Abstract
Many tropical organisms show large genetic differences among populations, yet the prevalent drivers of the underlying divergence processes are incompletely understood. We explored the effect of several habitat and natural history features (body size, macrohabitat, microhabitat, reproduction site, climatic heterogeneity, and topography) on population genetic divergence in tropical amphibians, based on a data set of 2680 DNA sequences of the mitochondrial cytochrome b gene in 39 widely distributed frog species from Brazil, Central America, Cuba, and Madagascar. Generalized linear models were implemented in an information-theoretic framework to evaluate the effects of the six predictors on genetic divergence among populations, measured as spatially corrected pairwise distances. Results indicate that topographic complexity and macrohabitat preferences have a strong effect on population divergence with species specialized to forest habitat and/or from topographically complex regions showing higher phylogeographic structure. This relationship changed after accounting for phylogenetic relatedness among taxa rendering macrohabitat preferences as the most important feature shaping genetic divergence. The remaining predictors showed negligible effects on the observed genetic divergence. A similar analysis performed using the population-scaled mutation rate (Θ) as response variable showed little effect of the predictors. Our results demonstrate greater evolutionary independence among populations of anurans from forested regions versus species from open habitats. This pattern may result from lower vagility and stringency in reproductive requirements of rainforest species. Conversely, open landscapes may offer ephemeral and unstable breeding sites suitable for vagile generalist species, resulting in reduced intraspecific divergence. Our results predict that, for a given period of time, there should be a higher chance of speciation in tropical anurans living in forests than in species adapted to open habitats.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
As genetic data from multiple taxa accumulates and sound statistical tools are developed, phylogeography is becoming one of the most integrative fields in evolutionary biology, targeting the processes that generated the observed distribution of biodiversity, as formerly envisioned by the architects of the field (Hickerson et al. 2010). Underlying this discipline is the observation that the genetic structure displayed by populations of organisms, over different spatial scales, radically differs among species. The pioneering comparative study of Avise et al. (1987), based mainly on mitochondrial DNA data (mtDNA), identified four distinct types of phylogeographic structure: (1) deep haplotype trees that are geographically structured, (2) deep haplotype trees that are unstructured, (3) shallow haplotype trees showing geographic structure, (4) shallow unstructured trees, each type corresponding to a particular historical setting. Despite many refinements of phylogeographic analysis, this categorization still remains valid.
A typical goal of comparative mtDNA phylogeography is to identify common sets of historical vicariant events that have geographically structured a group of ancestrally co-distributed organisms in a similar way (Avise et al. 1987; Arbogast and Kenagy 2001; Zink 2002; Rissler and Smith 2010) or resulted in concomitant assortment of genetic variation in space (Carnaval and Moritz 2008; Rocha et al. 2008; Fouquet et al. 2012). However, it is likely that the distribution of genetic variation in a species is not only shaped by historical events but also by ecological constraints (e.g. Endler 1992; Duminil et al. 2007). Intrinsic and extrinsic factors influencing rates and patterns of dispersal and survival of migrants act across a range of temporal and spatial scales and can together lead to either accruement or erosion of the genetic structure within and among groups of individuals (Grosberg and Cunningham 2001).
Correlating patterns of phylogeography or genetic variation with different species-specific traits offers a possibility to understand why the genetic structure of certain species responded in a similar way to climatic and geological history. Examples, often based on allozyme meta-analyses, include substrate type as a determinant of highly congruent spatial genetic structures in plant species (Alvarez et al. 2009), temperate versus boreal-temperate distribution and seed type influencing genetic differentiation in European trees (Aguinagalde et al. 2005), mating system, seed dispersal mode and geographic range size influencing population genetic structure in seed plants (Duminil et al. 2007), life-history traits determining connectivity in marine fishes (Galarza et al. 2009; Luiz et al. 2012), dispersal ability influencing isolation-by-distance in insects (Peterson and Denno 1998), and reduction of genetic variation on islands (Frankham 1997).
Amphibians are a classic example of organisms typically displaying deep phylogeographic structure of type I, presumably because of their relatively low dispersal ability (Avise 2000, 2009). Although most amphibians studied to date show such deep phylogeographic structure (Avise 2000, 2009; Vences and Wake 2007; Wells 2007; Zeisset and Beebe 2008), numerous examples for amphibian species of shallow, unstructured phylogeography exist, often corresponding to temperate species characterized by fast postglacial range expansions (Babik et al. 2004; Kuchta and Tan 2005; Crottini et al. 2007; Vásquez et al. 2013; Vences et al. 2013) but also species for which such glacial influences are more difficult to invoke (Carnaval 2002; Burns et al. 2007; Rabemananjara et al. 2007; Makowsky et al. 2009). Furthermore, examples of deep haplotype lineages that are poorly geographically structured and even occur in sympatry have been reported for some tropical species such as Agalychnis callidryas (Robertson et al. 2009) and Oophaga pumilio (Hauswaldt et al. 2011). Phylogeographically unstructured amphibian species are often generalists (Carnaval 2002), live in open habitats (e.g., Makowsky et al. 2009), and reproduce in lentic water (summary in Vences and Wake 2007), but these presumed correlations still require proper testing.
Here we determine the phylogeographic structure of 39 tropical anuran species characterized by relatively wide geographic distributions. We use homologous fragments of the mitochondrial cytochrome b gene and estimate the average genetic divergence among populations, corrected by geographical distance, as the main indicator of phylogeographic structure. As many alternative hypotheses can be drawn to explain the extant genetic diversity observed between populations of a given species, we used an information-theoretic framework to evaluate the strength of evidence supporting alternative models relating several natural history features, topography and climate as predictors of the observed genetic divergence. We find that genetic divergence is mainly influenced by macrohabitat type (forested vs. open) and topographic complexity, suggesting that ecological predictors may substantially determine phylogeographic structure in tropical anurans.
Materials and methods
Target species
For this analysis we selected 39 species of tropical frogs from three regions: Madagascar, the Atlantic coast of Brazil, and Cuba, plus one species from Central America (Table 1). The species spanned a wide array of body sizes, habitat specializations, and major phylogenetic clades. Our selection was largely based on the feasibility of obtaining samples but especially, on the availability of precise geographic information for each locality and expert knowledge on the natural history and taxonomy of the target species. All species occupy rather wide ranges [4858–11,635,995 km2, mean 721,768 km2 (IUCN 2013)] and our sampling is representative of their distribution within the study regions. We used an integrative taxonomic approach and considered as species only those mitochondrial lineages for which additional evidence is available and thus far does not suggest subdivision into unrecognized cryptic species. We note that the additional available evidence, besides morphometry and bioacoustics, also encompasses lack of haplotype sharing in nuclear gene DNA sequences; data completeness and genes sequenced are however inconsistent among taxa, hampering comparisons of nuclear gene phylogeographic structure and leading us to restrict our study to mtDNA only.
Molecular methods
We used proteinase K (10 mg/ml) digestion followed by a standard salt extraction protocol (Bruford et al. 1992) to extract genomic DNA from ethanol-preserved tissue samples. We performed standard polymerase chain reactions in a final volume of 10 μl and using 0.3 μl each of 10 μM primer, 0.25 μl of total dNTP (10 mM), 0.08 μl of 5 u/μl GoTaq and 2.5 μl 5× GoTaq Reaction Buffer (Promega). For the majority of the species we PCR-amplified a fragment of the mitochondrial cob gene using primers Cytb-a and Cytb-c of Bossuyt and Milinkovitch (2000). Specific primers were designed for some species that showed low efficiency in PCRs (Table SM1). All successfully amplified PCR products were purified using Exonuclease I and Shrimp Alkaline Phosphatase (SAP) or Antarctic Phosphatase (AP) according to the manufacturer’s instructions (NEB). Forward strands of purified PCR templates were sequenced with the respective primers using dye-labelled dideoxy terminator cycle sequencing on an ABI 3130 automated DNA sequencer. Chromatograms were checked and base-calls were corrected by hand using CodonCode Aligner (v. 3.5.6, Codon Code Corporation). The 1724 newly obtained sequences were submitted to GenBank (accession numbers: KR347487-8487, KR907891-8643). For several taxa, sequences had been determined in previous projects and were retrieved from GenBank, a complete list of accession numbers is provided (Table SM2).
Explanatory and response variables
We considered sampling localities for each species as independent sample units because most localities were separated by substantial geographical distances (mean value = 336.98 km). Genetic divergence among populations was summarized by calculating a single value (average geographically-corrected genetic distance; see below) using all cob sequences available per sampling locality. Although a few sampling localities were geographically proximate (with a minimum distance of 1 km in the miniaturized Eleutherodactylus limbatus; minimum distances of 2–3 km in E. atkinsi, E. auriculatus, E. cuneatus, Oophaga pumilio, Ptychadena mascareniensis, and Pseudis tocantins; >5 km in all others species), mean geographic distances for each of these species were much higher (78.3–456.0 km, averaged over all pairs of localities, Table SM2). We therefore treated each locality as a separate sample and refrained from defining criteria to lump them into population-level units, given that such criteria would have necessarily remained species-specific and subjective due to a dearth of information on dispersal in tropical frogs.
To obtain a response variable summarizing phylogeographic structure of each species we first calculated average uncorrected genetic distance (P) between sampling localities, with pair-wise deletion of missing sites, for each species in MEGA 5 (Tamura et al. 2011). This measure of genetic differentiation is more consistent than F ST and related estimates, which can be biased when the number of individuals sampled per population is small or uneven (Meirmans and Hedrick 2011). The isolation by distance model predicts an increase in genetic differentiation with geographic distance (Wright 1943); to account for this effect we applied a scaling correction and calculated the adjusted genetic distance (Padj) as the ratio between the average genetic distance (P, expressed in percentage) and the average geographic distance among populations (measured in kilometers from the original collection coordinates in WGS84 projection using ArcGIS software). As devised, Padj measures the genetic divergence scaled on the unit of geographic distance. This metric represents the strength of phylogeographic structuring observed across the sampled region and is comparable to the scaled genetic distance applied by Guarnizo and Cannatella (2013).
A number of explanatory (predictor) variables, representing natural history features with putative effects on genetic divergence at the species level, were compiled from the literature, our field experience, or calculated from available data. These included the following:
-
1.
Body size (continuous: expressed in mm). We used the maximum value reported in the literature for each species. Maximum values of size, rather than averages, can be a good indicator of dispersal ability and thus be the most relevant size measure influencing genetic structure. Furthermore, for the set of species considered, maximum snout-vent length was the more consistent estimate of body size that could be extracted from the literature. We log-transformed the original values to approximate the normal distribution.
-
2.
Macrohabitat (categorical: open/forest). We classified each species according to its large scale habitat preference based on author experiences and literature data. For this purpose, we considered as open habitat those species exclusively or mainly found in open areas such as savannas and pastures, whereas taxa usually occurring in densely covered areas were classified as forest species. Those occurring in forested regions but typically found near roads, clearings or human settlements were classified as open habitat species.
-
3.
Microhabitat (categorical: semi-aquatic/arboreal/terrestrial). This classification was based on author experience and literature data. Species were classified according to the microhabitat where they are most commonly found in nature (throughout their activity period).
-
4.
Reproduction site (pond/stream/direct). Species were classified according to their characteristic oviposition site. A specific category was created to accommodate direct-developing (or nidicolous, or phytotelmic) species whose eggs are laid in a variety of humid places but not in water.
-
5.
Climatic heterogeneity (continuous: non-dimensional). We performed a multidimensional ordination of the set of populations sampled in climatic space. The values of 19 climatic variables at the sampled localities were extracted from the WorldClim database (Hijmans et al. 2005), with approximately 1 km resolution, using the intersect function in ArcGIS 10 software (ESRI Inc., Redlands, CA). These values were normalized and submitted to a principal components analysis (PCA) using the “prcomp” function in R (R-Development Core Team 2014). Scores of the first four components (with eigenvalues >1 and representing 93.2 % of total variation) were used to compute a per-species hypervolume in climatic space using the R package “hypervolume” (Blonde et al. 2014). We performed 1000 replications of the algorithm and estimated the bandwidth value using the “estimate_bandwidth” function, other parameters were kept at default settings. Climatic hypervolumes thus constructed should summarize the multi-dimensional range of bioclimatic variables observed among the localities sampled and therefore represent the effects of a multitude of spatial and geomorphologic factors that ultimately shape the climate (geographic location, elevation, orientation, etc.).
-
6.
Topographic complexity (continuous: expressed in percent). We used the geographical coordinates of the localities sampled to derive a minimum convex polygon (MCP) per species. Topographic complexity was calculated as the standard deviation of the slopes observed within a species’ MCP. Standard deviation of slope remains the single most effective measure of surface roughness due to the simplicity of calculation, detection of fine scale/regional relief, and performance at a variety of scales (Grohmann et al. 2011). Slope calculations and zonal statistics were performed in ArcGIS 10 software using altitude data from the NASA Shuttle Radar Topographic Mission (SRTM) with a spatial resolution of approximately 1 km (available at http://srtm.usgs.gov). As a MCP spans the whole area between the sampled localities of a species, our topographic complexity measure is a simplified estimate that characterizes the entire region where dispersal events might have occurred in the past, regardless of range discontinuities and the current distribution of suitable habitats. However, considering the limited number of locality records available for most species, the calculations using MCPs may be more realistic than fine-scale mapping or ecological niche modeling (Wollenberg et al. 2011) and from a biogeographic perspective still constitute an adequate estimate of habitat heterogeneity and potential barriers to gene flow.
The selected predictors represent expected drivers of genetic differentiation but were deliberately restricted to features that can be unambiguously coded and result in uncorrelated variables (as determined by Pearson’s test: −0.22 < r < 0.26; 0.35 < P < 0.99). Each of them probably constitutes a generalization of a multitude of underlying factors potentially determining the spatial genetic structure in each particular species.
To account for the possible influence of demographic history on the observed results, we estimated theta, the population-scaled mutation rate (Θ = 2Neμ), as an alternative response variable and conducted identical analyses using the same set of predictors. For each species alignment we used phangorn (Schliep 2011), and ape (Paradis et al. 2004) R packages to construct UPGMA genealogies from which Θ was estimated using the theta.tree function of the pegas R package (Paradis 2010).
Statistical analyses
We log-transformed the response variable and two of the continuous predictors (size and climatic heterogeneity) to improve normality fit, which was assessed by Shapiro–Wilk’s test in R. We used an information-theoretic approach for model selection based on information criteria (Burnham and Anderson 2002) to evaluate the importance of different predictors in determining genetic divergence among populations. The dataset containing the values of the five predictors and response variable for each species was analyzed with the R package glmulti (Calcagno and de Mazancourt 2010). This statistical procedure generates all possible linear models relating predictors and the response variable and then conducts automated model selection by ranking the models by a chosen information criterion, the corrected Akaike Information Criteria (AICc) in our case. We implemented the procedure using linear models and deliberately excluded interaction terms because the sample size available for some combinations of factors was low.
Model selection uncertainty was assessed by comparing ∆AICc values, Akaike weights and evidence ratios (Burnham and Anderson 2002). We used a multi-model averaging approach that incorporates the information from all models in the a priori set. This procedure accounts for selection uncertainty and provides more robust estimates by including information not captured by the best model (Burnham et al. 2011). Using model-averaging procedures in glmulti, we estimated the relative importance of predictors (ratio of the cumulative weights of all models including the variable to those not including the variable) and calculated the model-averaged regression coefficients (b) and their associated 95 % confidence intervals. Based on the simulation tests of Calcagno and de Mazancourt (2010), we considered as important, those predictors with importance values above 80 %.
Some of the included species occur on offshore islands close to the mainland, and populations from these islands were included in our analysis (Oophaga pumilio: Bocas del Toro archipelago; Hylodes asper, Ischnocnema sp. CS3, I. parva: Ilhabela; Eleutherodactylus auriculatus: Isla de la Juventud; Blommersia wittei, Boophis tephraeomystax: Nosy Be; Gephyromantis sculpturatus: Nosy Boraha, Nosy Mangabe; and G. redimitus, Mantidactylus guttulatus: Nosy Mangabe). As we will discuss below, we do not see these short-distance recent saltwater barriers as qualitatively different from other barriers to dispersal; yet, to test a possible bias caused by their inclusion, we (1) repeated the model-averaging analysis after excluding the data from the respective localities and (2) compared the genetic distances of island to mainland populations to those of mainland populations.
We assessed if the phylogenetic relationships among our target species correlate with Padj, Θ, or predict similarity in any of the explanatory variables by estimating the proportion of phylogenetic signal in each variable compared to that expected under Brownian motion (the K statistic of Blomberg et al. (2003)) and determined its statistical significance according to a permutation-based procedure in the R package picante (Kembel et al. 2010). The input phylogeny was estimated by maximum likelihood based on nucleotide variation in our cob alignment, enforcing a topology that conformed to the multigene phylogenies of Pyron and Wiens (2011) and Wollenberg et al. (2011). This tree (Figure SM1) was computed in PAUP 4.10 (Swofford 2003). We used the chronos function of the ape R package (setting the age of the whole tree to 1 with λ = 0) to fit a chronogram to this tree. We repeated the model selection procedure using phylogenetic contrasts extracted from each variable. Finally, to evaluate the relationship between the phylogenetically-corrected Padj and Θ, we performed a Pearson’s correlation test in R.
Results
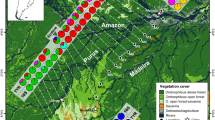
The final dataset included 2680 cob sequences from 39 species of tropical anurans from Brazil, Cuba, and Madagascar (and including one species from Central America), averaging 66 sequences per species (range 9–270) from an average of 13 distinct localities per species (3–34), representative of their distributions within the study areas (Table 1, Figure SM2). The observed phylogeographic structures varied substantially and values of geographically-adjusted genetic divergence (Padj) ranged from 0.001 % × km−1 in Peltophryne peltocephala to 0.111 % × km−1 in Hylodes asper (Table 1). Six examples are illustrated in Fig. 1; the gene trees obtained from the cob alignments for each species are also provided (Figure SM2).
Phylogeographic differences among representative open habitat and forest specialist anurans. In each panel a topographic map illustrates the localities sampled, progressively colored by their geographic position, and a Neighbour-Joining phylogenetic tree, constructed from P distances of the DNA sequences of the cob gene of the corresponding samples. Open habitat species (left) are compared to forest habitat species (right) in Brazil (a), Cuba (b), and Madagascar (c); species were selected to show particularly pronounced differences in phylogeographic pattern. Note the different scales between left and right panels indicating up to ten-fold differences in levels of genetic differentiation (average values of geographically-corrected genetic divergences were three-fold higher in forest vs. open habitat species: 0.029 vs. 0.009 % × km−1). (Color figure online)
A total of 64 first-order models relating the five predictors and the genetic distances were compared. The information metrics of the 18 best ranked models included in the 95 % confidence set, with AICc weights >0.01, are shown in Table 2. A graphical summary of the information-theoretic ranking of models is presented in Fig. 2a, b. The best-fitting model included two predictors, macrohabitat and topographic complexity, and explained half of the variance observed in the data (R 2 = 0.53, ANOVA F 2,36 = 22.41, p < 0.001) (Fig. 2). Inspection of the evidence ratios indicated that this model was two times more likely than the next two competing models, which showed evidence ratios within two AICc units of the best-fitting model. These two models also included macrohabitat and topographic complexity but with the additional influence of body size (second best model) or microhabitat (third best model) as predictors of genetic distances (Table 2, Fig. 2). Mean geographically-adjusted genetic divergence (Padj) in species from forested habitat was three-fold higher than in those from open habitat (0.029 vs. 0.009 % × km−1; see original data in Table 1).
Correlates of genetic divergence among populations of 39 tropical amphibian species. a Plot of the ranked AICc values (AIC profile) of the 64 models analyzed, the horizontal red line indicates the top ranked models (within two AICc units from the best-fitting model). b Estimated importance of predictors computed across all models, the red line illustrates an 80 % threshold for importance. c Graphical representation of best-fitting model relating the log-transformed adjusted genetic distance (log(Padj)) with macrohabitat type and topographic complexity (slopeSD), and d box-plot of log(Padj) values by macrohabitat type. (Color figure online)
Multi-model averages indicate that topographic complexity and macrohabitat preferences have higher relative importance values than the other predictors (Fig. 2). The multi-model estimates indicate a strong positive effect of topographic complexity (b = 0.08; 95 %CI 0.03–0.14) and a strong negative effect of “open” macrohabitat preferences (b = −0.29; −0.56 to −0.02) on the observed genetic divergences (Fig. 2). The other two categorical predictors, microhabitat and reproduction site, showed negligible effects on the observed genetic divergence. Other continuous predictors showed very weak effects on the genetic divergence as the model-averaged confidence intervals of b included zero in all cases: body size (−0.08, −0.34–0.19), climatic heterogeneity (−0.02, −0.15–0.10).
We evaluated whether the observed pattern could derive from regional differences. To this effect we conducted an additional analysis in which we included “region” (as a factor with three levels: “Brazil”, “Cuba”, and “Madagascar”; after excluding Oophaga pumilio) in addition to the other six predictors of genetic diversity. The results of this analysis were practically identical to those reported above with “region” ranked as the least important factor in explaining the variation in genetic diversity (Table SM3, Figure SM3a). Additionally, in order to assess the effect of potential sampling bias, we conducted an additional analysis excluding poorly sampled species: Ischnocnema parva (3 localities, 9 sequences), Blommersia wittei (3, 12), and Heterixalus carbonei (3, 16). The results of this analysis (Table SM4, Figure SM3b) were also very similar to those obtained using the full dataset. A further analysis carried out after excluding the data derived from samples collected on offshore islands produced very similar results to those obtained using the full dataset (Table SM5, Figure SM3c).
We found that Padj was not influenced by phylogeny (Bloomberg’s K = 0.060, P = 0.052), neither was Θ (K = 0.046, P = 0.206) or the climatic heterogeneity index (K = 0.046, P = 0.661). All the other predictors had small but significant levels of phylogenetic signal: log-transformed body size (K = 0.078, P = 0.019), reproduction site (K = 0.094, P = 0.003), microhabitat (K = 0.153, P = 0.001), macrohabitat (K = 0.057, P = 0.047), and topographic complexity (K = 0.087, P = 0.004). Model selection analysis on the phylogenetically-corrected data also indicated a stronger effect of macrohabitat preferences and topographic complexity. The best ranked model included only these two explanatory variables, followed by another model including only macrohabitat (Table SM6). Multi-model averages indicated that macrohabitat effect has much higher relative importance than the other predictors (Figure SM4) and model-averaged estimates indicate a strong effect of the contrasts of macrohabitat preferences on the contrasts of Padj (b = 0.416, 95 %CI 0.166–0.665) while all the other predictors showed negligible effects.
There was a weak and nonsignificant negative correlation between Θ values and Padj distances (Pearson’s r = −0.30, p = 0.062, CI 0.56 to −0.01). Also, the same set of predictors showed a much weaker effect on Θ and the best-fitting model, including the effects of topographic complexity and climatic heterogeneity (Table SM7), explained only 18 % of the variance (R 2 = 0.179, ANOVA F 2,36 = 5.15, p = 0.011). The topographic complexity ranked first in terms of relative importance among predictors (Figure SM4). Similarly, the phylogenetic contrasts of Θ were weakly influenced by the explanatory variables with the null model ranking as the second best model in the candidate set (Table SM8) and none of the variables were of high predictive importance (Figure SM4).
Discussion
We found evidence for a greater evolutionary independence among populations of tropical anurans, as measured by mtDNA divergence, occurring in forest habitat and/or in topographically complex areas, than among populations from open habitat and/or smoother landscapes. This result is derived from data encompassing three tropical biomes (Cuba, Madagascar, South America) and holds for a set of species that vary with respect to a wide array of life history and ecological traits. More importantly, the macrohabitat preference was the only important effect observed when phylogenetic relationship was controlled for and thus may be considered a general determinant of differentiation among populations of tropical anurans. Although genetic divergence among populations does not determine a speciation event, it is correlated with factors that promote speciation (Turelli et al. 2001; Coyne and Orr 2004), and can be expected to be especially important in clades in which allopatric speciation predominates. In this respect, our results predict that, for a given period of time, there should be a higher chance of speciation in anurans living in montane regions and/or forest habitat than for species adapted to open habitats and/or lowlands. This prediction is consistent with higher speciation rates in taxa restricted to mountainous rainforest habitat (Wollenberg et al. 2008).
The most important trend detected by our analysis is related to habitat preferences and not to reproductive mode or adult body size as previous hypotheses suggested (Dubois 2004; Pabijan et al. 2012). Previous analyses of the relationship between genetic differentiation and natural history features of amphibians have been either restricted in geographic span or taxonomic coverage. Nevo and Beiles (1991) analyzed published allozyme diversity data from 189 amphibian species and concluded that ecological rather than demographic variables predominate in explaining and predicting genetic diversity and divergence in amphibian populations. This interpretation of results was later contested due to a lack of proper phylogenetic correction, uneven sampling of amphibian families, and correlation among the chosen ecological predictors (Wells 2007). Pabijan et al. (2012) analyzed mitochondrial DNA divergence in 40 species of mantellid frogs inhabiting two rainforest communities in Madagascar. Their findings indicate an inverse association between body size and nucleotide divergence between populations but no influence of other life-history variables (reproductive mode, range size, microhabitat and skin texture) on genetic variation. According to these results, lower dispersal capacity in small-bodied frogs may explain the size-specific differences in regional differentiation observed in mantellids (Pabijan et al. 2012).
Proximate causes of population divergence
One straightforward explanation for the observed pattern is that forest-adapted anuran species living in climatically buffered microhabitats (Scheffers et al. 2014) are less tolerant to climatic fluctuations and therefore less dispersal-prone. On the other hand, species from open landscapes are adapted to ephemeral and unstable breeding sites and might evolve higher vagility to reach spatially shifting locations of breeding sites (Phillips et al. 2006) resulting in a higher degree of gene flow over large distances (Vences et al. 2002; Burns et al. 2007; Van Bocxlaer et al. 2010). Dispersal, and hence gene flow, may furthermore be limited if either valleys or ridges form barriers in montane areas (Wollenberg et al. 2008; Gehring et al. 2012; Guarnizo and Cannatella 2013).
Our results align perfectly with those of a comparatively similar study involving 27 tropical bird species (Tilston-Smith et al. 2014). These authors found that understorey-adapted species showed higher values of genetic diversity than canopy-adapted species and hypothesize that the principal drivers of speciation are the organism-specific abilities to persist and disperse in the landscape. Also in beetles, species that are adapted to historically less stable habitats, i.e., sand dunes versus stable compact-soil habitats, have higher dispersal abilities and consequently a lower degree of between-population genetic differentiation (Papadopoulou et al. 2009, 2011). In salmonid fishes, the interaction of landscape characteristics and life history traits (e.g. differing dispersal capabilities) may determine the degree of genetic structure among species (Gomez-Uchida et al. 2009). Taken as a whole, these results lend strong support to the important role of dispersal capacity and ecological specialization as drivers of speciation.
Studies in co-occurring amphibian species have shown that the genetic structure differs in ways that can be predicted by their dispersal capacities and life history attributes (Robertson et al. 2009; Richardson 2012). Landscape homogeneity and stochasticity in available breeding sites have been linked to increased population connectivity and lower genetic differentiation in desert-adapted amphibians (Chan and Zamudio 2009) and higher levels of habitat heterogeneity have been related to elevated diversification rates and increased regional differentiation within amphibian species in mountainous areas (Guarnizo and Cannatella 2013, 2014). Our results emphasize the role of habitat characteristics in determining the population genetic structure of tropical amphibians providing one possible explanation for the higher rates of speciation observed in tropical forest-dwelling amphibians inhabiting predominantly mountainous areas which have been considered as “species pumps” (Smith et al. 2007).
Concurrent changes in long-term effective population sizes among open-habitat, lowland species, or alternatively, forest-dwelling species, may offer another potential explanation for the disparate levels of genetic structure between these groups. For example, climate driven reductions of forest cover in the Quaternary, broadly synchronous in the Old World and New World tropics (Servant et al. 1993; Thomas 2008), could have induced range expansions or contractions in open-habitat and forest species, respectively. These historic events could potentially produce the observed pattern in our study. However, the mitochondrial population-scaled mutation parameter (Θ) was not associated with any of the predictor variables, nor was it significantly correlated with average genetic distances among populations, suggesting that effective population size is insufficient to explain the observed patterns. Nevertheless, this lack of correlation should be viewed with care because population size estimates based on a single locus may be inaccurate and the assumption of a link between population size and mtDNA diversity has been criticized on various grounds (e.g. Bazin et al. 2006; Nabholz et al. 2009). We herein used mtDNA as a first approximation to genetic diversity, as done in other studies of poorly known tropical organisms (e.g., Carnaval et al. 2009; Tilston-Smith et al. 2014) but we suggest that variation in the nuclear genome be used to further explore the influence of ecological factors on population parameters.
Recent land-use changes and clearings of forested regions may have increased the functional landscape connectivity for some generalist open-habitat species (Youngquist and Boone 2014) which could have also benefited from novel breeding sites (Knutson et al. 2004). If selection for increased dispersal capability and low site-fidelity occurs in open-habitat, then these species would advantageously exploit the newly-available habitat and show reduced phylogeographic structure (Chan and Zamudio 2009). An example of rapid adaptive change with increase of dispersal capacity has been documented in invasive populations of Rhinella marina in Australia (Phillips et al. 2006) and could be more frequent than currently acknowledged. Additionally, some human activities (agriculture, construction) might also result in the accidental translocation and establishment of individuals with a concomitant reduction in the observed genetic divergence between populations, and these events should be more likely among open-habitat species than among forest-restricted species.
Although we are convinced that our results are a robust representation of a real biological pattern, some caveats remain. Our approach assumes that each species, as defined herein, is a single independent evolutionary lineage (de Queiroz 2007). We considered species for which a sufficient amount of biological data (advertisement calls, morphology, and differentiation in nuclear genes) is available, and where none of these data suggest the existence of sympatric or allopatric cryptic species. However, depending on the species criteria used, future studies might result in a taxonomic subdivision of taxa such as Mantidactylus betsileanus into a northern and a southern species (Fig. 1), distinguished by a substantial mitochondrial subdivision. Each of these units would then have a much lower phylogeographic structure than the widespread unit herein considered as M. betsileanus.
The Mantidactylus example points to the methodological dilemma of using species as units for comparative analyses of inter-lineage divergences, as already pointed out by Riddle and Hafner (1999). Because factors such as forested macrohabitat, high elevation, or small body size (Pabijan et al. 2012) reduce gene flow among amphibian populations, they might simultaneously lead to deeper phylogeographic structure within species and to elevated rates of allopatric speciation. Therefore only a few genuinely widespread species might exist under factors favoring allopatric speciation, such as in extensive forest cover or higher elevation (this study) or among small-sized species (Pabijan et al. 2012). Distribution ranges are known to be smaller in small-sized mantellids (Wollenberg et al. 2011; Pabijan et al. 2012) and it is likely that this pattern applies to amphibians in general, as it does to other organisms (Gaston and Blackburn 1996). So far, it remains however untested whether amphibian species distributed in open habitats and at low elevations have statistically larger ranges, taking body size into account.
The interdependence of phylogeographic differentiation, speciation and range size might also account for the conspicuous lack of predictive power of body size in our data set, contrary to results of a previous study (Pabijan et al. 2012). The experimental design of that study excluded precisely those factors (elevation and macrohabitat) that turned out most influential in the analysis herein, suggesting that the influence of body size on phylogeographic pattern might overall be comparatively minor, yet influential in environmentally homogeneous settings. Furthermore, the two sites compared in Pabijan et al. (2012) are at a distance of roughly 250 km from each other, and the analysis thus included several species of rather narrow distribution that would not have qualified for our analysis herein. Given that small species are expected to have smaller ranges, the under-representation of such small sized species might have further reduced the influence of body size in our analysis.
Our analysis employed a population-based approach, and considered each sampling site as a sampling unit for the calculation of genetic distances. This could potentially lead to low values of genetic divergence if geographically proximate sampling sites were actually non-independent samples of the same population. Given the overall limited dispersal ability and high philopatry of amphibians (summary in Vences and Wake 2007; Zeisset and Beebe 2008) we consider our approach justified, and are convinced that sampling sites separated by 1–3 km can harbor distinct genetic demes (see also Jehle et al. 2005). In any case it is highly unlikely that this issue has biased our results because both minimum and mean geographical distances in our data set were on average lower in forest versus open-area species (21 vs. 110 km and 267 vs. 457 km) yet forest species had higher average genetic divergences.
For several species our analysis includes samples from offshore islands such as the Bocas del Toro archipelago in Panama, the Isla de la Juventud in Cuba, and Nosy Be, Nosy Boraha, and Nosy Mangabe in Madagascar. These islands are separated by shallow depths from the mainland, and given current knowledge of Pleistocene sea level changes they all have been connected to the mainland during the Last Glacial Maximum 20,000 years ago (e.g., Fleming et al. 1998; Iturralde-Vinent 2006; Gehara et al. 2013a). Saltwater barriers obviously constitute a formidable barrier to gene flow in frogs (but see Vences et al. 2003; Measey et al. 2007). The temporal discontinuity of saltwater barriers separating the island populations of our target species led us to consider them as not qualitatively different from other barriers such as large rivers or past incursions of salt water straits. Analyses without the affected species did not result in important deviations from the main pattern observed, suggesting that inclusion of the island samples did not bias our results. Because in many cases, our study species are co-distributed over large parts of their ranges, we expect their phylogeographic patterns to be affected by the same major barriers (e.g. rivers, volcanic activity). However, currently available methods cannot reliably quantify such barriers over large time and geographic scales. Although unlikely, we cannot rule out that our results were influenced by a higher incidence of extrinsic barriers in the ranges of the forest species included, rather than intrinsic dispersal capacity of the species themselves. Given that our approach compares the products of differentiation over vast geographic and temporal scales, we recommend that future refinements should include: (1) calculating geographical distances based on dispersal paths of suitable habitat (e.g., Brown and Yoder 2015); to account for the deep genetic divergences observed, these should be modeled into deeper time (Pliocene) than currently possible with available paleo-climatic layers; as well as (2) quantification of barriers not usually captured by such models, such as rivers.
Our preferred hypothesis interprets the encountered differences in genetic divergence between open versus forest habitat species by contrasting dispersal ability, leading to higher population connectivity and gene flow in the open habitat species. Similar differences in genetic variation among species could however also be caused by disparities in mutation rate (Nabholz et al. 2009) which in turn is determined by generation time and fecundity (Romiguier et al. 2014), and metabolic rate (Santos 2012). Although these variables are not reliably known for most of the species in our data set, there is no indication for lower fecundity and/or lower metabolic rate in tropical amphibians dwelling in open habitat, and we therefore do not expect a strong influence of differences in mutation rate on either the phylogeographic structure or the demographic estimates of our target species.
To complement the emerging picture of ecological and morphological drivers of phylogeographic differentiation in amphibians, and based on results herein, we identify potentially instructive avenues of further research: on the one hand, a more detailed analysis of amphibian range sizes is overdue and should simultaneously assess the influences of latitude, elevation, body size, macrohabitat, and other factors on the global diversity of amphibians. Secondly, the use of species as units for analysis has numerous advantages if species are defined as groups of metapopulations characterized by ongoing gene flow. However, it is well known that gene flow can extend beyond species boundaries in amphibians, especially among closely-related forms (e.g. Liu et al. 2010; Hedrick 2013; Zieliński et al. 2013) and might not be accurately described by mitochondrial markers (Irwin 2002; Currat et al. 2008). Hence, phylogeographic comparative analyses of units defined by equivalent divergence times, ideally determined from multiple unlinked nuclear loci, will outperform analyses based on species units and ultimately reveal the effects of natural history and ecological variables on population genetic divergence.
References
Aguinagalde I, Hampe A, Mohanty A, Martín JP, Duminil J, Petit RJ (2005) Effects of life-history traits and species distribution on genetic structure at maternally inherited markers in European trees and shrubs. J Biogeogr 32:329–339
Alvarez N, Thiel-Egenter C, Tribsch A, Holderegger R, Manel S, Schönswetter P, Taberlet P, Brodbeck S, Gaudeul M, Gielly L, Küpfer P, Mansion G, Negrini R, Paun O, Pellecchia M, Rioux D, Schüpfer F, Loo MV, Winkler M, Gugerli F, Consortium I (2009) History or ecology? Substrate type as a major driver of spatial genetic structure in Alpine plants. Ecol Lett 12:632–640
Arbogast BS, Kenagy GJ (2001) Comparative phylogeography as an integrative approach to historical biogeography. J Biogeogr 28:819–825
Avise JC (2000) Phylogeography: the history and formation of species. Harvard University Press, Cambridge
Avise JC (2009) Phylogeography: retrospect and prospect. J Biogeogr 36:3–15
Avise JC, Arnoldt J, Ball RM, Bermingham E, Lamb T, Neigel JE, Reeb CA, Saunders NC (1987) Intraspecific phylogeography: the mitochondrial DNA bridge between population genetics and systematics. Annu Rev Ecol Syst 18:489–522
Babik W, Branicki W, Sandera M, Litvinchuk SN, Borkin LJ, Irwin JT, Rafiński J (2004) Mitochondrial phylogeography of the moor frog, Rana arvalis. Mol Ecol 13:1469–1480
Bazin E, Glémin S, Galtier N (2006) Population size does not influence mitochondrial genetic diversity in animals. Science 312:570–572
Blomberg SP, Garland T, Ives AR (2003) Testing for phylogenetic signal in comparative data: behavioral traits are more labile. Evolution 57:717–745
Blonde B, Lamanna C, Violle C, Enquist BJ (2014) The n-dimensional hypervolume. Global Ecol Biogeogr 23:595–609
Bossuyt F, Milinkovitch MC (2000) Convergent adaptive radiations in Madagascan and Asian ranid frogs reveal covariation between larval and adult traits. Proc Natl Acad Sci USA 97:6585–6590
Brown JL, Yoder AD (2015) Shifting ranges and conservation challenges for lemurs in the face of climate change. Ecol Evol 5:1131–1142
Bruford MW, Hanotte O, Brookfield JFY, Burke T (1992) Single-locus and multilocus DNA fingerprinting. In: Hoelzel AR (ed) Molecular genetic analysis of populations: a practical approach. IRL Press, Oxford, pp 225–270
Burnham KP, Anderson DR (2002) Model selection and multimodel inference: a practical information-theoretic approach, 2nd edn. Springer, New York
Burnham KP, Anderson DR, Huyvaert KP (2011) AIC model selection and multimodel inference in behavioral ecology: some background, observations, and comparisons. Behav Ecol Sociobiol 65:23–35
Burns EL, Eldridge MDB, Crayn DM, Houlden BA (2007) Low phylogeographic structure in a widespread endangered Australian frog Litoria aurea (Anura: Hylidae). Conserv Genet 8:17–32
Calcagno V, de Mazancourt C (2010) glmulti: an R package for easy automated model selection with (generalized) linear models. J Stat Softw 34:1–29
Carnaval AC (2002) Phylogeography of four frog species in forest fragments of northeastern Brazil—a preliminary study. Integr Comp Biol 42:913–921
Carnaval AC, Moritz C (2008) Historical climate modelling predicts patterns of current biodiversity in the Brazilian atlantic forest. J Biogeogr 35:1187–1201
Carnaval AC, Hickerson MJ, Haddad CFB, Rodrigues MT, Moritz C (2009) Stability predicts genetic diversity in the Brazilian Atlantic Forest hotspot. Science 323:785–789
Chan LM, Zamudio KR (2009) Population differentiation of temperate amphibians in unpredictable environments. Mol Ecol 18:3185–3200
Coyne JA, Orr HA (2004) Speciation. Sinauer Associates, Sunderland
Crottini A, Andreone F, Kosuch J, Borkin LJ, Litvinchuk SN, Eggert C, Veith M (2007) Fossorial but widespread: the phylogeography of the common spadefoot toad (Pelobates fuscus). Mol Ecol 16:2374–2754
Currat M, Ruedi M, Petit RJ, Excoffier L (2008) The hidden side of invasions: massive introgression by local genes. Evolution 62:1908–1920
de Queiroz K (2007) Species concepts and species delimitation. Sys Biol 56:879–886
Dubois A (2004) Developmental pathway, speciation and supraspecific taxonomy in amphibians 1. Why are there so many frog species in Sir Lanka? Alytes 22:19–37
Duminil J, Fineschi S, Hampe A, Jordano P, Salvini D, Vendramin GG, Petit RJ (2007) Can population genetic structure be predicted from life-history traits? Am Nat 169:662–672
Endler JA (1992) Genetic heterogeneity and ecology. In: Berry RJ, Crawford TJ, Hewitt GM (eds) Genes in ecology. Blackwell Science, Oxford, pp 315–334
Fleming K, Johnston P, Zwartz D, Yokoyama Y, Lambeck K, Chappell J (1998) Refining the eustatic sea-level curve since the Last Glacial Maximum using far- and intermediate-field sites. Earth Planet Sci Lett 163:327–342
Fouquet A, Noonan BP, Rodriguez MT, Pech N, Gilles A, Gemmell NJ (2012) Multiple quaternary refugia in the eastern Guiana Shield revealed by comparative phylogeography of 12 frog species. Sys Biol 61:461–489
Frankham R (1997) Do island populations have less genetic variation than mainland populations? Heredity 78:311–327
Galarza JA, Carreras-Carbonell J, Macpherson E, Pascual M, Roques S, Turner GF, Rico C (2009) The influence of oceanographic fronts and early-life-history traits on connectivity among littoral fish species. Proc Natl Acad Sci USA 106:1473–1478
Gaston KJ, Blackburn TM (1996) Range size-body size relationships: evidence of scale dependence. Oikos 75:479–485
Gehara M, Summers K, Brown JL (2013a) Population expansion, isolation and selection: novel insights on the evolution of color diversity in the strawberry poison frog. Evol Ecol 27:797–824
Gehara M, Canedo C, Haddad CFB, Vences M (2013b) From widespread to microendemic: molecular and acoustic analyses show that Ischnocnema guentheri (Amphibia: Brachycephalidae) is endemic to Rio de Janeiro, Brazil. Conserv Genet 14:973–982
Gehring P-S, Pabijan M, Randrianirina JE, Glaw F, Vences M (2012) The influence of riverine barriers on phylogeographic patterns of Malagasy reed frogs (Heterixalus). Mol Phylogenet Evol 64:618–632
Gomez-Uchida D, Knight TW, Riuzzante DE (2009) Interaction of landscape and life history attributes on genetic diversity, neutral divergence and gene flow in a pristine community of salmonids. Mol Ecol 18:4854–4869
Grohmann CH, Smith MJ, Riccomini C (2011) Multiscale analysis of topographic surface roughness in the Midland Valley, Scotland. IEEE Trans Geosci Remote Sens 49:1200–1213
Grosberg RK, Cunningham CW (2001) Genetic structure in the sea: from populations to communities. In: Bertness MD, Gaines S, Hay ME (eds) Marine community ecology. Sinauer Associates, Sunderland, pp 61–84
Guarnizo CE, Cannatella DC (2013) Genetic divergence within frog species is greater in topographically more complex regions. J Zool Syst Evol Res 51:333–340
Guarnizo CE, Cannatella DC (2014) Geographic determinants of gene flow in two sister species of tropical Andean frogs. J Hered 105:216–225
Hauswaldt JS, Ludewig A-K, Vences M, Pröhl H (2011) Widespread co-occurrence of divergent mitochondrial haplotype lineages in a Central American species of poison frog (Oophaga pumilio). J Biogeogr 38:711–726
Hedrick PW (2013) Adaptive introgression in animals: examples and comparison to new mutation and standing variation as sources of adaptive variation. Mol Ecol 22:4606–4618
Hickerson MJ, Carstens BC, Cavender-Bares J, Crandall KA, Graham CH, Johnson JB, Rissler LJ, Victoriano PF, Yoder AD (2010) Phylogeography’s past, present, and future: 10 years after Avise, 2000. Mol Phylogenet Evol 54:291–301
Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A (2005) Very high resolution interpolated climate surfaces for global land areas. Int J Climatol 25:1965–1978
Irwin DE (2002) Phylogeographic breaks without geographic barriers to gene flow. Evolution 56:2383–2394
Iturralde-Vinent MA (2006) Meso-cenozoic Caribbean paleogeography: implications for the historical biogeography of the region. Int Geol Rev 48:791–827
IUCN (2013) The IUCN red list of threatened species. In: International Union for Conservation of Nature and Natural Resources
Jehle R, Burke T, Arntzen JW (2005) Delineating fine-scale genetic units in amphibians: probing the primacy of ponds. Conserv Genet 6:227–234
Kembel SW, Cowan PD, Helmus MR, Cornwell WK, Morlon H, Ackerly DD, Blomberg SP, Webb CO (2010) Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26:1463–1464
Knutson MG, Richardson WB, Reineke DM, Gray BR, Parmelee JR, Weick SE (2004) Agricultural ponds support amphibian populations. Ecol Appl 14:669–684
Kuchta SR, Tan A-M (2005) Isolation by distance and post-glacial range expansion in the rough-skinned newt, Taricha granulosa. Mol Ecol 14:225–244
Liu K, Wang F, Chen W, Tu L, Min M-S, Bi K, Fu J (2010) Rampant historical mitochondrial genome introgression between two species of green pond frogs, Pelophylax nigromaculatus and P. plancyi. BMC Evol Biol 10:201
Luiz OJ, Madin JS, Robertson DR, Rocha LA, Wirtz P, Floeter SR (2012) Ecological traits influencing range expansion across large oceanic dispersal barriers: insights from tropical Atlantic reef fishes. Proc R Soc B 279:1033–1040
Makowsky R, Chesser J, Rissler LJ (2009) A striking lack of genetic diversity across the wide-ranging amphibian Gastrophryne carolinensis (Anura, Microhylidae). Genetica 135:169–183
Measey GJ, Vences M, Drewes RC, Chiari Y, Melo M, Bourles B (2007) Freshwater paths across the ocean: molecular phylogeny of the frog Ptychadena newtoni gives insights into amphibian colonization of oceanic islands. J Biogeogr 34:7–20
Meirmans PG, Hedrick PW (2011) Assessing population structure: Fst and related measures. Mol Ecol Res 11:5–18
Nabholz B, Glémin S, Galtier N (2009) The erratic mitochondrial clock: variations of mutation rate, not population size, affect mtDNA diversity across birds and mammals. BMC Evol Biol 9:54
Nevo E, Beiles A (1991) Genetic diversity and ecological heterogeneity in amphibian evolution. Copeia 1991:565–592
Pabijan M, Wollenberg KC, Vences M (2012) Small body size increases the regional differentiation of populations of tropical mantellid frogs (Anura: Mantellidae). J Evol Biol 25:2310–2324
Papadopoulou A, Anastasiou I, Keskin B, Vogler AP (2009) Comparative phylogeography of tenebrionid beetles in the Aegean archipelago: the effect of dispersal ability and habitat preference. Mol Ecol 18:2503–2517
Papadopoulou A, Anastasiou I, Spagopoulou F, Stalimerou M, Terzopoulou S, Legakis A, Vogler AP (2011) Testing the species–genetic diversity correlation in the Aegean archipelago: toward a haplotype-based macroecology? Am Nat 178:241–255
Paradis E (2010) pegas: an R package for population genetics with an integrated-modular approach. Bioinformatics 26:419–420
Paradis E, Claude J, Strimmer K (2004) APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20:289–290
Peterson MA, Denno RF (1998) The influence of dispersal and diet breadth on patterns of genetic isolation by distance in phytophagous insects. Am Nat 152:428–446
Phillips BL, Brown GP, Webb JK, Shine R (2006) Invasion and the evolution of speed in toads. Nature 439:803
Pyron RA, Wiens JJ (2011) A large-scale phylogeny of Amphibia including over 2,800 species, and a revised classification of extant frogs, salamanders, and caecilians. Mol Phylogenet Evol 61:543–583
Rabemananjara FCE, Chiari Y, Ramilijaona OR, Vences M (2007) Evidence for recent gene flow between north-eastern and south-eastern Madagascan poison frogs from a phylogeography of the Mantella cowani group. Front Zool 4:1
R-Development Core Team (2014) R: A language and environment for statistical computing. Version 3.1.1. R Foundation for Statistical Computing, Vienna, Austria
Richardson JL (2012) Divergent landscape effects on population connectivity in two co-occurring amphibian species. Mol Ecol 21:4437–4451
Riddle BR, Hafner DJ (1999) Species as units of analysis in ecology and biogeography: time to take the blinders off. Global Ecol Biogeogr 8:433–441
Rissler LJ, Smith WH (2010) Mapping amphibian contact zones and phylogeographical break hotspots across the United States. Mol Ecol 19:5404–5416
Robertson JM, Duryea MC, Zamudio KR (2009) Discordant patterns of evolutionary differentiation in two Neotropical treefrogs. Mol Ecol 18:1375–1395
Rocha LA, Rocha CR, Robertson DR, Bowen BW (2008) Comparative phylogeography of Atlantic reef fishes indicates both origin and accumulation of diversity in the Caribbean. BMC Evol Biol 8:e157
Romiguier J, Gayral P, Ballenghien M, Bernard A, Cahais V, Chenuil A, Chiari Y, Dernat R, Duret L, Faivre N, Loire E, Lourenco JM, Nabholz B, Roux C, Tsagkogeorga G, Weber AA, Weinert LA, Belkhir K, Bierne N, Glémin S, Galtier N (2014) Comparative population genomics in animals uncovers the determinants of genetic diversity. Nature 515:261–263
Santos JC (2012) Fast molecular evolution associated with high active metabolic rates in poison frogs. Mol Biol Evol 29:2001–2018
Scheffers BR, Edwards DP, Diesmos A, Williams SE, Evans TA (2014) Microhabitats reduce animal’s exposure to climate extremes. Glob Change Biol 20:495–503
Schliep KP (2011) phangorn: phylogenetic analysis in R. Bioinformatics 27:592–593
Servant M, Maley J, Turcq B, Absy M-L, Brenac P, Fournier M, Ledru M-P (1993) Tropical forest changes during the Late Quaternary in African and South American lowlands. Global Planet Change 7:25–40
Smith SA, Oca ANMd, Reeder TW, Wiens JJ (2007) A phylogenetic perspective on elevational species richness patterns in Middle American treefrogs: why so few species in lowland tropical rainforests? Evolution 61:1188–1207
Swofford DL (2003) PAUP* Phylogenetic analysis using parsimony (*and other methods). Version 4. Sinauer Associates, Sunderland, Massachusetts
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Thomas MF (2008) Understanding the impacts of Late Quaternary climate change in tropical and sub-tropical regions. Geomorphology 101:146–158
Tilston-Smith B, McCormack JE, Cuervo AM, Hickerson MJ, Aleixo A, Cadena CD, Pérez-Emán J, Burney CW, Xie X, Harvey MG, Faircloth BC, Glenn TC, Derryberry EP, Prejean J, Fields S, Brumfield RT (2014) The drivers of tropical speciation. Nature 515:406–409
Turelli M, Barton NH, Coyne JA (2001) Theory and speciation. Trends Ecol Evol 16:330–343
Van Bocxlaer I, Loader SP, Roelants K, Biju SD, Menegon M, Bossuyt F (2010) Gradual adaptation toward a range-expansion phenotype initiated the global radiation of toads. Science 372:679–682
Vásquez D, Correa C, Pastenes L, Palma RE, Méndez MA (2013) Low phylogeographic structure of Rhinella arunco (Anura: Bufonidae), an endemic amphibian from the Chilean Mediterranean hotspot. Zool Stud 52:35e
Vences M, Wake DB (2007) Speciation, species boundaries and phylogeography of amphibians. In: Heatwole H (ed) Amphibian Biology. Surrey Beatty & Sons, Chipping Norton, pp 2613–2671
Vences M, Aprea G, Capriglione T, Andreone F, Odierna G (2002) Ancient tetraploidy and slow molecular evolution in Scaphiophryne: ecological correlates of speciation mode in Malagasy relict amphibians. Chromosome Res 10:127–136
Vences M, Vieites DR, Glaw F, Brinkmann H, Kosuch J, Veith M, Meyer A (2003) Multiple overseas dispersal in amphibians. Proc R Soc B 270:2435–2442
Vences M, Hauswaldt JS, Steinfartz S, Rupp O, Goesmann A, Künzel S, Orozco-terWengel P, Vieites DR, Nieto-Roman S, Haas S, Laugsch C, Gehara M, Bruchmann S, Pabijan M, Ludewig A-K, Rudert D, Angelini C, Borkin LJ, Crochet P-A, Crottini A, Dubois A, Ficetola GF, Galán P, Geniez P, Hachtel M, Jovanovic O, Litvinchuk SN, Lymberakis P, Ohler A, Smirnov NA (2013) Radically different phylogeographies and patterns of genetic variation in two European brown frogs, genus Rana. Mol Phylogenet Evol 68:657–670
Wells KD (2007) The ecology and behavior of amphibians. The University of Chicago Press, Chicago
Wollenberg KC, Vieites DR, van der Meijden A, Glaw F, Cannatella DC, Vences M (2008) Patterns of endemism and species richness in Malagasy cophyline frogs support a key role of mountainous areas for speciation. Evolution 62:1890–1907
Wollenberg KC, Vieites DR, Glaw F, Vences M (2011) Speciation in little: the role of range and body size in the diversification of Malagasy mantellid frogs. BMC Evol Biol 11:217. doi:10.1186/1471-2148-11-217
Wright S (1943) Isolation by distance. Genetics 28:114–138
Youngquist MB, Boone MD (2014) Movement of amphibians through agricultural landscapes: the role of habitat on edge permeability. Biol Conserv 175:148–155
Zeisset I, Beebe TJC (2008) Amphibian phylogeography: a model for understanding historical aspects of species distributions. Heredity 101:109–119
Zieliński P, Nadachowska-Brzyska K, Wielstra B, Szkotak R, Covaciu-Marcov SD, Cogălniceanu D, Babik W (2013) No evidence for nuclear introgression despite complete mtDNA replacement in the Carpathian newt (Lissotriton montandoni). Mol Ecol 22:1884–1903
Zink RM (2002) Methods in comparative phylogeography, and their application to studying evolution in the North American aridlands. Integr Comp Biol 42:953–959
Acknowledgments
We are grateful to numerous friends and colleagues who have contributed samples and sequences for the present project, or provided assistance during fieldwork. In particular we would like to thank (in alphabetic order) Roberto Alonso, Franco Andreone, Adrian Garda, Sebastian Gehring, Frank Glaw, Jörn Köhler, Roger-Daniel Randrianiaina, Fanomezana Ratsoavina, Leslie Rissler, Axel Strauß, Chris Thawley, and David R. Vieites. Thanks are also due to Meike Kondermann and Gabi Keunecke for help with labwork. We are grateful to authorities in Brazil, Cuba and Madagascar for research, collection, and export permits. This study was carried with funds of the Deutsche Forschungsgemeinschaft (Grant VE247/7-1 to MV). AR and MP were supported by postdoctoral fellowships of the Humboldt Foundation. MG was supported by the Katholischer Akademischer Austauschdienst. CFBH was supported by Grants #2008/50928-1 and #2013/50741-7, São Paulo Research Foundation (FAPESP), and Grant #302518/2013-4, Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Rodríguez, A., Börner, M., Pabijan, M. et al. Genetic divergence in tropical anurans: deeper phylogeographic structure in forest specialists and in topographically complex regions. Evol Ecol 29, 765–785 (2015). https://doi.org/10.1007/s10682-015-9774-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10682-015-9774-7