Abstract
The present study focuses prudent elucidation of microbial pollution and antibiotic sensitivity profiling of the fecal coliforms isolated from River Cauvery, a major drinking water source in Karnataka, India. Water samples were collected from ten hotspots during the year 2011–2012. The physiochemical characteristics and microbial count of water samples collected from most of the hotspots exhibited greater biological oxygen demand and bacterial count especially coliforms in comparison with control samples (p ≤ 0.01). The antibiotic sensitivity testing was performed using 48 antibiotics against the bacterial isolates by disk-diffusion assay. The current study showed that out of 848 bacterial isolates, 93.51 % (n = 793) of the isolates were found to be multidrug-resistant to most of the current generation antibiotics. Among the major isolates, 96.46 % (n = 273) of the isolates were found to be multidrug-resistant to 30 antibiotics and they were identified to be Escherichia coli by 16S rDNA gene sequencing. Similarly, 93.85 % (n = 107), 94.49 % (n = 103), and 90.22 % (n = 157) of the isolates exhibited multiple drug resistance to 32, 40, and 37 antibiotics, and they were identified to be Enterobacter cloacae, Pseudomonas trivialis, and Shigella sonnei, respectively. The molecular studies suggested the prevalence of bla TEM genes in all the four isolates and dhfr gene in Escherichia coli and Sh. sonnei. Analogously, most of the other Gram-negative bacteria were found to be multidrug-resistant and the Gram-positive bacteria, Staphylococcus spp. isolated from the water samples were found to be methicillin and vancomycin-resistant Staphylococcus aureus. This is probably the first study elucidating the bacterial pollution and antibiotic sensitivity profiling of fecal coliforms isolated from River Cauvery, Karnataka, India.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Rivers have always been an important source of drinking water, along which our ancient civilization flourished, and most developmental activities still depend on them (Gholami and Srikantaswamy 2009). Cauvery is one of the major rivers of India and is regarded as the “Ganga of South India.” The river originates at Talacauvery, Coorg district, Karnataka, India, and serves as the major source of water for the southern states of Tamil Nadu, Kerala, and the Union Territory of Puducherry draining an area of 81,155 sq. km (Vanham et al. 2011). There are many urban centers, industries, and hospitals that are situated along the banks of River Cauvery. Moreover, Bangalore City of India is located on the periphery of the river basin. The river is the primary source for drinking water to the surrounding urban and rural residents of Karnataka, India (Venkatesha Raju et al. 2012). Recent reports from State Pollution Control Boards of Karnataka and Tamil Nadu, India (2010) have indicated significant level of pollutants in the water of River Cauvery. In the recent past, the biological oxygen demand (BOD) and heavy metal contents in the riverine water bodies have increased dramatically. The river, being a natural dumping ground for the industries situated along the bank, discharge of sewage from various industries, hospitals, and pharmaceutical sectors to the water bodies lead to high turbidity, reduced transparency, and increased suspended solids and heavy metals. Such environments might have a direct influence on the proliferation of fecal coliforms which cause several health hazards (Gazzaz et al. 2012; Umamaheswari and Saravanan 2009).
Over time, the river Cauvery has been subjected to human interference on a regular basis, and its water quality has significantly deteriorated (Selvaraj et al. 2015). Major anthropogenic activities such as agriculture, extraction of water for drinking, domestic activities, discharge of sewage, sand dredging, boating, fishing, open defecation, religious rituals, and many other activities are posing a serious threat to the biota by altering the physicochemical and biological properties of the river system (Vignesh et al. 2013).
The enormous accumulation of domestic and industrial wastes along with pesticide residues resulted in the massive multiplication of various pathogenic microorganisms in River Cauvery. This paves way for many waterborne diseases (Sharma et al. 2012) such as typhoid, diarrhea, cholera, dysentery, and gastroenteritis (Moon et al. 2014). Recent reports have revealed that many bacteria mutate and acquire genes responsible for multiple drug resistance (MDR) to the present generation antibiotics and have emerged as “superbugs.” At present, many fresh water ecosystems have turned into reservoirs for antibiotic-resistant bacteria (Xu et al. 2014). Walsh et al. (2012) have reported the prevalence of carbapenem-resistant Escherichia coli harbored NDM-1 gene in drinking water samples from New Delhi, India. The superbugs carrying various drug-resistant genes in tap and spring waters in the coastal region of Turkey (Ozgumus et al. 2007) and surface and drinking waters in Mainz, Germany were also reported (Schwartz et al. 2003). In addition, many pathogenic bacteria and their drug-resistant genes towards β-lactams, Amoxicillin/Ampicillin (bla TEM ), Streptomycin/Spectinomycin (aadA), Tetracycline (tet), Chloramphenicol (cmlA), and Vancomycin (van) from various aquatic ecosystem were also reported (Thevenon et al. 2012). Similarly, Gao et al. (2012) reported prevalence of sulfonamide-resistant genes in various aquatic isolates in Tianjin, China, and Diwan et al. (2012) reported occurrence of Cefotaxime and Ciprofloxacin-resistant genes in hospital-associated waste water isolates in Madhya Pradesh, India.
The bacteria have become multidrug-resistant by natural or by acquired means. The natural resistance (intrinsic resistance) is the ability to resist a particular antimicrobial agent through its inherent structural or functional characteristics. The acquired resistance is caused either by mutation in chromosomal genes, or by the acquisition of mobile genetic elements, such as plasmids or transposons, carrying the antibiotic resistance genes (Lupo et al. 2012). The resistance can be transferred between bacteria by recombination via transformation, conjugation, and transduction. Thus, gene responsible for resistance to a single antibiotic may be shared among different bacteria (Lupo et al. 2012).
Our previous study reported high occurrence of multidrug-resistant fecal coliforms in the water samples collected from a drinking water reservoir which receives water from Vrishabhavathi River, a sub-tributary of the river Cauvery (Skariyachan et al. 2013). Hence, there is an urgent need to assess the physiochemical and bacteriological status of the parent river which acts as major drinking water source for the urban and rural residents in Karnataka, India. As the information available about microbial flora present in River Cauvery is sparse and limited, it is important to analyze the microbial populations, especially with the prevalence of multidrug-resistant bacteria such as fecal coliforms, fecal streptococci, and staphylococci. This is probably the first study reporting on the physiochemical characteristics, fecal microbial population, and predominance of multidrug resistance in the water samples collected from Cauvery River.
Methodology
Description of the study area
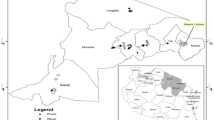
The area under study is River Cauvery which is one of the largest rivers of India. The river flows between 75°27′ to 79°54′ east longitude and 10°9′ to 13°30′ north latitude. This study was carried out at ten hotspots along the banks of River Cauvery, covering about 210 km in Karnataka through the districts of Coorg, Mysore, and Mandya. The sampling spots for the study were Talacauvery (12.38°N 75.52°E), Napokulu (12.18oN 75.41oE), Cauvery Nisargadhama (12.20oN 75.48oE), Balmurikshetra (12.17oN 76.38oE), Ranganthittu (12.25oN 76.39oE), Srirangapattana (12.25oN 76.41oE), Nanjanagudu (12.70oN 76.40oE), Tirumakudal Narsipura (Kapila) (12.13oN 76.54oE), Tirumakudal Narsipura (Cauvery; 12.13oN 76.54oE), and Talakadu (13.44oN 75.22oE) as indicated by the State pollution control board, Govt of Karnataka, India on August 2010 (Fig. 1a). All these places are highly populated and are tourist destinations of South India. Moreover, many urban centers, temples, hospitals, and large-scale industries are located in these areas. Most of them discharge waste materials, such as ritual waste from temples (places of worship), domestic, household and hospital waste waters, and municipal and industrial sewages, into the river in high magnitude, which has turned into a cesspool of organic wastes (Fig. 1b).
a The map showing the course of Cauvery River and selected hotspot for water sample collection. b The drastic water pollution observed in various hotspots of River Cauvery during the survey and sampling. A Napoklu, B Ranganathittu, C Balmurikshetra, D Srirangapattana, E Nanjanagud, F Tirumakudal Narasipura
Sample collection and transportation
The water samples were collected from the ten hot spots as per the specific protocol mentioned by American Public Health Association ( 2005). One bottle of the 2-l water samples were collected at a depth of 1 m from the surface per site per sampling in suitable sterile containers from the selected hotspots during January 2011, December 2011, and June 2012. Similarly, 2 l of water samples each from River Cauvery being supplied to household drinking water, from water treatment plant, and from local drinking water tank were collected and used as control. The water samples were tightly sealed using plastic stoppers under aseptic conditions. These sampled bottles were transported to the laboratory in ice boxes and stored at 4 °C. All the water samples were processed within 12 h after collection.
Physiochemical characterization—estimation of biological oxygen demand
The time, temperature, pH, and other physiological parameters during the time of sample collection were recorded (Supplementary material, Table 1). Further, the dissolved oxygen (DO) and BOD were estimated by Winkler’s method (Shriwastav et al. 2010) for 5 days.
Isolation of bacteria and determination of total count
One-milliliter water sample from each of the sets were serially diluted and plated to nutrient agar, MacConkey’s agar, thiosulfate citrate bile salts sucrose agar, Salmonella–Shigella agar, Hichrome MeResa agar, selective Strep agar, bromothymol blue lactose agar, milk agar with centrimide, mannitol salt agar, xylose lysine deoxycholate agar, deoxycholate citrate agar, and blood agar (Hi-Media, India) by standard plate techniques (Geldreich et al. 1972). The plates were incubated at 37 °C for 24 h. The experiment by nutrient agar was replicated as three independent trials. The morphological characteristics of each colony were analyzed, and the viable bacterial count (colony-forming units (CFU) per milliliter) was enumerated by digital colony counter (Labtronics, India).
Determination of fecal coliform count
The estimation of fecal coliform count was carried out by most probable number (MPN) techniques (Highsmith and Abshire 1975). The media used was single and double strength lauryl sulfate tryptose broth (Hi-Media, India) for presumptive test, brilliant green bile broth for confirmed test, and eosin methylene blue agar (Hi-Media, India) for completed test along with comparative controls. The positive results were characterized by acid and gas formation. The procedure was replicated as three independent trails and the results were compared with standard MPN table (Tillett 1987).
Microbial characterization of isolated bacteria
The morphological characteristics of isolated bacteria were studied by Gram staining (Beveridge 2001). The motility of bacteria was determined by hanging drop technique (Luna et al. 2005). The isolates were further characterized by standard biochemical tests. The biochemical tests performed were IMViC (Murray et al. 2007), catalase (MacFaddin 1980), urease (MacFaddin 1980), hydrogen sulfide production (Roser et al. 2005), oxidase (Gordon and McLeod 1928), nitrate reduction (Moreno-Vivián et al. 1999), coagulase (Sperber and Tatini 1975), starch hydrolysis (Colonna et al. 1992), triple sugar iron (Sulkin and Willett 1940), lysine decarboxylase (Falkow 1958), arginine decarboxylase (Moeller 1955), and ornithine decarboxylase (MacFaddin 2000).
Antibiotic susceptibility testing
The antibiotic sensitivity testing was performed on isolated bacteria using Mueller–Hinton agar (Hi-Media, India) by disk-diffusion method (Bauer et al. 1966). The total numbers of antibiotics are 48 which included in the families of cephalosporins and other β-lactams, sulfonamides, quinolones/fluoroquinolone, macrolids, and aminoglycosides. Mueller–Hinton agar plates were aseptically inoculated with the test organism by lawn culture technique and filter paper disks, impregnated with a specific concentration of antibiotic, was placed on the medium. The plates were incubated at 37 °C for 24 h. The procedure was performed as three independents replicates, and the mean value of diameter of the zone of inhibition was measured and compared with standard (Clinical and Laboratory Standards Institute 2013) and their resistant, intermediate, and sensitive patterns were recorded.
Statistical analysis
The BOD values and viable bacterial counts (CFU/ml) observed in all the three samplings were tested for statistical significance. The analysis of variance (ANOVA) was performed at 0.01 significance level to analyze the variations among ten sampling sites (n = 3; January 2011, December 2011, and June 2012) for BOD and CFU/ml. The ANOVA was performed using the “stats” module available in R package (R Core Team 2013).
Molecular characterization of multidrug-resistant bacteria and selected drug-resistant genes
The four major bacterial isolates that exhibited maximum drug resistances to most of the conventionally used antibiotics were further selected for 16S rDNA gene sequencing. The pure bacterial isolates were used for molecular characterization which was carried out at BioAxis DNA Research Centre Pvt. Ltd., Hyderabad, India. The bacterial DNA was extracted from the pure cultures of four selected isolates, and the 16S rDNA genes were amplified using the primers 16SF AGAGTTTGATCCTGGCTCAG and 16SR ACGGCTACCTTGTTACGACTT. The amplified PCR product was subjected to electrophoresis using agarose gel (1.8 %) in TAE buffer and visualized by staining with ethidium bromide. The forward and reverse DNA sequencing reaction of PCR amplicon was carried out on ABI 3730xl Genetic Analyzer to obtain the sequence. The final gene sequence obtained was further analyzed for best homology search by Basic Local Alignment Search Tool (BLAST; http://www.ncbi.nlm.nih.gov/BLAST). Similarly, the resistance genes for β-lactam (bla TEM ) and Trimethoprim (dhfr) were selected based on the antibiotic sensitivity results. The complete open-reading frames along with promoter sequence of these genes were amplified by PCR using the primers 5′-GCTTACTATGCCATTAT-3′ and 5′-ATTATATTTTGTATTATCG-3′. The gene sequences were further obtained by sequencing (ABI 3730xl Genetic Analyzer). The 16S rDNA sequences of four MDR bacterial isolates, gene sequences of bla TEM from four isolates, and dhfr from two isolates were submitted to GenBank database (http://www.ncbi.nlm.nih.gov/GenBank).
Results and discussion
Physiochemical analysis
The physiochemical parameters recorded at the time of sample collection are given in supplementary materials (Supplementary material, Table 1). The temperature and pH were found to be ideal for the bacterial growth in most of the sampling spots. The BOD was found to be greater in most of the sampling areas ranging from 1.1 to 2.8, 0.8–4.6, and 2.0–5.9 mg/L in January 2011, December 2011, and June 2012, respectively, in comparison with the control water samples. The control water samples showed the BOD values within the permissible limits. The samples collected from Talacauvery (3.5–4.6 mg/L), Srirangapattana (2.0–2.2 mg/L), Nanjanagud (2.1–3.2 mg/L), Tirumakudal Narasipura (Cauvery; 2.4–5.9 mg/L), and Talkadu (2.7–3.9 mg/L) showed increase in the BOD content at each time of sample collection (Table 1). The statistical analysis indicated that variations observed in the mean BOD values at each time of the sample collections were found to be significant (p ≤ 0.01; Table 1). The BOD values estimated during June 2012 was slightly higher than that of the values obtained from other sampling time (Table 1). This increase in the biological oxygen demand was probably due to the sewage accumulated from temples, tourists, organic and agricultural farms, residents, hospitals, and municipal and industrial sectors along the river basins during the month of March, April, and May which was visible during the survey of this study. These 3 months are the summer season in Karnataka which are expected to contribute high loads of waste materials from all these sectors compared to other seasons. However, the waste garbage generated during other three seasons is also contributing to high level of pollutants. The mean BOD levels in most of the water samples collected were exceeding the permissible limits suggested by European Union for aquatic life (Chapman 1996). The BOD values of polluted water systems are estimated to be greater than 2 mg/L, and such BOD values were observed in many places along the banks of River Cauvery in this study. Such kind of bulk liquid BOD concentration has direct influence over the microbial community in the water bodies especially the growth of coliforms (Downing and Nerenberg 2008).
Bacteriological analysis
The number of viable bacterial count (CFU/ml) estimated from most of the sampling sites were high when compared with the quality of drinking water prescribed by WHO (Aryal et al. 2012; Table 1). The viable bacterial count estimated at different time points were found to be 1.7 × 105 to 8.5 × 106 CFU/ml, 1.8 × 105 to 8.7 × 106 CFU/ml, and 2.2 × 105 to 9.2 × 106 CFU/ml during Januray 2011, December 2011, and June 2012, respectively. The total bacterial count from the control water samples were found to be within the permissible range as per American Public Health Association (2005). Among the ten sampling areas studied, Talacauvery (2.1 × 106 to 3.6 × 106 CFU/ml), Srirangapattana (8.5 × 106 to 9.5 × 106 CFU/ml), Nanjanagud (8.1 × 106 to 8.4 × 106 CFU/ml), Tirumakudal Narasipura (Cauvery; 3.1 × 106 to 3.7 × 106 CFU/ml), and Talkadu (4.4 × 106 to 5.6 × 106 CFU/ml) displayed the highest bacterial count. The variations observed in the bacterial count during the sampling replicates from ten different hot spots were found to be significant (p ≤ 0.01; Table 1). This study suggests that the water samples collected from Talkadu showed maximum bacterial count in comparison with other sampling spots. It is clear that there was high relation with flow gradient towards the total count as Talkadu is at the lowest sampling point in the study. The current study suggests that the bacterial counts in these hot spots are increasing with respect to time. These observations were made based on one time samplings at three different time points. Hence, further studies for a longer term need to be performed to conclude the relationship with increasing mode of bacterial count, BOD, and time.
The bacterial count (CFU/ml) estimated in this study was beyond the permissible limits described by WHO, rendering the water as non-potable (Aryal et al. 2012). The current study also estimated the most probable number count which is the measure of the fecal coliforms limit (MPN/100 ml). The MPN indices of most of the water samples were found to be very high indicating higher fecal contamination (Table 1). The fecal coliform counts in the sampling spots were in the range of 79 to >2,400 MPN/100 ml, 280 to >2,400 MPN/100 ml, and 1,600 to >2,400 MPN/100 ml during January 2011, December 2011, and June 2012, respectively. The MPN count of the control water samples found to be in the permissible limit (~8 MPN/100 ml) as per American Public Health Association ( 2005). The BOD level in the water samples collected from Talacauvery, though high, showed less number of fecal coli forms during January 2011 and December 2011 when compared with other sampling spots. This is probably due to the low temperature (18–20 °C) and pH (5.8–6.0) observed at the time of water collection from Talacauvery during the months of January and December 2011. It is assumed that the coliform counts obtained from Talacauvery during June 2012 was considerably high due to the variation in temperature and pH. During June 2012, the temperature (26 °C) and pH (6.1) of the water samples were found to be high compared with January and December 2011. Thus, one of the main parameters required for proliferations of fecal coliforms is the favorable conditions in water environment, especially the temperature and pH (Ramírez Castillo et al. 2013). The total and fecal coliform counts are one of the main parameters for assessing the quality of drinking water (Kolawole et al. 2011). The consumptions of such polluted water may lead to hazardous outbreaks of various waterborne diseases (Bessong et al. 2009).
This study further focused on the isolation and characterization of the bacteria present in the collected water samples by standard microbiology procedures. The cultural and other physiological characteristics were studied and most of the bacteria isolated were found to be coliforms (Gram-negative, aerobic, or facultative anaerobic bacilli) which imply the presence of enteric pathogens. This study also identified the presence of fecal bacteria in majority of the water samples. There are reports on the existence of such morphologically and physiologically related pathogenic bacteria in water samples collected from South Africa (Hysko et al. 2010) and other parts of the world (Catry et al. 2014). The bacteria isolated from various hotspots were further characterized based on Bergy’s Manual of Systematic Bacteriology (Supplementary materials, Table 2). The current study identified a total of 848 isolates from 30 water samples collected from the selected hotspot of Cauvery River in three different time points, among which 33.37 % (n = 283) were found to be E. coli, 20.51 % (n = 174) were Shigella spp., 13.44 % (n = 114) were Enterobacter spp., and 12.85 % (n = 109) were found to be Pseudomonas spp. (Fig. 2). In addition, this study also identified lower percentages of Salmonella spp. (4.71 %, n = 40), Citrobacter spp. (1.27 %, n = 11), Yokanella spp. (1.43 %, n = 12) and Proteus spp. (1.11 %, n = 9), Edwersiella spp. (0.81 %, n = 7), Morganella spp. (1.05 %, n = 9) and Klebsiella spp. (1.08 %, n = 9), Alcaligenes spp. (0.73 %, n = 6), Vibrio spp. (0.80 %, n = 7), Serratia spp. (0.47 %, n = 5), Staphylococcus spp. (4.95 %, n = 42), and Streptococcus spp. (1.29 %, n = 11; Fig. 2). Occurrences of such microorganisms in many fresh water ecosystems create critical problems worldwide; and most of them lead to severe gastrointestinal diseases (Bessong et al. 2009; Hysko et al. 2010; Kolawole et al. 2011).
The total percentage of each bacterial isolates from River Cauvery and their percentage of antibiotics resistance patterns during the sample collection (2011–2012). The figure clearly shows that most of the bacterial isolates are emerged as multidrug-resistant strains. Among the total 848 isolates of bacteria, 32.19 % (n = 273) of E. coli, 12.61 % (n = 157) of Shigella spp., 12.61 % (n = 107) of Enterobacter spp., 12.14 % (n = 103) of Psuedomonas spp., 4.12 % (n = 35) of Salmonella spp., 0.82 % (n = 7) Citrobacter spp., 1.06 % (n = 9) of Yokanella spp., 1.11 % (n = 9) of Proteus spp., 0.70 % (n = 6) of Edwersiella spp., 1.05 % (n = 9) of Morganella spp., 1.08 % (n = 9) of Klebsiella spp., 0.73 % (n = 6) of Alcaligenes spp., 0.60 % (n = 7) of Vibrio spp., 0.35 % (n = 5) % of Serratia spp, 4.95 % (n = 42) of Staphylococcus areus, and 1.06 % (n = 9) of Streptococcus spp. showed multiple drug resistance to most of the tested antibiotics. All the isolates of Proteus spp., Morganella spp., Klebsiella spp., Alcaligenes spp., Vibrio, and Staphylococcus spp. (especially against vancomycin and methicillin) showed multidrug resistance to majority of the tested antibiotics
Antibiotic sensitivity testing
The present study demonstrated that 93.51 % (n = 793) of the bacterial isolates out of 848 were found to be multidrug resistance to most of the current generation antibiotics (Fig. 2). The study revealed that all the four main isolates were found to be drug resistant to at least 30 different antibiotics out of 48. Out of 283 E. coli isolates, 96.46 % (n = 273) of E. coli were found to be fully resistant to 30 antibiotics in which most of them were expected to be acquired resistance. Similarly, out of 114 Enterobacter isolates, 93.85 % (n = 107) of Enterobacter spp. were showed full resistance to 32 antibiotics. Analogously, out of 109 Pseudomonas isolates, 94.49 % (n = 103) of Pseudomonas spp. were found to be fully resistant to 40 antibiotics. Similarly, out of 174 Shigella isolates, 90.22 % (n = 157) of Shigella spp. were illustrated full resistance to 37 antibiotics (Fig. 3). From the antibiotic sensitivity testing, it is clear that isolates, such as E. coli, Enterobacter spp., Pseudomonas spp., and Shigella spp., have acquired drug resistance to most of the carbapenam groups which includes Faropenem, Doripenem, Imipenem, Meropenem, and Ertapenem (Fig. 3). These antibiotics are regarded as the strongest β-lactams and last line drugs currently available against these Gram-negative enteric pathogens.
The antimicrobial resistance patterns of four major bacteria (Escherichia coli, Enterobacter spp., Psuedomonas spp. and Shigella spp.) isolated from various hotspots of River Cauvery during 2011–2012 by disc diffusion method. The assay was performed as three independent replicates, and the mean value of the diameter of the zone of inhibition was compared with standard chart (CLSI 2013). Figure shows that 96.46 % (n = 273) of E. coli, 93.85 % (n = 107) of Enterobacter spp., 94.49 % (n = 103) of Psuedomonas spp., and 90.22 % (n = 157) of Shigella spp. were fully resistant to various antibiotics that emerged as superbugs
The current study also demonstrated that most of the minor isolates were also found to be resistant to β-lactams, third and fourth generation cephalosporins, aminoglycosides, macrolides, and nitrofurans. From this study, it is clear that the isolates of Salmonella spp. (n = 35), Cirobacter spp. (n = 7), Proteus spp. (n = 9) and Klebsiella spp. (n = 9) showed resistance to 29, 40, 19, and 26 antibiotics, respectively (Fig. 4). Similarly, the isolates of Morganella spp. (n = 9), Yokenella spp. (n = 9), Edwersiella spp. (n = 6), and Serratia spp. (n = 5) showed resistance to 24, 13, 16, and 27 antibiotics, respectively (Fig. 5). Furthermore, the isolates of Alcaligenes spp. (n = 6), Vibrio spp. (n = 7), Staphylococcus aureus (n = 42), and Streptococcus spp. (n = 9) showed resistance to 21, 12, 24, and 20 antibiotics, respectively (Fig. 6). An important finding of this study is that although some intrinsic resistances were common among the isolates, most of the isolates emerged as multidrug-resistant strains probably by horizontal transfer. Furthermore, the preliminary antimicrobial testing showed that all the isolates of S. aureus were found to be resistant to both methicillin and vancomycin (Fig. 6). To the best of our knowledge, this is probably the first study showing the occurrence of methicillin and vancomycin-resistant S. aureus (MRSA and VRSA) in River Cauvery. However, further molecular analysis is essential to confirm the presence of drug-resistant genes in these isolates. The presence of MRSA and VRSA is very often found in clinical samples (Catry et al. 2014); however, their persistence in many fresh and marine water ecosystems were recently reported (Plano et al. 2013).
The antimicrobial resistance patterns of the minor bacterial isolates (Salmonella spp., Citrobacter spp., Proteus spp., and Klebsiella spp.) obtained from various hotspots of River Cauvery during 2011–2012. The assay was performed as three independent replicates, and the mean value of the diameter of the zone of inhibition was compared with standard chart (CLSI, 2013). Figure shows that the isolates of Salmonella spp. (n = 35), Cirobacter spp. (n = 7), Proteus spp. (n = 9), and Klebsiella spp. (n = 9) showed resistance to 29, 40, 19, and 26 antibiotics, respectively, in which most of them are expected to be acquired resistance
The antimicrobial resistance patterns of the minor bacterial isolates (Morganella spp., Yokenella spp., Edwersiella spp., and Serratia spp.) obtained from various hotspots of River Cauvery during 2011–2012. The assay was performed as three independent replicates, and the mean value of the diameter of the zone of inhibition was compared with standard chart (CLSI, 2013). Figure shows that the isolates of Morganella spp. (n = 9), Yokenella spp. (n = 9), Edwersiella spp. (n = 6), and Serratia spp. (n = 5) showed resistance to 24, 13, 16, and 27 antibiotics, respectively, in which most of them are expected to be acquired resistance
The antimicrobial resistance patterns of the minor bacterial isolates (Alcaligenes spp., Vibrio spp., Staphylococcus aureus, and Streptococcus spp.) obtained from various hotspots of River Cauvery during 2011–2012. The assay was performed as three independent replicates, and the mean value of the diameter of the zone of inhibition was compared with the standard chart (CLSI, 2013). Figure shows that the isolates of Alcaligenes spp. (n = 6), Vibrio spp. (n = 7), Staphylococcus aureus (n = 42), and Streptococcus spp. (n = 9) showed resistance to 21, 12, 24, and 20 antibiotics, respectively, in which most of them are expected to be acquired resistance
The E. coli isolates which illustrated extensive drug resistance to broad-spectrum β-lactams, resistance to third and fourth generation antibiotics were further characterized by 16S rDNA sequencing. The BLAST search revealed that the 16S rDNA gene sequence of the isolate was 99 % identical to E. coli which belongs to a pathogenic serovar type, Enterohemorrhagic O157: H7. (GenBank Accession NR_074891). This strain causes severe, acute hemorrhagic diarrhea, and abdominal cramps (Karch et al. 2005). Hence, there may be a possibility that the strain also belongs to the same serovar type. Similarly, the other three isolates which showed extensive resistances towards present generation antibiotics were also characterized. The16S rDNA gene sequences of two isolates showed 100 % identity to Shigella sonnei (Accession NR_074894) and Pseudomonas trivialis (Accession NR_028987) and 99 % identity to Enterobacter cloacae (Accession NR_028912).
Recent reports have revealed that many pathogenic E. coli can survive in soil environment which resulted in the contamination of fresh water system in East China (Wang et al. 2014). Studies from California and Arizona, USA showed the ecological habitat and persistence of Enterohemorrhagic E. coli in leafy green producing soil (Ma et al. 2012). The major portion of the Cauvery river covers leafy green producing soils which could be one of the reasons for predominance of these bacteria in the banks of the River. Recently it was reported that E. coli isolated from selected surface water sources in Zaria, Nigeria was found to be multidrug-resistant to most of the β-lactams, fourth generation cephalosporins and fluoroquinolones The antimicrobial resistance, virulent factors and genetic diversity of different isolates of E. coli from household water supply in Dhaka, Bangladesh (Talukdar et al. 2013) and San Pedro River, Mexico were also reported (Ramírez Castillo et al. 2013). The current study revealed that most of the Shigella spp., isolated from River Cauvery has become extremely resistant to even fourth generation cephalosporins and aminoglycosides. Similar types of extended spectrum of β-lactamase mediated third generation cephalosporin resistance were reported in Southern Vietnam (Vinh et al. 2009), Bangladesh (Rahman et al. 2004), Israel (Vasilev et al. 2007), Iran (Tajbakhsh et al. 2012) and Spain (Arias et al. 2006). This study also showed that river Cauvery harbors high prevalence of P. trivialis, a phytopathogen commonly present in the phyllosphere of grasses. The banks of the river are mainly covered with vegetations, especially green grasses which may be one of the reasons for such high density of P. trivialis. The current study thus provides an insight into the antimicrobial susceptibility pattern of this organism as there are limited reports about MDR P. trivialis. The emergence of MDR En. cloacae against carbapenem and related antibiotics are considered as major public health concern in Spain (Fernández et al. 2011) and Northern Iran (Bayani et al. 2013). This study also emphasizes the massive occurrence of MDR En. cloacae in River Cauvery which can become a critical health issue in India in the near future.
The present study also addresses the identification of one of the β-lactam resistant genes (bla TEM ) and trimethoprim resistant genes (dhfr) from these isolates based on their antibiotic sensitivity patterns. The genes were sequenced and are deposited in NCBI GenBank. This study identified bla TEM gene (540 bp) coding for β-lactamase from E. coli (accession no. KF225562), Sh. sonnei (accession no. KF225561), En. cloacae (accession no. KF225564), and P. trivialis (accession no. KF225563). Similarly, this study also identified the presence of dhfr gene (564 bp), gene responsible for trimethoprim resistance, from E. coli (accession no. KF225560) and Sh. sonnei (accession no. KF225559).
The massive discharges of sewage from pharmaceutical industries, chemical and electroplating industries, animal farm houses, hospitals, and temples have drastically polluted the river and contributed to the high content of organic matters in the water leading to the change in the physiochemical status of water and in turn favored the growth of coliforms (Supplementary material, Table 1). The aquatic ecosystems such as rivers and streams are ideal reservoirs for antibiotic resistant bacteria (Ramírez Castillo et al. 2013). The misuse and overuse of antibiotics contribute to the selection and persistence of antibiotic resistant bacteria (Amábile-Cuevas 2010). The acquisition of resistance genes in the bacteria by various methods minimize therapeutic options and lead to frequent treatment failures. The prevention and control of MDRs have become a global priority and one that requires all healthcare facilities and agencies to assume responsibilities. By considering the entire medical, social, economical, and environmental relevance, there is a necessity to understand the antimicrobial resistance profile and undertake all precautionary measures to maintain the natural status of river Cauvery.
Conclusion
The course of River Cauvery is deteriorating due to the disposal of sewage from pharmaceutical industries, chemical and electroplating industries, animal farm houses, agricultural sectors, hospitals, and temples. The physiochemical environment of the river has become an ideal ground for the rapid multiplication of many gastrointestinal pathogens. The results showed that DO, BOD, total bacterial count, and fecal coliform count have exceeded permissible limits. The antibiotic sensitivity profiling indicated that most of the isolates have acquired multidrug resistance against present generation antibiotics and emerged as superbugs. This study also emphasizes the importance of environmental studies for understanding the spread, evolution, and public health relevance of antibiotic resistance factors. The current study shows that the predominance of many multidrug-resistant bacteria is apparent in River Cauvery. This study clearly reveals that the prevention and control of MDRs is a global priority and addresses this issue and undertakes all necessary actions to preserve the natural status of the river. However, more sampling procedures for longer terms, in different seasons, are required to confirm the prevalence of various MDR genes in the bacterial population of the river.
References
Amábile-Cuevas, C. (2010). Antibiotic resistance in Mexico: a brief overview of the current status and its causes. Journal of Infection in Developing Countries, 4, 126–131.
American Public Health Association (APHA). (2005). In A. D. Eaton, L. S. Clesceri, E. W. Rice, A. E. Greenberg, & M. A. H. Franson (Eds.), Standard methods for the examination of water & wastewater. Washington: APHA.
Arias, C., Sala, M. R., Domínguez, A., Bartolomé, R., Benavente, A., Veciana, P., Pedrol, A., & Hoyo, G. (2006). Outbreak working group, waterborne epidemic outbreak of Shigella sonnei gastroenteritis in Santa Maria de Palautordera, Catalonia, Spain. Epidemiology Infection, 134, 598–604.
Aryal, J., Gautam, B., & Sapkota, N. (2012). Drinking water quality assessment. Journal of Nepal Health Research Council, 10, 192–196.
Bauer, A. W., Kirby, W. M., Sherris, J. C., & Turck, M. (1966). Antibiotic susceptibility testing by a standardized single disk method. American Journal of Clinical Pathology, 45, 493–496.
Bayani, M., Siadati, S., Rajabnia, R., & Taher, A. A. (2013). Drug resistance of Pseudomonas aeruginosa and Enterobacter cloacae isolated from ICU, Babol. Northern Iran. International Journal of Molecular and Cellular Medecine, 2, 204–209.
Bessong, P. O., Odiyo, J. O., Musekene, J. N., & Tessema, A. (2009). Spatial distribution of diarrhoea and microbial quality of domestic water during an outbreak of diarrhoea in the Tshikuwi community in Venda, South Africa. Journal of Health, Population and Nutrition, 27, 652–659.
Beveridge, T. J. (2001). Use of the Gram stain in microbiology. Biotechnic & Histochemistry, 76, 111–118.
Catry, B., Latour, K., Jans, B., Vandendriessche, S., Preal, R., Mertens, K., & Denis, O. (2014). Risk factors for methicillin resistant Staphylococcus aureus: a multi-laboratory study. PloS One, 9, e89579.
Chapman, D. (1996). Water quality assessments: a guide to the use of biota, sediments and water in environmental monitoring. London: Chapman & Hall.
Clinical and Laboratory Standards Institute. (2013). M100-S23. Performance standards for antimicrobial susceptibility testing: 23rd informational supplement. Wayne: Clinical and Laboratory Standards Institute.
Colonna, P., Leloup, V., & Buléon, A. (1992). Limiting factors of starch hydrolysis. European Journal of Clinical Nutrition, 46(Suppl 2), S17–S32.
Diwan, V., Chandran, S. P., Tamhankar, A. J., Stålsby Lundborg, C., & Macaden, R. (2012). Identification of extended-spectrum β-lactamase and quinolone resistance genes in Escherichia coli isolated from hospital wastewater from central India. The Journal of Antimicrobial Chemotherapy, 67, 857–859.
Downing, L. S., & Nerenberg, R. (2008). Effect of bulk liquid BOD concentration on activity and microbial community structure of a nitrifying, membrane-aerated biofilm. Applied Microbiology and Biotechnology, 81, 153–162.
Falkow, S. (1958). Activity of lysine decarboxlase as an aid in the identification of Salmonellae and Shigellae. American Journal of Clinical Pathology, 29, 598–600.
Fernández, A., Pereira, M. J., Suárez, J. M., Poza, M., Treviño, M., Villalón, P., Sáez-Nieto, J. A., Regueiro, B. J., Villanueva, R., & Bou, G. (2011). Emergence in Spain of multidrug-resistant Enterobacter cloacae clinical isolates producing SFO-1 extended-spectrum beta-lactamase. Journal of Clinical Microbiology, 49, 822–828.
Gao, P., Mao, D., Luo, Y., Wanga, L., Xu, B., & Xu, L. (2012). Occurrence of sulfonamide and tetracycline-resistant bacteria and resistance genes in aquaculture environment. Water Research, 46, 2355–2364.
Gazzaz, M. N., Yusoff, K. M., Ramli, M. F., Aris, A. Z., & Juahir, H. A. (2012). Characterization of special patterns in river water quality using chemo metric pattern recognition techniques. Marine Pollution Bulletin, 6, 688–698.
Geldreich, E. E., Nash, H. D., Reasoner, D. J., & Taylor, R. H. (1972). The necessity of controlling bacterial populations in potable waters; community water supply. Journal of the American Water Works Association, 64, 596–602.
Gholami, S., & Srikantaswamy, S. (2009). Statistical multivariate analysis in the assessment of river water quality in the vicinity of KRS Dam, Karnataka, India. Natural Resources Research, 3, 235–247.
Gordon, J., & McLeod, J. W. (1928). Practical application of the direct oxidase reaction in bacteriology. The Journal of Pathology and Bacteriology, 31, 185–190.
Highsmith, A. K., & Abshire, R. L. (1975). Evaluation of a most probable number technique for the enumeration of Pseudomonas aeruginosa. Applied Microbiology, 30, 596–601.
Hysko, M., Gace, B., & Puto, K. (2010). Microbial water pollution of Vjosa River and the vegetable contamination by irrigation. Republic of Macedonia: BALWOIS: Water Observation and Information System for Decision Support—Ohrid. May 25-29.
Karch, H., Tarr, P. I., & Bielaszewska, M. (2005). Enterohaemorrhagic Escherichia coli in human medicine. International Journal of Medical Microbiology, 295, 405–418.
Kolawole, O. M., Ajayi, K. T., Olayemi, A. B., & Okoh, A. I. (2011). Assessment of water quality in Asa River (Nigeria) and its indigenous Clarias gariepinus fish. International Journal of Environmental Research and Public Health, 8, 4332–4352.
Luna, V. A., Peak, K. K., Veguilla, W. O., Reeves, F., Heberlein-Larson, L., Cannons, A. C., Amuso, P., & Cattani, J. (2005). Use of two selective media and a broth motility test can aid in identification or exclusion of Bacillus anthracis. Journal of Clinical Microbiology, 43, 4336–4341.
Lupo, A., Coyne, S., & Berendonk, T. U. (2012). Origin and evolution of antibiotic resistance: the common mechanisms of emergence and spread in water bodies. Frontiers in Microbiology, 3, 1–13.
Ma, J., Ibekwe, A. M., Crowley, D. E., & Yang, C. H. (2012). Persistence of Escherichia coli O157: H7 in major leafy green producing soils. Environmental Science & Technology, 46, 12154–12161.
MacFaddin, J. F. (1980). Biochemical tests for identification of medical bacteria. pp. 173–183. Baltimore: Williams & Wilkins.
MacFaddin, J. F. (2000). Biochemical tests for identification of medical bacteria (3rd ed.). Baltimore: Lippincott. Williams and Wilkins.
Moeller, V. (1955). Simplified tests for some amino acid decarboxylases and for the arginine dihydrolase system. Acta Pathologica et Microbiologica Scandinavica, 36, 158–172.
Moon, S., Sohn, I. W., Hong, Y., Lee, H., Park, J. H., Kwon, G. Y., Lee, S1., & Youn, S. K. (2014). Emerging pathogens and vehicles of food- and water-borne disease outbreaks in Korea, 2007-2012. Osong Public Health Research Perspectives, 5, 34–39.
Moreno-Vivián, C., Cabello, P., Martínez-Luque, M., Blasco, R., & Castillo, F. (1999). Prokaryotic nitrate reduction: molecular properties and functional distinction among bacterial nitrate reductases. Journal of Bacteriology, 181, 6573–6584.
Murray, P. R., Baron, E. J., Jorgensen, J. H., Landry, M. L., Pfaller, M. A. (Eds.). (2007). Manual of clinical microbiology. 9th Ed. ASM Press. 1: 335.
Ozgumus, O. B., Celik-Sevim, E., Alpay-Karaoglu, S., Sandalli, C., & Sevim, A. (2007). Molecular characterization of antibiotic resistant Escherichia coli strains isolated from tap and spring waters in a coastal region in Turkey. Journal of Microbiology, 45, 379–387.
Plano, L. R., Shibata, T., Garza, A. C., Kish, J., Fleisher, J. M., Sinigalliano, C. D., Gidley, M. L., Withum, K., Elmir, S. M., Hower, S., Jackson, C. R., Barrett, J. B., Cleary, T., Davidson, M., Davis, J., Mukherjee, S., Fleming, L. E., & Solo-Gabriele, H. M. (2013). Human-associated methicillin-resistant Staphylococcus aureus from a subtropical recreational marine beach. Microbial Ecology, 65, 1039–1051.
Core Team, R. (2013). R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing.
Rahman, M., Shoma, S., Rashid, H., Siddique, A. K., Nair, G. B., & Sack, D. A. (2004). Extended-spectrum beta-lactamase-mediated third-generation cephalosporin resistance in Shigella isolates in Bangladesh. The Journal of Antimicrobial Chemotherapy, 54, 846–847.
Ramírez Castillo, F. Y., Avelar González, F. J., Garneau, P., Márquez Díaz, F., Guerrero Barrera, A. L., & Harel, J. (2013). Presence of multi-drug resistant pathogenic Escherichia coli in the San Pedro River located in the State of Aguascalientes, Mexico. Frontiers of Microbiology, 4, 147.
Roser, D. J., Ashbolt, N. H. G., Mathew, K., Nair, J., Ryken-Rapp, D., & Toze, S. (2005). Hydrogen sulphide production tests and the detection of groundwater faecal contamination by septic seepage. Water Science and Technology, 51, 291–300.
Schwartz, T., Kohnen, W., Jansen, B., & Obst, U. (2003). Detection of antibiotic-resistant bacteria and their resistance genes in wastewater, surface water, and drinking water biofilms. FEMS Microbiology Ecology, 43, 325–335.
Selvaraj, K. K., Sundaramoorthy, G., Ravichandran, P. K., Girijan, G. K., Sampath, S., & Ramaswamy, B. R. (2015). Phthalate esters in water and sediments of the Kaveri River, India: environmental levels and ecotoxicological evaluations. Environmental Geochemistry and Health, 37, 83–96.
Sharma, A., Bhattacharya, A., Bora, C. R., Shukla, V., & Parihar, P. (2012). Diversity of enteropathogens in River Narmada and their environmental and health implications. Microorganisms in environmental management. Microbes and environment (pp. 35–60). Netherlands: Springer.
Shriwastav, A., Sudarsan, G., Bose, P., & Tare, V. (2010). Modification of Winkler's method for determination of dissolved oxygen concentration in small sample volumes. Analytical Methods, 2, 1618–1622.
Skariyachan, S., Lokesh, P., Rao, R., Kumar, A. U., Vasist, K., & Narayanappa, R. (2013). A pilot study on water pollution and characterization of multidrug resistant superbugs from Byramangala tank, Ramanagara district, Karnataka, India. Environmental Monitor Assessment, 185, 5483–5495.
Sperber, W. H., & Tatini, S. R. (1975). Interpretation of the tube coagulase test for identification of Staphylococcus aureus. Applied Microbiology, 29, 502–505.
Sulkin, S. E., & Willett, J. C. (1940). A triple sugar-ferrous sulfate medium for use in identification of enteric organisms. The Journal of Laboratory and Clinical Medicine, 25, 649–653.
Tajbakhsh, M., García Migura, L., Rahbar, M., Svendsen, C. A., Mohammadzadeh, M., Zali, M. R., Aarestrup, F. M., & Hendriksen, R. S. (2012). Antimicrobial-resistant Shigella infections from Iran: an overlooked problem? The Journal of Antimicrobial Chemotherapy, 67, 1128–1133.
Talukdar, P. K., Rahman, M., Rahman, M., Nabi, A., Islam, Z., Hoque, M. M., Endtz, H. P., & Islam, M. A. (2013). Antimicrobial resistance, virulence factors and genetic diversity of Escherichia coli isolates from household water supply in Dhaka, Bangladesh. PLoS ONE, 8, e61090. doi:10.1371/journal.pone.0061090.
Thevenon, F., Adatte, T., Wildi, W., & Poté, J. (2012). Antibiotic resistant bacteria/genes dissemination in lacustrine sediments highly increased following cultural eutrophication of Lake Geneva (Switzerland). Chemosphere, 86, 468–476.
Tillett, H. E. (1987). Most probable numbers of organisms: revised tables for the multiple tube method. Epidemiology and Infection, 99, 471–476.
Umamaheswari, S., & Saravanan, A. N. (2009). Water quality of Cauvery river basin in Trichirappalli, India. International Journal and Lakes and Rivers, 2, 1–20.
Vanham, D., Weingartner, R., & Rauch, W. (2011). The Cauvery river basin in southern India: major challenges and possible solutions in the 21st century. Water Science and Technology, 64, 122–131.
Vasilev, V., Japheth, R., Yishai, R., Andorn, N., Valinsky, L., Navon-Venezia, S., Chmelnitsky, I., Carmeli, Y., & Cohen, D. (2007). Extended-spectrum beta-lactamase-producing Shigella strains in Israel, 2000–2004. European Journal of Clinical Microbiology & Infectious Diseases, 26, 189–194.
Venkatesha Raju, K., Somashekar, R. K., & Prakash, K. L. (2012). Heavy metal status of sediment in river Cauvery, Karnataka. Environmental Monitoring and Assessment, 184, 361–373.
Vignesh, S., Muthukumar, K., Santhosh Gokul, M., & James, R. A. (2013). Microbial pollution indicators in the Cauvery river, Southern India. On a sustainable future of the earth's natural resources, Springer earth system sciences (pp. 363–376). Berlin: Springer.
Vinh, H., Baker, S., Campbell, J., Hoang, N. V., Loan, H. T., Chinh, M. T., Anh, V. T., Diep, T. S., le Phuong, T., Schultsz, C., & Farrar, J. (2009). Rapid emergence of third generation cephalosporin resistant Shigella spp. in Southern Vietnam. Journal of Medical Microbiology, 58, 281–283.
Walsh, T. R., Weeks, J., Livermore, D. M., & Toleman, M. A. (2012). Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. The Lancet Infectious Diseases, 1, 355–362.
Wang, H., Ibekwe, A. M., Ma, J., Wu, L., Lou, J., Wu, Z., Liu, R., Xu, J., & Yates, S. R. (2014). A glimpse of Escherichia coli O157: H7 survival in soils from eastern China. The Science of the Total Environment, 476–477, 49–56.
Xu, Z. Q., Flavin, M. T., & Flavin, J. (2014). Combating multidrug-resistant Gram-negative bacterial infections. Expert Opinion on Investigational Drugs, 23, 163–182.
Acknowledgments
The authors would like to thank Dr. Jeffery F. Miller, University of California, Los Angeles for his encouragement throughout the study. The authors also acknowledge Dr. Murugan Sevanan, Assistant Professor (SG), School of Biotechnology, Karunya University, Coimbatore, Tamil Nadu; and Dr. Shinomol K George, Assistant Professor, Department of Biotechnology, DSCE and Saurabh Yadav, Senior Research Fellow, MBU, IISc, Bangalore for their critical reading of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Skariyachan, S., Mahajanakatti, A.B., Grandhi, N.J. et al. Environmental monitoring of bacterial contamination and antibiotic resistance patterns of the fecal coliforms isolated from Cauvery River, a major drinking water source in Karnataka, India. Environ Monit Assess 187, 279 (2015). https://doi.org/10.1007/s10661-015-4488-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10661-015-4488-4