Abstract
Cucumber is an important member of Cucurbitaceae family which is grown and consumed globally. It is consumed in the form of salads or pickles and has abundant supply of minerals and vitamins. It has been a focus of extensive research due to its publically available genome sequences. Marker assisted breeding has been employed to generate improved varieties of cucumber with resistance to various diseases besides desirable horticultural traits. One such trait is the production of parthenocarpic fruits i.e. seedless fruits developed in the absence of pollination. Naturally, parthenocarpy occurs under unfavourable environmental conditions. However, it is possible to artificially induce parthenocarpy by applying plant growth regulators. At molecular level, parthenocarpy is a result of complex role-play of genes and hormones with tight regulation. Many researchers aim at developing parthenocarpic varieties by either transferring the trait into existing varieties with resistance to specific diseases or by knocking off genes controlling parthenocarpy. A comprehensive knowledge of background and genetics of parthenocarpy is required in both cases. This review includes an-inclusive study of the molecular basis of parthenocarpy. The genes and hormones involved and their functions, inheritance of parthenocarpy and markers associated with it have been given in detail.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Vegetables are an indispensable component of human diet. They can be consumed either raw or cooked or preserved in the form of pickles. Cucumber (Cucumis sativus L.) is one of the major horticultural crops which offer great economic benefits to the cultivators and nutritional benefits to the consumers. The world production of cucumber increased from 67.9 M tonnes in 2011 to 93.5 M tonnes in 2021 (www.fao.org/faostat/en/) with Asia alone accounting for 85.4% of the total production. It is a Cucurbit and is known to have its origin in foothill regions of the Himalayas in India, China and Burma (Sebastian et al. 2010). Modern day cucumber (C. sativus var. sativus) is known to originate from three wild and semi-wild varieties, C. sativus L. var. sikkimensis, C. sativus L. var. hardwickii and C. sativus L. var. xishuangbannanesis. The cultivated cucumber is divided into four geographical groups based on their genomic analysis: India; Eurasia along with the West; Eastern Asia along with China; and Xishuangbanna from South-western China (Lv et al. 2012). The largest proportion of the genetic diversity belongs to the Indian group which also includes C. sativus L. var. hardwickii. The cucumber collection centers of the US, the Netherlands and China collectively comprise 3342 cucumber accessions (Lv et al. 2012). The widely available germplasm is beneficial to breeders for development of improved varieties of cucumbers having beneficial traits via marker assisted breeding. Production of seedless cucumbers also known as parthenocarpy is one such trait favoured by cultivators and consumers globally. In cucumber, parthenocarpy process has not been completely understood due to its complex mechanism under tight regulation by several environmental and genetic factors (Kaur et al. 2023). In-depth knowledge of hormones and genes regulating parthenocarpic fruit-set formation is essential for the development of parthenocarpic varieties either via conventional breeding or via genetic engineering approaches. The complete genomic sequences of cucumber are available at http://cucurbitgenomics.org/v2/ in public domain which prove beneficial for the study of genes. The updated database comprises five genome assemblies Cucumis sativus Chinese Long 9930 v3 (Li et al. 2019), Cucumis sativus Gy-14 v2 (Weng Lab, USDA-ARS Vegetable Crops Research Unit, Wisconsin), and Cucumis sativus B-10 v3 (Osipowski et al. 2020; Turek et al. 2023), Cucumis sativus var. hardwickii cv. PI 183967 v1 (Qi et al. 2013), and Cucumis hystrix v1 (Qin et al. 2021).
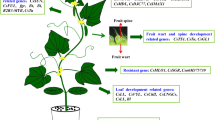
Parthenocarpy (Parthenos, virgin; karpos, fruit) is one of the most significant characters in cucumber which is the biological phenomenon of natural or artificial induction of fruit formation in the absence of pollination or any other kind of stimuli of the ovary (Gustafson 1939) (Fig. 1) and the plant is said to exhibit the trait of parthenocarpy when its fruits are either totally lacking seeds or have seeds of insignificant size (Pandolfini 2009). Cucumber is considered a favourable model plant to study the parthenocarpy as the trait is prevalent in its germplasm resources. Parthenocarpic cucumber fruits are bitterness free and juicier as compared to the seeded fruits and have the ability to upsurge the crop yields under unfavourable environmental circumstances such as its cultivation under protected conditions (Fabrice et al. 2000). The phenomenon of parthenocarpy in cucumber was first studied by Noll (1902) who coined the term ‘Parthenocarpy’. During early researches, reports for parthenocarpy were given by Ewert-Proskau (1909), Strong (1921, 1932), Wellington and Hawthorn (1928) and Hawthorn and Wellington (1930). It has been reported that in cucumber the facultative parthenocarpy varies quantitatively (Cao et al. 1997; El-Shawaf and Baker 1981a, b; Sun et al. 2006a, b). For the first time, De Ponti (1976) developed the parthenocarpic hybrid of cucumber by crossing parthenocarpic slicing cucumber line with gynoecious pickling cucumber line.
Parthenocarpy could be affected by abiotic, physiological, and genomic factors. Abiotic factors such as low temperature and short day length are known to enhance parthenocarpy (Rudich et al. 1977). Such environmental factors along with their interaction with genotype also effect the degree of parthenocarpy (Sun et al. 2006a), which makes selection of parthenocarpic progenies difficult during breeding programs. Certain hormones tend to regulate parthenocarpy. For example, the endogenous levels of indole-3-acetic acid (IAA) in parthenocarpic ovaries and fruits were higher in comparison to their pollinated counterparts (Boonkorkaew et al. 2008; Kim et al. 1992a, b). Various studies have demonstrated the role of exogenously applied growth-promoting chemicals which include auxins and auxin transport inhibitors, gibberellins, cytokinins, and brassinosteroid in inducing parthenocarpy (Cantliffe 1972; Fu et al. 2008; Kim et al. 1994; Quebedeaux and Beyer1972). Thus, fruit setting could be induced by applying compatible foreign pollen to the stigma in cucurbits (Yasuda 1935) because pollens have the above mentioned phytohormones (Gustafson 1937, 1942). Moreover, parthenocarpy can be stimulated in cucumbers by introduction of the DefH9-iaaM auxin-synthesizing gene (Yin et al. 2006).
The parthenocarpic varieties are also not suitable for outdoor cultivation as they develop fruits with seeds which render them unmarketable. Parthenocarpy was used for both protected and open-field production after Peterson (1960) developed a gynoecious cucumber line (MSU 713-5) and Peterson and Andher (1960) discovered a method for maintenance of such lines by inducing staminate flowers via gibberellic acid application. In case of greenhouse hybrids, the fruits set at initial five nodes are not vital as such fruits are removed. But in case of open cultivation, it is important that fruit setting at the initial nodes starts as early as possible. This provides early harvest opportunity, which is especially important for early yield (Kushnereva 2008). Thus, in order to develop parthenocarpic cucumber hybrids which are suitable for outdoor conditions, it is essential that the parental genotypes are selected such that their parthenocarpic potential is less reliant on environment.
Role of phyto-hormones in parthenocarpy
The formation of parthenocarpic fruits is thought to be initiated by gibberellic acid (GA) biosynthesis pathway (Fos et al. 2000). Furthermore, because auxin can be applied exogenously to promote parthenocarpy, auxins are essential for the formation of parthenocarpic fruits. Normally, auxin helps in maintaining the concentration of enzyme gibberellic acid oxidase which in turn regulates synthesis of GA (Fig. 2). However precise interactions between gibberellins and auxins might differ depending upon the plant type and tissue (Nitsch 1972). However, the role of endogenous levels of GAs in development of seedless fruit might not necessarily be required during the entire period of fruit development as high levels of GAs produced from 3 to 4 weeks after anthesis might not be required in later stages. Contrarily, low levels of gibberellins from 1 to 4 weeks after anthesis might result in inhibition of fruit development (Kataoka et al. 2004).
The first hormone known to cause parthenocarpy in plants was auxin. Cucurbitaceae crops, including watermelon, cucumber, and zucchini, undergo parthenocarpy when exposed to analogs of auxin (Wong 1938, 1939). Cucumbers have been successfully induced to exhibit parthenocarpy by Qian et al. (2018) using naphthaleneacetic acid (NAA), an exogenous plant growth regulator, one day prior to and on the day of flowering. There have been reports of a number of auxin biosynthetic routes involved in induction of parthenocarpy, including IAOx (indole-3-acetaldoxime), IAM (indole-3-acetamide), and Trp-IPyA (tryptophan-indole-3-pyruvic acid). In case of Trp-IPyA pathway, which is the most widely studied pathway, TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS (TAA1) genes convert tryptophan into indole-pyruvic acid which undergoes decarboxylation to form indole acetic acid in an irreversible reaction catalysed by YUCCA genes (Homayouni and Strader 2020).
Another essential plant hormone that regulates plant parthenocarpy after auxins is GA (Davison 1960). The ability of various gibberellins to produce parthenocarpy varies throughout species. For example, Qian et al. (2018) found that although GA3 does not successfully promote parthenocarpy in cucumbers, it robustly stimulates the production of seedless fruit in tomatoes. However, gibberellin GA4+7 treatment resulted in parthenocarpic cucumbers. There was no discernible weight difference between GA4+7-treated and pollinated cucumbers. According to Qian et al. (2018), GA4+7-treated cucumbers exhibited less firm flesh and higher amounts of total flavonoids and proteins during storage in contrast to pollinated cucumbers.
Another essential plant hormone that encourages parthenocarpy in plants is cytokinin. Cell division is stimulated by spraying N-(2-chloro-4-pyridyl)-N′-phenylurea (CPPU), an exogenous cytokinin growth regulator. CPPU was initially used by Hayata et al. (1995) to treat watermelon ovaries that were not pollinated, which led to the development of seedless fruit. Furthermore, the form, soluble solids content, and rind thickness of the watermelon fruit were unaffected by a 200 g/m3 CPPU concentration, indicating its direct application in production (Hayata et al. 1995; Maroto et al. 2005). When compared to pollinated watermelons, watermelons treated with CPPU might have a little lower sugar content; however, this could be due to varietal differences (Huitrón et al. 2007, Lopez-Galarza et al. 2004). In the production of Cucurbitaceae crops, such as cucumber (Qian et al. 2018), melon (Hayata et al. 2000; Liu et al. 2023), pumpkin (Ogawa and Takisawa 2022), and watermelon (Maroto et al. 2005), CPPU is frequently used to stimulate or augment parthenocarpy. This treatment positively impacts fruit setting, promotes fruit enlargement, and increases production overall. According to Sakakibara (2005), the primary roles of cytokinins are to suppress leaf senescence, promote plant germination and bud differentiation, and regulate meristem cell proliferation. Adenosine phosphate-isopentenyl transferase (IPT) catalyzes the initial stage of the production of cytokines. Nucleotide precursors are transformed into the active form by the LONELY GUY (LOG) enzyme (El-Showk et al. 2013). Cytokinin oxidases (CKXs) mediate the breakdown of cytokinins (Köllmer et al. 2014).
The exogenous application of phyto-hormones greatly influences parthenocarpic fruits in various crops. The hormones include auxins {NAA 2,4-dichlorophenoxyacetic acid (2,4-D)}, cytokinins {CPPU, 6-benzylaminopurine (BAP)}, GAs, brassinosteroids, ethylene and melatonin (De Jong et al. 2009; Kumar et al. 2014; Sharif et al. 2022). 2,4-D is a synthetic auxin which had been proven to induce parthenocarpy in tomato and pear (Cong et al. 2019; Serrani et al. 2007). Similarly, NAA effectively induced parthenocarpy in cucumber (Qian et al. 2018) and strawberry (Kang et al. 2013). Application of zeatin (cytokinin) to the unpollinated ovaries resulted in parthenocarpic fruits in case of tomato with a fruit setting percentage of 80% (Ding et al. 2013). Exogenous application of CPPU in pear cultivar resulted in parthenocarpy by accumulating IAA and reducing abscisic acid levels (Cong et al. 2020). GA positively regulates various fruit-related traits (Garmendia et al. 2019). Treating cucumber with combination of different GAs (GA4+7) successfully induced parthenocarpy (Qian et al. 2018). Brassinosteroids belong to group of steroid hormones playing a critical role in plant growth. However, fewer reports are available for brassinosteroids induced parthenocarpy as compared to other hormones. An analog of brassinosteroids i.e. 24-epibrassinolide was able to induce parthenocarpy in cucumber (Fu et al. 2008).
Environmental factors affecting parthenocarpy
Parthenocarpy is induced by a number of environmental conditions, including temperature, photoperiod, light intensity, and nutrient availability (Gou et al. 2022; Rylski 1974; Smith and Cochran 1935; Sun et al. 2016). According to genetic studies, most of a crop's natural parthenocarpic qualities are quantitative traits that are influenced by both environmental and genetic variables (Beraldi et al. 2004; Fos et al. 2000; Leitzow et al. 2016; Wu et al. 2016). A healthy supply of nutrients is essential for fruit growth and is particularly important for parthenocarpic fruit development (Gou et al. 2022). It has been documented that parthenocarpy can be induced by low temperatures in Cucurbitaceae crops, such as cucumber, melon, and zucchini (De Ponti 1976; Hao et al. 2009; Rylski 1974).
Short day conditions in cucumbers increase auxin activity, which in turn promotes parthenocarpy (Rudich et al. 1977). Cucumber ovary auxin level rises in response to low night-time temperatures, causing parthenocarpy. On the other hand, high temperatures prevent the beginning of parthenocarpy in cucumber ovaries by inhibiting the manufacture of auxin and gibberellin (Matlob and Kelly 1975). Rudich et al. (1977) reported that under high temperature conditions, parthenocarpic fruit development was greatly reduced in non-parthenocarpic Gy irrespective of day length. However, parthenocarpic line produced significant number of fruits under high temperature with long-day or short-day conditions. Cucumber parthenocarpy was shown to be more common when ovaries were found to have twice as much auxin at 15 °C as at 25 °C (Kim et al. 1992a, b). Meng et al. (2023) examined the impact of hormones on the low-temperature parthenocarpic fruit's development and discovered a small number of differentially expressed genes (230–460 DEGs) among the ovaries prior to anthesis According to endogenous hormone determination, the IAA and ACC accumulation throughout fruit development displayed an inverted curve: IAA accumulated continuously whereas ACC synthesis was suppressed during parthenocarpy at low temperatures.
Inheritance of parthenocarpy
The genetic basis of cucumber has been narrowed to 3–8% polymorphism among cultivated varieties and 10–25% among botanical cultivars (Behera et al. 2011). India has a huge genetic diversification as well as variability in case of fruit traits that have not been completely utilized for improving the crop’s genetic makeup. The development of gynoecious cucumbers possessing parthenocarpy has emerged a significant hurdle for breeders because these are used as parent in F1 hybrid development (Dhall et al. 2023). Although, the genetics of parthenocarpy have been a focus of research in previously published literature, the studies differ from each other due to the variations on genetic background, experimental designs as well as environmental factors. Table 1 exhibits the summary of the studies on the inheritance of parthenocarpy in cucumber. In cucumber, the genetic study of parthenocarpy started in 1930 and Pike and Peterson were the first to report inheritance of parthenocarpy in 1968. They reported that parthenocarpy was conditioned by a gene P which showed incomplete dominance. When occurring as homozygous, this gene formed parthenocarpic fruits at early nodes (usually at 5th node); while in heterozygous condition, it produced late fruits with parthenocarpy and also fewer in number than homozygous genotypes. Similar reports on the inheritance of parthenocarpy via incomplete dominant gene were given by Kim et al. (1992a); Juldasheva (1973) and Jat et al. (2017) (Table 1). However, According to Hawthorn and Wellington (1930) and Meshcherov and Juldasheva (1974), there is single recessive gene that controls parthenocarpy. Kvasnikov et al. (1970) used a European type of processing cucumber to propose that multiple genes showing incomplete recessiveness were responsible for controlling parthenocarpy. De Ponti and Garretsen (1976) studied the inheritance of parthenocarpy in the pickling cucumber by crossing eight pure gynoecious lines having different degrees of parthenocarpy (43–90% fruit set) with two plants (72421a, 72421b) from non-parthenocarpic gynoecious (NPG) line in incomplete diallel fashion and also developed six generations (P1, P2, B11, B12, F1, F2) of above crosses to study the presence of non-allelic gene interaction (epistasis) by using six generation mean analysis. They reported that the inheritance of parthenocarpy could be explained by three independent, isomeric major genes with additive action, together with additive × dominant (j) interaction. Independent studies by Castle (1921) and Wright (1968) report that all alleles controlling parthenocarpy are unlinked and exhibit equal additive effects, and that both parthenocarpic and non-parthenocarpic parental lines are homozygous for alternative alleles at all loci affecting the parthenocarpy. Since then, numerous studies have been carried out in regard to parthenocarpy in cucumber.
Numerous researches have suggested that parthenocarpy inheritance is congruent with qualities that are quantitative in nature, utilizing a variety of analytical techniques. The techniques include variance component analysis (Sun et al. 2006a; Yan et al. 2008), generation means analysis (Cao et al. 1997; Sun et al. 2006b), diallel analysis, and North Carolina Design II (El-Shawaf and Baker 1981a, b). Other genes are also involved in controlling the parthenocarpic behaviour resulting in parthenocarpic fruit set with a range of 0.33–0.62 with narrow-sense heritability as well as 5–13 effective factors (Sun et al. 2006a). The earliest case of parthenocarpy was observed in greenhouse slicers, followed by greenhouse Beit Alpha type and pickling type for production in high tunnels and fields respectively. The trait of parthenocarpy could be transferred from a donor to genotypes via few backcrosses (Sun et al. 2006c). They also suggested that 10 QTLs distributed across four genomic regions and 8 linked AFLP markers were responsible for controlling parthenocarpy in cucumber. Yan et al. (2008) reported parthenocarpy as quantitative trait under the control of two major genes and additional minor genes and suggested that, for the breeding of high parthenocarpic gynoecious cucumber, both parents used for crossing should have high parthenocarpy. While in another study by Yan et al. (2010, 2012a) on the inheritance of parthenocarpy in same backgrounds in cucumber using the mixed major and polygene model for inheritance of quantitative traits suggested that two additive-dominant-epistatic major genes and additive-dominant minor genes controlled parthenocarpy in gynoecious cucumber while in monoecious cucumber additive-dominant-epistatic polygenes were responsible instead of additive-dominant polygenes along with additive-dominant-epistatic major genes. Kumar et al. (2016) reported the predominant role of non-additive gene action governing the parthenocarpy in monoecious and gynoecious cucumber crosses. Devi et al. (2022) developed six generations by crossing parthenocarpic line PPC-6 with non-parthenocarpic genotype (Pusa Uday) and observed that a single incomplete dominant gene controlled parthenocarpy in the PPC-6 and QTL-seq analysis identified two major genomic regions, one each on chromosomes 3 and 6 spanning over a region of 2.7 Mb and 7.8 Mb, respectively. Gou et al. (2022) used 31 parthenocarpic cucumber lines for Genome-wide association study (GWAS) and observed the incompletely dominant control of parthenocarpy in cucumber and detected 5324 SNPs associated with parthenocarpy, from which six parthenocarpic loci, including two novel loci (Pfs1.1 and Pfs4.1) were identified.
Gene expression studies for parthenocarpy
Important details regarding the levels of gene expression connected to different plant metabolic processes can be found in the transcriptome of plants. The study of expression profiles of the genes becomes important in understanding the complete picture behind parthenocarpy. The endogenous levels of IAA modify the expression of parthenocarpy. The first report on the effect of endogenous level of auxins modifying the parthenocarpy in cucumber was given by Kim et al. (1992b). They stated that accumulation of IAA in the ovary was closely related with parthenocarpy and revealed that IAA is the major hormone which regulates parthenocarpy in cucumber and also stated that exogenous applied hormones promoted IAA accumulation in ovary. The actual elucidation of mechanism of regulating parthenocarpy remains uncertain. But Li et al. (2017) provided some evidences on the mechanism of parthenocarpy and revealed that the mechanism might be supported by a ‘parallel switch’ which consists of hormone dependent as well as hormone independent pathways. In case of pathway independent of hormones, process of fruit set is stimulated by certain regulatory proteins which are hormone insensitive known as the NP (Natural Parthenocarpy)-specialized proteins in ‘EC1’ (environmentally stable parthenocarpic cucumber cultivar). However, when sufficient amount of hormones are present, the young fruits which are formed through both pathways continue to grow till maturity. In hormone independent pathway, hormone sensitive fruits are developed which ultimately leads to fruit abortion. However, in case of hormone dependent pathway, the fruits became unresponsive to hormones and remains in a resting state because of increased expression of proteins which inhibits abortion. However, in such case, the fate of dormant fruits remains unknown. The genes involved in regulation of parthenocarpy include those involved in phytohormone, biosynthesis, signalling and transduction (Sharif et al. 2018). The known auxin transporter gene families include PIN-FORMED (PIN), AUX1/LAX, and the B subfamily of ATP-binding cassette (ABCB) transporters, of which PIN genes constitute the major auxin efflux transporters (Hu et al. 2021). The auxin signalling gene families which regulate parthenocarpy include auxin/IAA (AUX/IAA), transport inhibitor response 1/auxin signalling f-box proteins (TIR1/AFB), and auxin-responsive factor (ARF) families (Luo et al. 2018). The auxin signalling genes (AUX 22A-like-1, AUX22B-like-2, and AUX 28-like) were expressed higher in parthenocarpic cucumber as compared to non-parthenocarpic cucumber (Su et al. 2021). Other auxin signalling genes (TIR1 and AFB2 genes) have also been reported in cucumber by Xu et al. (2017). They investigated the heterologous overexpression of CsTIR1 and CsAFB2 in tomato resulting in an early fruit set.
The genes involved in GA synthesis also regulate parthenocarpy. In case of pear, GA20ox (gibberellic acid oxidase) was found be highly expressed in pollinated fruits (Wang et al. 2020). Similarly in tomato, fruit set was accompanied with the increased expressed of GA biosynthesis genes (SlGA20ox1, SlGA20ox2, and SlGA20ox3) along with decreased expression of GA deactivation gene SlGA2ox1 (Okabe et al. 2019). In case of cucumber, the genes CsGA3ox3, CsGA3ox4, CsGA2ox1, CsGA2ox2, CsGA2ox3, and CsGA2ox5 were feebly expressed in the parthenocarpic genotypes which indicated their negative role in inducing parthenocarpy (Mandal et al. 2022). In addition to these, gibberellin signalling genes DELLA and GID1 (GA INSENSITIVE DWARF1) also regulate parthenocarpy. It was observed that the DELLA and GID1arenegative and positive regulators of gibberellic acid biosynthesis, respectively (Hedden 2020). In addition to genes regulating auxin and gibberellin synthesis, cytokinin genes also regulate parthenocarpic fruit development. The cytokinin biosynthesis genes (CYP735A1, CYP735A2, and LOG1) were highly expressed in parthenocarpic cultivar of cucumber in contrast to cytokinin dehydrogenase genes (CKX1 and CKX3) which were down-regulated (Su et al. 2021). Similarly, Mandal et al. (2022) reported the elevated expression of the cytokinin biosynthesis genes (CsIPT, CsIPT1, CsIPT3, CsLOG1, CsLOG2, CsCYP735A1, and CsCYP735A2), and decreased expression of CsCKX1, CsCKX2, and CsCKX3 in parthenocarpic cucumbers. The cytokinin signalling genes (CsRR3/4a, CsRR3/4b, CsRR8/9a, and CsRR8/9c) negatively regulate parthenocarpy in cucumber as they expressed strongly in non-parthenocarpic genotype (Su et al. 2021). Negative regulation of parthenocarpy by CsRR3/4a, CsRR3/4b, CsRR8/9a, and CsRR8/9c and positive regulation by CsRR8/9b, CsRR8/9d, CsRR8/9e, and CsRR16/17 in cucumber has also been reported (Mandal et al. 2023). In case of tomato, increased expression of cytokinin biosynthesis genes SlIPT1, SlIPT2, SlCYP735A1, SlCYP735A2, and SlLOG2 was observed in ovaries after anthesis (Matsuo et al. 2012).
Genetic engineering for parthenocarpy
The field of biotechnology has provided us with varied opportunities with easy technologies for obtaining parthenocarpic varieties as compared to conventional breeding approaches (Rotino et al.1997; Varoquaux et al. 2000). The developments of seeds and fruits are inter-connected and are regulated by plant hormones (Gillaspy et al. 1993). These processes are important to achieve parthenocarpic fruits and to guarantee expression of parthenocarpy without affecting the growth of vegetative parts of the plant. The gene of interest contains certain regulatory region(s) that exhibits highly valuable genetic information for controlling their expression. Thus, an abundance or decline in the expression of such phytohormone regulating genes might result in either development of parthenocarpic fruit with altered morphology or an inefficient fruit setting (Falavigna and Rotino 2006). Hence, the approach to generate transgenics via silencing a particular gene through RNA interference (RNAi) and antisense RNA technology is dominant tool to achieve parthenocarpy in fruits. The gene regulation for parthenocarpy in cucumber reported by various researchers is mentioned in the Table 2. Yin et al. (2006) introduced chimeric DefH9-iaaM construct in cucumber genome to generate parthenocarpic cucumber fruits. The construct had DefH9 promoter and iaam coding sequence which produces parthenocarpic fruits by synthesis of auxins without pollination and the non-parthenocarpic or seeded fruits are obtained after pollination. The parthenocarpy achieved though was facultative (Rotino et al. 1997). To date, the use of DefH9-iaaM chimeric gene for parthenocarpic fruit development has been reported in tobacco (Rotino et al. 1997), eggplant (Rotino et al. 1997), tomato (Ficcadenti et al. 1995; Pandolfini 2009), strawberry and raspberry (Mezzetti et al. 2000).
Apart from DefH9-iaaM, other genes have also been targeted for inducing parthenocarpy in different crops. The techniques could be used to achieve high yielding parthenocapic cucumber genotypes. The down-regulation or loss-of-function mutation of SlIAA9, an Aux/IAA transcription factor which negatively regulates auxin response, induced parthenocarpy in tomato (Kim et al. 2019; Klap et al 2016; Saito et al. 2011; Wang et al. 2005). Auxin biosynthesis genes SlTAR1, ToFZY2, and ToFZY3 were found to be up-regulated in genome edited parthenocarpic tomato SlAGL6mutants (Gupta et al. 2021). The RNAi-mediated silencing of the SlDELLA increased the GA biosynthesis genes’ expression, thereby resulting in parthenocarpy (Marti et al. 2007). Down-regulation of SlTPL1 led to facultative parthenocarpy in tomato (He et al. 2021). In tomato, RNAi mediated gene suppression of Aucsia resulted in auxin-related phenotypes which included parthenocarpy (Molesini et al. 2009). The PARENTAL ADVICE-1 (PAD-1) is known to coordinate the reverse reaction of Trp-IPyA in the IAA biosynthesis pathway and its loss-of-function mutation tend to increase IAA accumulation in un-pollinated ovaries, thereby producing parthenocarpic fruits in case of eggplant (Matsuo et al. 2020).
Although, various studies of gene silencing/loss-of-function mutation have been successfully utilized to induce parthenocarpy, some genes can be over-expressed to yield the same results. The transient expression of CYP735A resulted in parthenocarpy in non-parthenocarpic cucumber (Cui 2013). Carmi et al. (2003) utilized ovary-specific expression of the Agrobacterium rhizogenes-derived gene rolB for inducing parthenocarpy in tomato.
Molecular breeding for parthenocarpy
In the initial studies it was exhibited that parthenocarpy in cucumber had been regulated by single or dual incomplete or recessive genes. But, recent studies suggested that the trait was controlled by several QTLs. The first molecular marker (ISSR) linked to parthenocarpy was developed by Chen et al. (2008). They found marker N92 showed polymorphism for non-parthenocarpic and parthenocarpic cucumber lines and estimated that there is genetic distance of 18.9 cM between this primer and cucumber parthenocarpy trait. Report for first QTL-linked marker associated with the parthenocarpy in cucumber was given by Sun et al. (2006c). They developed eight fluorescent amplified length polymorphism (AFLP) markers that are linked to parthenocarpy through QTL mapping and identified 10 QTLs related to parthenocarpy which were distributed over four genomic regions in cucumber. Selection for phenotypic bulks (high and low parthenocarpic) in small population of F2:3 (126) likely lead to the early generation fixation of alleles conditioning parthenocarpy. In addition, as the bulks used for BSA were derived by selfing instead of random mating in F3 generations, the assortment of all alleles for parthenocarpy into individual F4 plants would not be expected unless the ancestral F2 plant possessed all favourable alleles. Another development of AFLP markers associated with parthenocarpy in cucumber is defined by Yan et al. (2012b) and they reported AGG/CAA325 marker linked to non-parthenocarpy locus at distance of 9.7 cM. The name of the marker is based on 325 bp specific fragments that were amplified with the help of primer E41/M47 and no bands were observed in the parthenocarpic genotypes pool. Several attempts were also made to develop markers linked to parthenocarpy in pickling or processing types of cucumber. Wu et al. (2016) identified 7 QTLs linked with parthenocarpy including Parth2.1 as major QTL along with two candidate genes. Wu et al. (2016) discovered four related markers (SSR16226, Indel-T-32, Indel T-34, and Indel-T-39) to parthenocarpy in addition to reporting a major-effect QTL Parth2.1 and six minor-effect QTLs that contribute to the phenotypic variance of parthenocarpy in cucumbers. Minor effect QTL were designated as Parth1 at 101.0 cM on chromosome 1, Parth3.1 at 93.8 cM on chromosome 3, Parth5 at 58.0 cM on chromosome 5, Parth7 at 23.4 cM on chromosome 7, Parth2.2 at 50.3 cM on chromosome 2 and Parth3.1 at 57.5 cM on chromosome 3. The marker closely associated with Parth2.1 could be used for efficient marker assisted selection of parthenocarpy in cucumber. Lietzow et al. (2016) detected 7 QTLs related to parthenocarpic fruit set assigned as parth2.1 (on chromosome 2), parth4.1 (on chromosome 4), parth5.1 (on chromosome 5), parth6.1, parth6.2, parth6.3 (on chromosome 6) and parth7.1 (on chromosome 7). The QTLs parth5.1, parth6.1, parth6.2 and parth7.1 contributed equally in expressing parthenocarpy at commercially appropriate harvest stage while, parth7.1 was responsible for early parthenocarpy fruit set. This study is crucial in understanding parthenocarpy in cucumber for breeders as well as for researchers. Teams led by Wu et al. (2016) and Lietzow et al. (2016) detected 12 QTLs in two sources, although only 2 QTLs (parth2.1 and parth7.1) were found to be co-localized between them. This inconsistency in results reflects the problems in phenotyping parthenocarpy with accuracy, a trait which is hard to distinguish from yield. He et al. (2016) identified 9 genes including Csa2M349050.1, Csa2M350390.1, Csa2M351550.1, Csa2M354670.1, Csa2M354750.1, Csa2M355020.1, Csa2M353460.1, Csa2M354910.1, and Csa2M354980.1 which were expected to be potential genes involved in parthenocarpy of cucumber. All the genes except one were responsible for biosynthesis, regulation, and/or signal transduction for phytohormones and were expected to be involved in cell division. Zhihong et al. (2020) discovered 4 QTLs on chromosomes 1, 3 and 6, as well as one minor-effect QTL on chromosome 3 which occurred consistently throughout years. Gou et al. (2022) established that several QTLs and genes governed the parthenocarpy, which was inherited in an incompletely dominant way. The summarised information on development of QTLs for parthenocarpy in cucumber is provided in Table 3.
Thus validated identified molecular markers ease the incorporation of parthenocarpy in varied cucumber background that are accompanied by other desirable genes which might take longer time and more tedious ways if done through conventional breeding techniques.
Conclusion
Parthenocarpy is a phenomenon that results in the formation of seedless fruits without pollination. Such fruits are preferred over seeded ones by consumers. Thus, it becomes important for breeders to generate seedless varieties of cucumber with high resilience. The publically available genome sequences of cucumber accessions make it easier for breeders for generating improved varieties of cucumber. The QTLs and molecular markers associated with parthenocarpy further ease the processes. The major-effect QTLs parth2.1 and parth6.1 have been identified on chromosomes 2 and 6. Various genes have been identified which are directly involved in regulation of parthenocarpy. These genes form part of biosynthesis, signalling and perception of plant hormones such as auxins, cytokinins and gibberellins. These genes could be targeted via genetic engineering approaches for production of parthenocarpic fruits in cucumber.
References
Behera TK, Staub JE, Behera S, Delannay IY, Chen JF (2011) Marker-assisted backcross selection in an interspecific Cucumis population broadens the genetic base of cucumber (Cucumis sativus L.). Euphytica 178:261–272. https://doi.org/10.1007/s10681-010-0315-8
Beraldi D, Picarella ME, Soressi GP, Mazzucato A (2004) Fine mapping of the parthenocarpic fruit (pat) mutation in tomato. Theor Appl Genet 108:209–216
Boonkorkaew P, Hikosaka S, Sugiyama N (2008) Effect of pollination on cell division, cell enlargement, and endogenous hormones in fruit development in a gynoecious cucumber. Sci Hortic 116:1–7. https://doi.org/10.1016/j.scienta.2007.10.027
Cantliffe D (1972) Parthenocarpy in the cucumber induced by some plant growth regulating chemicals. Can J Plant Sci 52:781–785. https://doi.org/10.4141/cjps72-127
Cao BS, Chen XH, Xu Q, Gu CS (1997) The genetic effects of parthenocarpic generations of cucumber. Acta Hortic Sic 24:53–56
Carmi N, Salts Y, Dedicova B, Shabtai S, Barg R (2003) Induction of parthenocarpy in tomato via specific expression of the rolB gene in the ovary. Planta 217:726–735. https://doi.org/10.1007/s00425-003-1052-1
Castle WE (1921) An improved method of estimating the number of genetic factors concerned in cases of blending inheritance. Proc Natl Acad Sci USA 81:6904–6907
Chen XH, Wang J, Xu Q, Mi Y, Liang GH (2008) An ISSR marker linked to the parthenocarpic gene of cucumber. Mol Plant Breed 6:85–88
Cong L, Wu T, Liu H, Wang H, Zhang H, Zhao G, Wen Y, Shi Q, Xu L, Wang Z (2020) CPPU may induce gibberellin-independent parthenocarpy associated with PbRR9 in ‘Dangshansu’ pear. Hortic Res 7:68. https://doi.org/10.1038/s41438-020-0285-5
Cong L, Yue R, Wang H, Liu J, Zhai R, Yang J, Wu M, Si M, Zhang H, Yang C, Xu L, Wang Z (2019) 2,4-D-induced parthenocarpy in pear is mediated by enhancement of GA4 biosynthesis. Physiol Plant 166:812–820. https://doi.org/10.1111/ppl.12835
Cui L (2013) The study on molecular mechanism of cucumber parthenocarpy based on transcriptome. Dissertation, Nanjing Agricultural University, Nanjing
Davison R (1960) Fruit-setting of apples using gibberellic acid. Nature 188:681–682. https://doi.org/10.1038/188681b0
De Jong M, Mariani C, Vriezen WH (2009) The role of auxin and gibberellin in tomato fruit set. J Exp Bot 60:1523–1532. https://doi.org/10.1093/jxb/erp094
De Ponti OMB (1976) Breeding parthenocarpic pickling cucumbers (C. sativus L.): necessity, genetical possibilities environmental influences and selection criteria. Euphytica 25:29–40. https://doi.org/10.1007/BF00041526
De Ponti OMB, Garretsen F (1976) Inheritance of parthenocarpy in pickling cucumbers (Cucumis sativus L.) and linkage with other characters. Euphytica 25(1):633–642. https://doi.org/10.1007/BF00041600
Devi S, Sharma PK, Behera TK, Jaiswal S, Boopalakrishnan G, Kumari K, Mandal NK, Iquebal MA, Gopala Krishnan S, Bharti Ghosal C, Munshi AD, Dey SS (2022) Identification of a major QTL, Parth6.1 associated with parthenocarpic fruit development in slicing cucumber genotype, Pusa Parthenocarpic Cucumber-6. Front Plant Sci 13:1064556. https://doi.org/10.3389/fpls.2022.1064556
Dhall RK, Kaur H, Manchanda P, Eshanee (2023) Genetics and marker-assisted breeding for sex expression in cucumber. Front Genet 14:1180083. https://doi.org/10.3389/fgene.2023.1180083
Ding J, Chen B, Xia X, Mao W, Shi K, Zhao Y, Yu J (2013) Cytokinin-induced parthenocarpic fruit development in tomato is partly dependent on enhanced gibberellin and auxin biosynthesis. PLoS One 8(7):e70080. https://doi.org/10.1371/journal.pone.0070080
El-Shawaf IIS, Baker LR (1981) Inheritance of parthenocarpic yield in gynoecious pickling cucumber for once-over mechanical harvest by diallel analysis of six gynoecious lines. J Am Soc Hort Sci 106:359–364. https://doi.org/10.21273/JASHS.106.3.359
El-Shawaf IIS, Baker LR (1981) Combining ability and genetic variances of G×H F1 hybrids for parthenocarpic yield in gynoecious pickling cucumber form once-over mechanical harvest. J Am Soc Hort Sci 106:365–370. https://doi.org/10.21273/JASHS.106.3.365
El-Showk S, Ruonala R, Helariutta Y (2013) Crossing paths: cytokinin signalling and crosstalk. Development 140:1373–1383. https://doi.org/10.1242/dev.086371
Ewert-Proskau (1909) Neuere untersuchungen fiber parthenokarpie bei obstb/iumen und einigen anderen fruchttragenden gew/ichsen. Landw Jb 38:767–839
Fabrice RB, Michel D, Patrick G (2000) Less is better: new approaches for seedless fruit production. Biotopics 18:233–242. https://doi.org/10.1016/s0167-7799(00)01448-7
Falavigna A, Rotino GL (2006) Parthenocarpy, a strategy for fruit development under adverse environmental conditions. Seminar Nasional Pemanfaatan Bioteknologi untuk Mengatasi Cekaman Abiotik pada Tanaman, pp 42–51
Ficcadenti N, Sestili S, Pandolfini T, Cirillo C, Rotino GL, Spena A (1995) Genetic enginnering of parthenocarpic fruit development in tomato. Mol Breed 5:463–470. https://doi.org/10.1023/A:1009665409959
Fos M, Nuez F, García-Martínez JL (2000) The gene pat-2, which induces natural parthenocarpy, alters the gibberellin content in unpollinated tomato ovaries. Plant Physiol 122(2):471–480. https://doi.org/10.1104/pp.122.2.471
Fu FQ, Mao WH, Shi K, Zhou YH, Asami T, Yu JQ (2008) A role of brassinosteroids in early fruit development in cucumber. J Exp Bot 59:2299–2308. https://doi.org/10.1093/jxb/ern093
Garmendia A, Beltran R, Zornoza C, Garcia-Breijo FJ, Reig J, Merle H (2019) Gibberellic acid in citrus spp. flowering and fruiting: a systematic review. PLoS One 14:e0223147. https://doi.org/10.1371/journal.pone.0223147
Gillaspy G, Ben-David H, Gruissem W (1993) Fruits: a developmental perspective. Plant Cell 5:1439–1451. https://doi.org/10.1105/tpc.5.10.1439
Gou C, Zhu P, Meng Y, Yang F, Xu Y, Xia P, Chen J, Li J (2022) Evaluation and genetic analysis of parthenocarpic germplasms in cucumber. Genes 13:225. https://doi.org/10.3390/genes1302022
Gupta SK, Barg R, Arazi T (2021) Tomato agamous-like 6 parthenocarpy is facilitated by ovule integument reprogramming involving the growth regulator KLUH. Plant Physiol 185:969–984. https://doi.org/10.1093/plphys/kiaa078
Gustafson FG (1937) Parthenocarpy induced by pollen extracts. Am J Bot 24:102–107. https://doi.org/10.1002/j.1537-2197.1937.tb09075.x
Gustafson FG (1939) The cause of natural parthenocarpy. Am J Bot 26:135–138
Gustafson FG (1942) Parthenocarpy: natural and artificial. Bot Rev 8:599–654
Hao J, Li T, Kan S (2009) Study on the response of endogenous hormone to different night temperature and its relationship with fruit development in melon. Sci Agric Sin 42:2442–2448
Hawthorn LR, Wellington R (1930) Geneva, a greenhouse cucumber that develops fruit without pollination. N Y State Agric Exp Stn 2:3–11. https://doi.org/10.1007/BF02881046
Hayata Y, Niimi Y, Inoue K, Kondo S (2000) CPPU and BA, with and without pollination, affect set, growth, and quality of muskmelon fruit. HortSci 35:868–870. https://doi.org/10.21273/HORTSCI.35.5.868
Hayata Y, Niimi Y, Iwasaki N (1995) Synthetic cytokinin-1-(2=chloro=4=pyridyl)-3-phenylurea (CPPU)-promotes fruit set and induces parthenocarpy in watermelon. J Am Soc Hortic Sci 120:997–1000. https://doi.org/10.21273/JASHS.120.6.997
He M, Song S, Zhu X, Lin Y, Pan Z, Chen L, Chen D, Hu G, Huang B et al (2021) SlTPL1 silencing induces facultative parthenocarpy in tomato. Front Plant Sci. https://doi.org/10.3389/fpls.2021.672232
He M, Liu QA, Qi XH, Xu Q, Chen XH (2016) QTL mapping of cucumber parthenocarpy by QTL seq. Cucurbitaceae 2016. In: XIth Eucarpia meeting on cucurbit genetics & breeding, July 24–28, 2016, Warsaw, Poland 2016, pp 86–88
Hedden P (2020) The current status of research on gibberellin biosynthesis. Plant Cell Physiol 61:1832–1849. https://doi.org/10.1093/pcp/pcaa092
Homayouni AL, Strader LC (2020) Sugarrush: glucosylation of IPyA attenuates auxin levels. Proc Natl Acad Sci 117:7558–7560. https://doi.org/10.1073/pnas.2003305117
Hu Y, Omary M, Hu Y, Doron O, Hoermayer L, Chen Q, Megides O, Chekli O, Ding Z, Friml J, Zhao Y, Tsarfaty I, Shani E (2021) Cell kinetics of auxin transport and activity in Arabidopsis root growth and skewing. Nat Commun 12:1657. https://doi.org/10.1038/s41467-021-21802-3
Huitrón MV, Diaz M, Diánez F, Camacho F, Valverde A (2007) Effect of 2,4-D and CPPU on triploid watermelon production and quality. HortScience 42:559–564. https://doi.org/10.21273/HORTSCI.42.3.559
Jat GS, Munshi AD, Behera TK, Bhardwaj C (2017) Inheritance of parthenocarpy in gynoecious cucumber (C. sativus L.) cultivar PPC-2. J Hortic Sci 12(2):193–197
Juldasheva L (1973) Inheritance of the tendency towards parthenocarpy in cucumbers. Byull Vsesoyuznogo Ordena Lenina Inst Rastenievodstva Imeni NI Vavilova 32:58–59
Kang C, Darwish O, Geretz A, Shahan R, Alkharouf N, Liu Z (2013) Genome-scale transcriptomic insights into early-stage fruit development in woodland strawberry Fragaria vesca. Plant Cell 25:1960–1978. https://doi.org/10.1105/tpc.113.111732
Kataoka K, Uemachi A, Nonaka M, Yazawa S (2004) Effect of endogenous gibberellins in the early stages of fruit growth and development of the `Severianin’ tomato. J Hortic Sci Biotechnol 79(1):54–58. https://doi.org/10.1080/14620316.2004.11511736
Kaur H, Manchanda P, Kumar P, Dhall RK, Chhuneja P, Weng Y (2023) Genome-wide identification and characterization of parthenocarpic fruit set-related gene homologs in cucumber (Cucumis sativus L.). Sci Rep 13:2403. https://doi.org/10.1038/s41598-023-29660-3
Kim I, Okubo H, Fujieda K (1994) Studies on parthenocarpy in Cucumis sativus L.—effects of exogenous growth regulators on induction of parthenocarpy and endogenous hormone levels in cucumber ovaries. J Korean Soc Hortic Sci 35:187–196. https://doi.org/10.2503/hortj.MI-051
Kim IS, Okubo H, Fujieda K (1992a) Genetic and hormonal control of parthenocarpy in cucumber (Cucumis sativus L.). J Fat Agric 36:173–181
Kim IS, Okubo H, Fujieda K (1992b) Endogenous levels of IAA in relation to parthenocarpy in cucumber (Cucumis sativus L.). Sci Hortic 52:1–8. https://doi.org/10.1016/0304-4238(92)90002-T
Kim JS, Ezura K, Lee J, Ariizumi T, Eruza H (2019) Genetic engineering of parthenocarpic tomato plants using transient SlIAA9 knockdown by novel tissue-specific promoters. Sci Rep 9:18871. https://doi.org/10.1038/s41598-019-55400-7
Klap C, Yeshayahou E, Bolger AM, Arazi T, Gupta SK, Shabtai S, Salts Y, Barg R (2016) Tomato facultative parthenocarpy results from SlAGAMOUS-LIKE 6 loss of function. Plant Biotechnol J 15:634–647
Kollmer I, Novak O, Strnad M, Schmülling T, Werner T (2014) Overexpression of the cytosolic cytokinin oxidase/dehydrogenase (CKX7) from Arabidopsis causes specific changes in root growth and xylem differentiation. Plant J 78:359–371. https://doi.org/10.1111/tpj.12477
Kumar S, Kumar R, Kumar D, Gautam N, Dogra RK, Mehta DK, Sharma HD, Kansal S (2016) Parthenocarpic gynoecious lines parental lines of cucumber introduced from Netherlands for developing high-yielding, quality hybrids. J Crop Improv 30(3):352–369. https://doi.org/10.1080/15427528.2016.1163762
Kumar R, Khurana A, Sharma AK (2014) Role of plant hormones and their interplay in development and ripening of fleshy fruits. J Exp Bot 65:4561–4575. https://doi.org/10.1093/jxb/eru277
Kushnereva V (2008) Breeding of cucumber (C. sativus L.) for resistant to multiple diseases and other traits. Cucurbitaceae, In: Proceedings of the IXth EUCARPIA meeting on genetics and breeding of Cucurbitaceae, INRA, Avignon (France), pp 429–432
Kvasnikov BV, Rogova NT, Tarakanova SI, Ignatov SI (1970) Methods of breeding vegetable crops under the covered ground. Trudy Prikl Bot Genet Selek 42:45–57
Li J, Xu J, Guo QW, Wu Z, Zhang T, Zhang KJ, Cheng CY, Zhu PY, Lou QF, Cheng JF (2017) Proteomic insight into fruit set of cucumber (Cucumis sativus L.) suggests the cues of hormone-independent parthenocarpy. BMC Genom 18(896):1–18. https://doi.org/10.1186/s12864-017-4290-5
Li Q, Li H1, Huang W, Xu Y, Zhou Q, Wang S, Ruan J, Huang S, Zhang Z (2019) A chromosome-scale genome assembly of cucumber (Cucumis sativus L.). Gigasci 8:giz072
Lietzow CD, Zhu H, Pandey S, Havey MJ, Weng Y (2016) QTL mapping of parthenocarpic fruit set in North American processing cucumber. Theor Appl Genet 129:2387–2401. https://doi.org/10.1007/s00122-016-2778-z
Liu Y, Li Y, Guo H, Lv B, Feng J, Wang H, Zhang Z, Chai S (2023) Gibberellin biosynthesis is required for CPPU-induced parthenocarpy in melon. Hortic Res 10:uhad084. https://doi.org/10.1093/hr/uhad084
López-Galarza S, Bautista AS, Perez D, Miguel A, Baixauli C, Pascual B, Maroto J, Guardiola J (2004) Effects of grafting and cytokinin-induced fruit setting on colour and sugar-content traits in glasshouse-grown triploid watermelon. J Hortic Sci Biotechnol 79:971–976. https://doi.org/10.1080/14620316.2004.11511875
Luo L, Zeng J, Wu H, Tian Z, Zhao Z (2018) A molecular framework for auxin-controlled homeostasis of shoot stem cells in Arabidopsis. Mol Plant 11(7):899–913. https://doi.org/10.1016/j.molp.2018.04.006
Lv J, Qi J, Shi Q, Shen D, Zhang S et al (2012) Genetic diversity and population structure of cucumber (Cucumis sativus L.). PlosOne 7(10):e46919. https://doi.org/10.1371/journal.pone.0046919
Mandal NK, Kumari K, Kundu A, Arora A, Bhowmik PK, Iquebal MA, Jaiswal S, Behera TK, Munshi AD, Dey SS (2022) Cross-talk between the cytokinin, auxin, and gibberellin regulatory networks in determining parthenocarpy in cucumber. Front Genet. https://doi.org/10.3389/fgene.2022.957360
Maroto J, Miguel A, López-Galarza S, San Bautista A, Pascual B, Alagarda J, Guardiola J (2005) Parthenocarpic fruit set in triploid watermelon induced by CPPU and 2,4-D applications. Plant Growth Regul 45:209–213. https://doi.org/10.1007/s10725-005-3992-x
Marti C, Orzaez D, Ellul P, Moreno V, Carbonell J, Granell A (2007) Silencing of DELLA induces facultative parthenocarpy in tomato fruits. Plant J 52(5):865–876. https://doi.org/10.1111/j.1365-313X.2007.03282.x
Matlob AN, Kelly WC (1975) Growth regulator activity and parthenocarpic fruit production in snake melon and cucumber grown at high temperature. J Am Soc Hortic Sci 100:406–409. https://doi.org/10.21273/JASHS.100.4.406
Matsuo S, Kikuchi K, Fukuda M, Honda I, Imanishi S (2012) Roles and regulation of cytokinins in tomato fruit development. J Exp Bot 63:5569–5579. https://doi.org/10.1093/jxb/ers207
Matsuo S, Miyatake K, Endo M, Fukuoka H (2020) Loss of function of the Pad-1 aminotransferase gene, which is involved in auxin homeostasis, induces parthenocarpy in Solanaceae plants. Proc Natl Acad Sci 117:12784–12790. https://doi.org/10.1073/pnas.2001211117
Meng Y, Zhu P, Gou C et al (2023) Auxin and ethylene play important roles in parthenocarpy under low-temperature stress revealed by transcriptome analysis in cucumber (Cucumis sativus L.). J Plant Growth Regul. https://doi.org/10.1007/s00344-023-11172-z
Meshcherov E, Juldasheva L (1974) Parthenocarpy in cucumber. Trudy Prikl Bot Genet Selek 51:204–213
Mezzetti B, Landi L, Scortichini L, Rebori A, Spena A, Pandolfini T (2000) Genetic engineering of parthenocarpic fruit development in strawberry. Acta Hortic 567:101–104. https://doi.org/10.1111/pbi.12662
Molesini B, Pandolfini T, Rotino GL, Dani V, Spena A (2009) Aucsia gene silencing causes parthenocarpic fruit development in tomato. Plant Physiol 149(1):534–548. https://doi.org/10.1104/pp.108.131367
Nitsch JP (1972) Perenation through seeds and other structures. In: Steward FC (ed) Plant physiology, 6A: physiology of development: plant and their reproduction. Academic Press, New York
Niu Z, Song XF, Li XL, Guo XY, He SQ et al (2020) Inheritance and QTL mapping for parthenocarpy in cucumber. Sci Agric Sin 53(1):160–171. https://doi.org/10.3864/j.issn.0578-1752.2020.01.015
Noll F (1902) Fruchtbildung ohne vorausgegangene Bestaubung (Parthenokarpie) bei der Gurke. Sitzungsber Niederrhein Ges nat Heilk Bonn, pp 149–162
Ogawa M, Takisawa R (2022) Effect of exogenous plant hormones on parthenocarpy and fruit quality in tropical squash (Cucurbita moschata L.). Hortic J 91:508–513. https://doi.org/10.2503/hortj.UTD-368
Okabe Y, Yamaoka T, Ariizumi T, Ushijima K, Kojima M, Takebayashi Y et al (2019) Aberrant stamen development is associated with parthenocarpic fruit set through up-regulation of gibberellin biosynthesis in tomato. Plant Cell Physiol 60:38–51. https://doi.org/10.1093/pcp/pcy184
Osipowski P, Pawełkowicz M, Wojcieszek M et al (2020) A high-quality cucumber genome assembly enhances computational comparative genomics. Mol Genet Genomics 295:177–193. https://doi.org/10.1007/s00438-019-01614-3
Pandolfini T (2009) Seedless fruit production by hormonal regulation of fruit set. Nutrients 1:168–177. https://doi.org/10.3390/nu1020168
Peterson CE (1960) A gynoecious inbred line of cucumbers. Mich Agric Exp Stn Q Bull 43:40–42
Peterson CE, Andher LD (1960) Induction of staminate flowers on gynoecious cucumbers with gibberellin A3. Science 131:1673–1674. https://doi.org/10.1126/science.131.3414.1673
Pike LM, Peterson CE (1969) Inheritance of parthenocarpy in the cucumber (Cucumis sativus L.). Euphytica 18:101–105. https://doi.org/10.1007/BF00021987
Qi J, Liu X, Shen D et al (2013) A genomic variation map provides insights into the genetic basis of cucumber domestication and diversity. Nat Genet 45:1510–1515. https://doi.org/10.1038/ng.2801
Qian C, Ren N, Wang J, Xu Q, Chen X, Qi X (2018) Effects of exogenous application of CPPU, NAA and GA4+7 on parthenocarpy and fruit quality in cucumber (Cucumis sativus L.). Food Chem 243:410–413. https://doi.org/10.1016/j.foodchem.2017.09.150
Qin X, Zhang Z, Lou Q et al (2021) Chromosome-scale genome assembly of Cucumis hystrix-a wild species interspecifically cross-compatible with cultivated cucumber. Hortic Res 8:40. https://doi.org/10.1038/s41438-021-00475-5
Quebedeaux B, Beyer E (1972) Chemically-induced parthenocarpy cucumber by a new inhibitor of auxin transport. Hortic Sci 7:474–476. https://doi.org/10.21273/HORTSCI.7.5.474
Rotino GL, Perri E, Zottini M, Sommer H, Spena A (1997) Genetic engineering of parthenocarpic plants. Nat Biotech 15:1398–1401. https://doi.org/10.1038/nbt1297-1398
Rudich J, Baker LR, Sell HM (1977) Parthenocarpy in Cucumis sativus L. as affected by genetic parthenocarpy, thermo-photoperiod, and femaleness. J Am Soc Hortic Sci 102(2):225–228. https://doi.org/10.21273/JASHS.102.2.225
Rylski I (1974) Effects of season on parthenocarpic and fertilized summer squash (Cucumis pepo L.). Exp Agric 10:39–44. https://doi.org/10.1017/S0014479700006396
Saito T, Ariizumi T, Okabe Y, Asamizu E, Hiwasa-Tanase K, Fukuda N et al (2011) TOMATOMA: a novel tomato mutant database distributing Micro-Tom mutant collections. Plant Cell Physiol 52:283–296
Sakakibara H (2005) Cytokinin biosynthesis and regulation. In: Begley TP, Means AR, O’Malley BW, Riddiford L, Tashjian AH Jr (eds) Vitamins & hormones, 72nd edn. Elsevier, Amsterdam, pp 271–287
Sebastian P, Schaefer H, Telford IRH, Renner SS (2010) Cucumber (Cucumis sativus) and melon (C. melo) have numerous wild relatives in Asia and Australia, and the sister species of melon is from Australia. Proc Natl Acad Sci USA 107(32):14269–14273
Serrani JC, Fos M, Atarés A, Garcia-Martinez JL (2007) Effect of gibberellin and auxin on parthenocarpic fruit growth induction in the cv micro-tom of tomato. J Plant Growth Regul 26:211–221. https://doi.org/10.1007/s00344-007-9014-7
Sharif R, Su L, Chen X, Qi X (2022) Hormonal interactions underlying parthenocarpic fruit formation in horticultural crops. Hortic Res 9:uhab024. https://doi.org/10.1093/hr/uhab024
Sharif RXC, Zhang H, Arnao MB, Ali M, Ali Q, Muhammad I, Shalmani A, Nawaz MA, Chen P, Li Y (2018) Melatonin and its effects on plant systems. Molecules 23:2352. https://doi.org/10.3390/molecules23092352
Smith O, Cochran HL (1935) Effect of temperature on pollen germination and tube growth in the tomato. Cornell Univ Agr Exp Sta Mem 175
Strong WJ (1921) Greenhouse cucumber breeding. Proc Am Soc Hortic Sci 18:271–273
Strong WJ (1932) Parthenocarpy in cucumber. Sci Agric 12:665–669
Su L, Rahat S, Ren N, Kojima M, Takebayashi Y, Sakakibara H, Wang M, Chen X, Qi X (2021) Cytokinin and auxin modulate cucumber parthenocarpy fruit development. Sci Hortic 282:110026. https://doi.org/10.1016/j.scienta.2021.110026
Sun C, Li Y, Zhao W, Song X, Lu M, Li X, Li X, Liu R, Yan L, Zhang X (2016) Integration of hormonal and nutritional cues orchestrates progressive corolla opening. Plant Physiol 171:1209–1229. https://doi.org/10.1104/pp.16.00209
Sun ZY, Lower RL, Staub JE (2006a) Analysis of generation means and components of variance for parthenocarpy in cucumber (Cucumis sativus L.). Plant Breed 125:277–280. https://doi.org/10.1111/j.1439-0523.2006.01224.x
Sun ZY, Lower RL, Staub JE (2006b) Variance component analysis of parthenocarpy in elite US processing type cucumber (Cucumis sativus L.) lines. Euphytica 148:331–339. https://doi.org/10.1007/s10681-005-9041-z
Sun ZY, Staub JE, Chung SM, Lower RL (2006c) Identification and comparative analysis of quantitative trait loci associated with parthenocarpy in processing cucumber. Plant Breed 125:281–287. https://doi.org/10.1111/j.1439-0523.2006.01225.x
Turek S, Pląder W, Hoshi Y, Skarzyńska A, Pawełkowicz M (2023) Insight into the organization of the B10v3 cucumber genome by integration of biological and bioinformatic data. Int J Mol Sci 24(4):4011. https://doi.org/10.3390/ijms24044011
Varoquaux F, Blanvillain R, Delseny M, Gallois P (2000) Less is better: new approaches for seedless fruit production. Trends Biotechnol 18(6):233–242. https://doi.org/10.1016/s0167-7799(00)01448-7
Wang H, Jones B, Li Z, Frasse P, Delalande C, Regad F et al (2005) The tomato Aux/IAA transcription factor IAA9 is involved in fruit development and leaf morphogenesis. Plant Cell 17:2676–2692. https://doi.org/10.1105/tpc.105.033415
Wang X, Li H, Gao Z, Wang L, Ren Z (2020) Localization of quantitative trait loci for cucumber fruit shape by a population of chromosome segment substitution line. Sci Rep 10:11030. https://doi.org/10.1038/s41598-020-68312-8
Wellington R, Hawthorn R (1928) A parthenocarpic hybrid derived from a cross between an English forcing cucumber and the Arlington White Spine. Am Soc Hortic Sci 25:97–100
Wong CY (1938) Induced parthenocarpy of watermelon, cucumber and pepper by the use of growth promoting substances. Proc Amer Soc Hortic Sci 89:632–636
Wong CY (1939) Induced parthenocarpy of watermelon, cucumber and pepper. Sci 89:417–418. https://doi.org/10.1126/science.89.2314.417
Wright S (1968) Evolution and the genetics of populations. I. Genetic and biometric foundations. University of Chicago Press, Chicago
Wu Z, Zhang T, Li L, Xu J, Quin X, Tinglin Z, Cui L, Lou Q, Li J, Chen J (2016) Identification of stable major-effect QTL (Parth2.1) controlling parthenocarpy in cucumber and associated candidate gene analysis via whole genome re-sequencing. BMC Plant Biol 16(182):1–14. https://doi.org/10.1186/s12870-016-0873-6
Xu J, Li J, Cui L, Zhang T, Wu Z, Zhu PY, Meng YJ, Zhang KJ, Yu XQ, Lou QF, Chen JF (2017) New insights into the roles of cucumber TIR1 homologs and miR393 in regulating fruit/seed set development and leaf morphogenesis. BMC Plant Biol 17:130. https://doi.org/10.1186/s12870-017-1075-6
Yan LY, Lou LN, Li XL, Lou QF, Feng ZH, Chen JF (2010) Inheritance of parthenocarpy in monoecious cucumber. Sci Agric Sin 43(6):1295–1301
Yan LY, Lou LN, Lou QF, Chen JF (2008) Inheritance of parthenocarpy in gynoecious cucumber (Cucumis sativus L.). Acta Hortic Sin 35(10):1441–1446
Yan LY, Lou NL, Li LX, Feng ZH, Lou FQ, Chen FJ (2012) Inheritance of parthenocarpy in cucumber under the same background. Acta Hortic 935:55–60. https://doi.org/10.17660/ActaHortic.2012.935.7
Yan Y, He M, He M, Qi X, Xu Q, Xu X, Chen X (2022) Genetic analysis and QTL mapping of parthenocarpy in Cucumis sativus L. Mol Plant Breed. https://doi.org/10.5376/mpb.2022.13.0005
Yan LY, Song XF, Li XL, Feng ZH, Lou QF, Chen JF (2012) Identification of an AFLP molecular marker linked to parthenocarpy gene in monoecious cucumber. Acta Hortic 929:293–286. https://doi.org/10.17660/ActaHortic.2012.929.41
Yasuda S (1935) Parthenocarpy induced by the stimulation of pollination in some plants of the Cucurbitaceae. Agric Hortic 10:1385–1390
Yin Z, Malinowski R, Ziolkowska A, Sommer H, Plcader W, Malepszy S (2006) The DefH9-iaaM-containing construct efficiently induces parthenocarpy in cucumber. Cell Mol Biol Lett 11:279–290. https://doi.org/10.2478/s11658-006-0024-4
Zhihong N, Song XF, Li XL, Guo XU, He SQ, He LJZ, Feng ZH, Sun CZ, Yan LY (2020) Inheritance and QTL mapping for parthenocarpy in cucumber. Sci Agric Sin 53(1):160–171. https://doi.org/10.3864/j.issn.0578-1752.2020.01.015
Acknowledgements
The authors thank Director, School of Agricultural Biotechnology for providing the infrastructural facilities to carry out the research work.
Funding
The work is supported by the Department of Science and Technology under the Science and Engineering Research Board-POWER grant (SPG/2021/000999).
Author information
Authors and Affiliations
Contributions
RKD and PM conceptualized the idea of the manuscript. HK and E wrote original draft of the manuscript. RKD and PM wrote and critically reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Dhall, R.K., Kaur, H., Manchanda, P. et al. Recent advances in genetics and molecular breeding of parthenocarpic cucumber (Cucumis sativus L.) under protected conditions. Euphytica 220, 104 (2024). https://doi.org/10.1007/s10681-024-03366-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-024-03366-7