributions. Ecol Model 190:231–259
Pyron RA, Burbrink FT (2010) Hard and soft allopatry: physically and ecologically mediated modes of geographic speciation. J Biogeogr 37:2005–2015
Rader RB, Belk MC, Shiozawa DK, Crandall KA (2005) Empirical tests for ecological exchangeability. Anim Conserv 8:239–247
Rambaut A, Drummond AJ (2009) Tracer v.1.6. http://beast.bio.ed.ac.uk/Tracer
Rissler LJ, Smith WH (2010) Mapping amphibian contact zones and phylogeographic break hotspots across the United States. Mol Ecol 19:5404–5416
Semlitsch RD, O’Donnell KM, Thompson FR III (2014) Abundance, biomass production, nutrient content, and the possible role of terrestrial salamanders in Missouri Ozark forest ecosystems. Can J Zool 92:997–1004
Shepard DB, Burbrink FT (2008) Lineage diversification and historical demography of a sky island salamander, Plethodon ouachitae, from the Interior Highlands. Mol Ecol 17:5315–5335
Shepard DB, Burbrink FT (2009) Phylogeographic and demographic effects of Pleistocene climatic fluctuations in a montane salamander, Plethodon fourchensis. Mol Ecol 18:2243–2262
Shepard DB, Burbrink FT (2011) Local-scale environmental variation generates highly divergent lineages associated with stream drainages in a terrestrial salamander, Plethodon caddoensis. Mol Phylogenet Evol 59:399–411
Sites JW Jr, Morando M, Highton R, Huber F, Jung RE (2004) Phylogenetic relationships of the endangered Shenandoah Salamander (Plethodon shenandoah) and other salamanders of the Plethodon cinereus group (Caudata: Plethodontidae). J Herpetol 38:96–105
Soltis DE, Morris AB, McLachlan JS, Manos PS, Soltis PS (2006) Comparative phylogeography of unglaciated eastern North America. Mol Ecol 15:4261–4293
Spotila JR (1972) Temperature and water in the ecology of lungless salamanders. Ecol Monogr 42:95–125
Stockman AK, Bond JE (2007) Delimiting cohesion species: extreme population structuring and the role of ecological interchangeability. Mol Ecol 16:3374–3392
Strange RM, Burr BM (1997) Intraspecific phylogeography of North American highland fishes: a test of the Pleistocene vicariance hypothesis. Evolution 51:885–897
Swenson NG, Howard DJ (2005) Clustering of contact zones, hybrid zones, and phylogeographic breaks in North America. Am Nat 166:581–591
Swets JA (1988) Measuring the accuracy of diagnostic systems. Science 240:1285–1293
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Wake DB (2009) What salamanders have taught us about evolution. Annu Rev Ecol Evol Syst 40:333–352
Watts WA (1980) The Late Quaternary vegetation history of the southeastern United States. Annu Rev Ecol Syst 11:387–409
Webb T III, Bartlein PJ (1992) Global changes during the last 3 million years: climatic controls and biotic responses. Annu Rev Ecol Syst 23:141–173
Weisrock DW, Larson A (2006) Testing hypotheses of speciation in the Plethodon jordani species complex with allozymes and mitochondrial DNA sequences. Biol J Linn Soc 89:25–51
Werle E, Schneider C, Renner M, Völker M, Fiehn W (1994) Convenient single-step, one tube purification of PCR products for direct sequencing. Nucl Acids Res 22:4354–4355
Wiens JJ (2004) Speciation and ecology revisited: phylogenetic niche conservatism and the origin of species. Evolution 58:193–197
Wiens JJ, Graham CH (2005) Niche conservatism: integrating evolution, ecology, and conservation biology. Annu Rev Ecol Evol Syst 36:519–539
Wiens JJ, Engstrom TN, Chippendale PT (2006) Rapid diversification, incomplete isolation, and the “speciation clock” in North American salamanders (Genus: Plethodon): testing the hybrid swarm hypothesis of rapid radiation. Evolution 60:2585–2603
Zachos J, Pagani M, Sloan L, Thomas E, Billups K (2001) Trends, rhythms, and aberrations in global climate 65 Ma to present. Science 292:686–693
Acknowledgments
We acknowledge the Arkansas Game and Fish Commission (#092720102), Arkansas Natural Heritage Commission (S-NHCC-10-019), Missouri Department of Conservation (#14775), Louisiana Department of Wildlife and Fisheries (LNHP-10-089), Oklahoma Department of Wildlife Conservation (#4970), and University of Central Arkansas Animal Care and Use Committee (IACUC #11-002) for granting permits and permissions. Also, we also thank C. Austin, B. Fitzpatrick, K. Irwin, J. Briggler, J. Boundy, B. Cash, G. Adams, A. Lewis, K. Pyatt, and the graduate students at UCA for help in project development, field collections, and lab techniques. We thank the Louisiana Museum of Natural History at Louisiana State University (LSUMZ) and Bell Museum of Natural History at the University of Minnesota (JFBM) for sample loans. We thank K. Kozak for providing the alignment of DNA sequences from his 2009 paper and the University of Central Arkansas for funding.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Thesing, B.D., Noyes, R.D., Starkey, D.E. et al. Pleistocene climatic fluctuations explain the disjunct distribution and complex phylogeographic structure of the Southern Red-backed Salamander, Plethodon serratus . Evol Ecol 30, 89–104 (2016). https://doi.org/10.1007/s10682-015-9794-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10682-015-9794-3