Abstract
Background
Water pollution in densely populated urban areas, mainly from municipal wastewater, poses a significant threat. Pathogenic bacteria, such as Vibrio spp. and fecal coliform, endanger public health and the environment. Additionally, antibiotic-resistant bacteria in wastewater complicate treatment and heighten public health concerns.
Methods
The study sampled municipal wastewater from ten Dhaka neighborhoods, selecting treatment plants, sewage outlets, and various collection points using meticulous techniques for representative samples. Bacteriological and biochemical analyses were conducted using standardized methods. Antimicrobial susceptibility testing (AST) was performed with the disk diffusion method against 13 widely used antibiotics.
Results
All sampled areas exhibited positive results for Vibrio spp., fecal coliform, E. coli, and Salmonella spp. Varying bacterial concentrations were observed, with the highest concentration of TVC, total vibrio spp., and total fecal coliform, total E. coli count, and total Salmonella spp. were found in Uttara (1.9 × 104 CFU/ml), Bangshal (1.8 × 102 CFU/ml), and Lalbag (2.1 × 103 CFU/ml), Mirpur (3.70 × 102 CFU/ml), and Lalbag (6 × 102 CFU/ml) respectively. AST results revealed significant resistance among all bacterial species to various antibiotics. Specifically, Vibrio spp. showed 100% resistance to cefuroxime, fecal coliform exhibited 90% resistance to cephradine, E. coli demonstrated 60% resistance to cephradine, and Salmonella spp. displayed 90% resistance to ampicillin.
Conclusion
The study highlights the existence of multiple antibiotic-resistant bacteria in Dhaka's wastewater. Addressing antibiotic resistance is essential to manage the risks of multiple antibiotic-resistant infections and maintain antibiotic effectiveness. These implications are critical for various stakeholders, including public health officials, policymakers, environmentalists, and urban planners.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Water pollution continues to be a critical global issue, especially in densely populated urban areas where municipal wastewater serves as a major source of contamination [1]. Furthermore, waterborne diseases pose a substantial threat to public health in these urban settings, as municipal wastewater can become a breeding ground for pathogenic bacteria [2]. Among the various pathogens found in wastewater, Vibrio spp. and fecal coliform bacteria stand out due to their ability to cause severe illnesses, presenting challenges to both environmental safety and public health [3]. Vibrio is a genus of Gram-negative bacteria characterized by a curved-rod structure, some of which can cause foodborne infections [4, 5]. Certain species of Vibrio are known to be pathogenic, primarily causing gastroenteritis, but they can also infect open wounds and lead to sepsis [6]. Pathogenic Vibrio species include V. cholera, V. parahaemolyticus, and V. vulnificus [4].
Fecal coliforms are facultatively anaerobic, rod-shaped, gram-negative, and non-sporulating bacteria [7]. Fecal coliforms are widely found in large amounts in the lower digestive tracts and feces of humans and other warm-blooded animals [8]. If they are found in water, it usually means the water has been recently contaminated by feces, which suggests a higher risk of harmful germs being present compared to just finding total coliform bacteria [9, 10]. Fecal coliforms serve as markers for enteric pathogenic pathogens [11, 12]. Some fecal coliforms, such as E. coli, can cause severe gastrointestinal illnesses, including diarrhea, abdominal cramps, and vomiting [13]. Pathogenic strains may also lead to more serious conditions like urinary tract infections, respiratory illnesses, and even life-threatening conditions such as septicemia [14]. The pathogenic forms of E. coli can cause severe intra- and extraintestinal infections in mammals and birds [15, 16].
Salmonella is a genus of rod-shaped gram-negative bacteria of Enterobacteriaceae [17]. The two known species of Salmonella are Salmonella enterica and Salmonella bongori [18]. Salmonella species are non-spore-forming, predominantly motile enterobacteria. Salmonella species are intracellular pathogens, of which specific serotypes cause illness. Most infections result from ingesting food contaminated with feces and from drinking water contaminated with feces [19].
Dhaka city, home to over 22.4 million of residents, faces a dual challenge: ensuring adequate sanitation facilities for its citizens while grappling with escalating water pollution in its municipal wastewater [20, 21]. Rapid urbanization, inadequate sewage treatment, and improper disposal practices have contributed to the contamination of water bodies, creating an environment conducive to the proliferation of pathogenic bacteria [20, 22]. The emergence of these multiple antibiotic-resistant bacteria further complicates the situation, raising concerns about the effectiveness of conventional treatment methods and the potential implications for public health [23, 24].
Moreover, studying antibiotic resistance in municipal wastewater is crucial due to its direct connection with human households, serving as a primary source of contamination [25]. As the population grows and antibiotic usage increases, resistant bacteria enter wastewater, creating a reservoir of antibiotic-resistant strains [26]. Understanding these resistance patterns in municipal wastewater is vital because it reflects the antibiotic resistance landscape within communities. Monitoring wastewater helps identify emerging antibiotic-resistant strains, enabling proactive measures in healthcare and environmental policies [27]. By studying resistance in this context, scientists gain insights into the prevalence and dissemination of antibiotic-resistant genes, guiding the development of effective strategies to curb the spread of antibiotic-resistant bacteria and safeguard public health [27].
Additionally, our capacity to cure common diseases is still in danger due to the creation and spread of antibiotic-resistant organisms that have developed new resistance mechanisms, or antimicrobial resistance [28]. With the increasing global development of multi- and pan-resistant bacteria, it is quite impossible to tackle these in a limited-resourced country like Bangladesh.
Bacteria with multiple antibiotic resistance (MAR) is a major global public health concern [29]. Therefore, research on multiple antibiotic-resistant (MAR) microorganisms is crucial. The purpose of this study is to examine and identify several antibiotic-resistant pathogenic bacteria (fecal coliform and Vibrio spp.) in Dhaka city's municipal wastewater. To date, no study has studied multiple antibiotic-resistant pathogenic bacteria (Vibrio spp. and fecal coliform) in municipal wastewater of Dhaka city. Additionally, this research is essential for multiple stakeholders, including public health officials, policymakers, environmentalists, and urban planners [30]. By comprehending the prevalence and antibiotic resistance patterns of Vibrio spp. and fecal coliform bacteria in Dhaka's municipal wastewater, tailored interventions can be developed. The findings will aid in refining sewage treatment methods, enhancing water quality monitoring systems, and shaping evidence-based public health policies.
2 Materials and methods
2.1 Sampling area and period of study
The study area for our research comprises ten neighborhoods in Dhaka (five samples from Dhaka north region- Kallayanpur, Rampura, Mohammadpur, Mirpur, Uttara, five samples from Dhaka south region- Dhanmondi, Sayedabad, Lalbag, Sadarghat, Bangshal) which were identified to represent the municipal wastewater system scattered throughout whole Dhaka city (see Fig. 1). Locations such as wastewater treatment plants, sewage outlets, and other points of municipal wastewater collection or discharge were included in the selection. A meticulous sampling technique was used. This study is cross sectional in nature and samples were collected in August 2023.
2.2 Sample collection
Before sample collection, all necessary equipment was sterilized, including obtaining sterile containers with tight-fitting lids, wearing gloves, and having disinfectant (70% ethanol solution), labels, and data collection sheets ready. Precautions included wearing disposable gloves and additional personal protective equipment like lab coats. Sampling points near inflow/outflow pipes and areas with significant wastewater flow were chosen. Equipment was disinfected by autoclaving, and samples were collected by submerging sterile containers into the wastewater stream, ensuring no air bubbles. The sampling process was conducted at multiple locations within each study area (composite sampling), ensuring the samples were representative and accounted for any potential spatial variations in antimicrobial resistance [31]. From each composite sample, 40 ml was collected and transferred to sterile Falcon tubes, providing more than enough quantity for laboratory analyses. Samples were immediately stored in ice-filled cooler/ice box at 4 °C and transported to the laboratory within 12 h. Samples were tightly sealed, labeled, and transported with care [32, 33]. After reaching the lab, samples were immediately subjected to bacteriological analyses using validated protocols [34, 35]. Detailed records were maintained, ensuring each sample's clear chain of custody. These meticulous techniques were crucial for providing reliable and representative wastewater samples from Dhaka city for studying antibiotic-resistant pathogenic bacteria.
2.3 Sample processing
To prepare the water samples, 1 ml of the sample was mixed with 9 ml of normal saline solution in a test tube. This mixture underwent initial dilution, followed by a series of dilutions from 1 to 5. Each serial dilute was marked in different test tubes.
2.4 Bacteriological analyses
The tests were conducted following the International Organization for Standardization (ISO) 2023 guidelines for all the species [36]. In the procedure, selective agar plates were prepared according to the manufacturer's instructions. After dilution, using a glass spreader (spread plate method), the diluted sample was spread over Oxoid UK's Thiosulfate Citrate Bile Sucrose agar (TCBS) and incubated at 37 °C for 24 h. TCBS-agar served as a selective medium for Vibrio spp., and plates were inspected for typical colonies, usually yellow and green [37]. For fecal coliform isolation, diluted water samples were spread evenly onto the surface of Oxoid UK's m-FC agar plate using a glass spreader. The plate was then placed in an incubator at 44 °C for 24–48 h, allowing fecal coliforms to grow and produce characteristic colonies [35]. Following incubation, colonies on the m-FC agar plate were observed. Total viable count (TVC), total Vibrio count, total fecal coliform count, total E. coli count, and total Salmonella count from wastewater samples were counted using colony counter machine. These procedures ensured adherence to standardized testing methods as outlined in validated studies [34, 35]. The same procedure was followed for the E. coli and Salmonella spp. Isolates. EMB (Eosin methylene blue agar) and SS (Salmonella-Shigella) agar were used for selective media of E. coli and Salmonella spp., respectively [38]. See the figures of isolates plates at supplementary files.
2.5 Biochemical confirmation
In the wastewater analysis, a battery of tests was performed to identify the bacterial species. For Vibrio spp., fecal coliforms, E. coli, and Salmonella spp., gram staining revealed a pink/red color, confirming their gram-negative nature [39]. Catalase activity was detected in all isolates (Vibrio spp., fecal coliforms, E. coli, and Salmonella spp.), as indicated by the formation of bubbles upon the addition of hydrogen peroxide [39]. Positive results for oxidase were observed, signified by the development of a purple color when the oxidase reagent was applied to Vibrio spp [40]. On the other hand, fecal coliforms, E. coli, and Salmonella spp. were negative for oxidase, as evidenced by the absence of a color change with the oxidase reagent. Moreover, each bacterial isolate was examined under a microscope for detecting the motility and morphological characteristics. These tests conclusively confirmed the presence of Vibrio spp. and fecal coliforms in our wastewater samples, offering valuable insights for our study. Our principal investigator, a prominent authority in this field, ensured the quality of this work throughout the study.
2.6 Antimicrobial susceptibility testing (AST)
Several subcultures were performed to enhance the purification of specific bacteria. Bacterial susceptibility to antimicrobial agents was determined using the disk diffusion method, following the guidelines established by Bauer et al. [41]. Thirteen antibiotic discs (Oxoid Ltd., Basingstoke, Hampshire, UK) containing Meropenem (10 g), Cefuroxime (30 g), Chloramphenicol (30 g), Cefotaxime (30 g), Tetracycline (30 g), Colistin (30 g), Nalidixic Acid (30 g), Azithromycin (15 g), Erythromycin (15 g), Ampicillin (10 g), Cephradine (30 g), Ciprofloxacin (5 g), and Cotrimoxazole (25 g) were utilized. Mueller–Hinton agar (Merck®) was employed according to the manufacturer's instructions for the antimicrobial susceptibility testing medium. Within 15 min of the application of the discs, the plates were inverted and incubated at 37 °C. After 16–18 h of incubation, the plates were examined and the diameters of the zones of complete inhibition to the nearest whole millimeter were measured. The antimicrobial susceptibility testing (AST) was conducted three times, and the average zone diameters were calculated for final categorization. The mean zone diameters for each antimicrobial agent were then classified as susceptible, intermediate, or resistant based on the interpretation table provided by following CLSI guidelines in 2020 [42]. See the figures of the AST plates in the supplementary section.
2.7 Grouping of the multiple antibiotic resistant (MAR) isolates
Based on the occurrence of resistance to more than three antibiotics, the isolates of each sampling site were grouped as multiple antibiotic-resistant isolates [43].
2.8 Ethics declarations
We affirm that this manuscript is original and has not been previously presented or published within this university or institution. Furthermore, all methods outlined in the manuscript were conducted in accordance with relevant guidelines and regulations.
2.9 Statistical analysis
The analysis involved Microsoft Excel (version 2021) and the Statistical Package for Social Sciences (SPSS version 26.0). Microsoft Excel was utilized for data cleaning, coding, and organization, following which the processed data was imported into SPSS software to compute descriptive statistics including frequencies, percentages, and correlations.
3 Result
3.1 Bacterial concentration
All the samples (10 out of 10) were positive for Vibrio spp., fecal coliform, E. coli, and Salmonella spp. Table 1 presents the concentration of different bacteria in various areas. The highest concentration of TVC, total vibrio spp., and total fecal coliform, total E. coli count, and total Salmonella spp. were found in Uttara (1.9 × 104 CFU/ml), Bangshal (1.8 × 102 CFU/ml), and Lalbag (2.1 × 103 CFU/ml), Mirpur (3.70 × 102 CFU/ml), and Lalbag (6 × 102 CFU/ml) respectively.
3.2 AST results
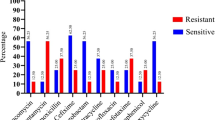
Figure 2 The prevalence of resistance among the tested Vibrio spp. strains was observed for selected antibiotics. Cefuroxime exhibited complete resistance (100%), while Cefotaxime faced intermediate in 90% of cases. Tetracycline showed an 80% intermediate rate, and Colistin was resistant in 50% of instances. Nalidixic Acid exhibited an 80% resistance rate, Ampicillin was resistant in 50% of cases, and Cephradine showed 30% resistance. Ciprofloxacin demonstrated 10% resistance, and Cotrimoxazole displayed a 10% resistance rate. These results highlight the presence of resistant Vibrio spp. strains, emphasizing the need for careful antibiotic selection to manage infections and mitigate antibiotic resistance effectively.
Figure 3 demonstrates the prevalence of resistance among the tested fecal coliform bacteria varied for the antibiotics. Cefuroxime (CXM-30) exhibited significant resistance, with 70% of isolates being resistant. Chloramphenicol (C30) encountered resistance in 10% of isolates, while Colistin (CL30) faced resistance in 40% of isolates. Ciprofloxacin (CIP5) displayed a resistance rate of 40%. Erythromycin (E15) exhibited 50% resistance, and Ampicillin (AM10) faced resistance in 80% of isolates. Cephradine (CE30) showed a high resistance rate of 90%. Conversely, Meropenem (MEM10), Cotrimoxazole (COT-25), and Tetracycline (TE30), Azithromycin (AZM15) displayed no or least resistance among the tested isolates.
The antibiotic resistance profile of the selected Escherichia coli (E. coli) isolates was assessed for various antibiotics, as depicted in Fig. 4. Notably, the isolates showed high sensitivity to Meropenem (MEM10) and Cefotaxime (CTX 30), with 80% and 60% susceptibility, respectively. Conversely, significant resistance was observed against certain antibiotics, including Cefuroxime (CXM-30) and Ampicillin (AM10), with 50% and 50% resistance rates, respectively. Intermediate resistance was noted for some antibiotics, such as Cefotaxime (CTX30) and Cotrimoxazole (COT-25), indicating a moderate level of susceptibility. The isolates exhibited an exceptionally high sensitivity to Erythromycin (E15) and Azithromycin (AZM15), with resistance rates of only 10% for both antibiotics.
Figure 5 demonstrates the antibiotic resistance profile of the selected Salmonella spp. isolates is presented in. The isolates showed varying degrees of resistance to the tested antibiotics. Notably, meropenem (MEM10) exhibited 100% sensitivity, while cefotaxime (CTX30) and ampicillin (AM10) showed high resistance rates of 80% and 90%, respectively. Ciprofloxacin (CIP5) displayed a balanced resistance pattern, with 20% resistance, 40% intermediate susceptibility, and 40% sensitivity. Colistin (CL30) revealed 50% resistance and 50% intermediate susceptibility. Overall, the isolates exhibited diverse antibiotic resistance patterns, underscoring the importance of prudent antibiotic use and continuous monitoring to address emerging resistance trends in Salmonella spp.
These findings emphasize the challenges of antibiotic resistance in microbial infections and underscore the importance of careful antibiotic use to combat resistance effectively [44]. See details in Figs. 2, 3, 4, and 5. The supplementary section provides the correlation matrixes for the antibiotics used among all four types of microbes.
3.3 MAR
Table 2 reveals the antibiotic resistance profiles of Vibrio spp. strains from different sources. Among them, Kallayanpur, Rampura, Mohammadpur, Dhanmondi, Sayedabad, Lalbag, Sadarghat, and Bangshal exhibit multiple antibiotic-resistant (MAR) patterns, indicating resistance to multiple antibiotic classes, posing significant challenges for effective treatment. In contrast, Mirpur and Uttara display non-multiple antibiotic-resistant (NMAR) patterns, denoting resistance to specific antibiotics. Mohammadpur, Lalbag, and Sadarghat strains have particularly high MAR Indices (0.46 and 0.54), suggesting widespread antibiotic resistance. Additionally, Table 3 demonstrates the antibiotic resistance profiles of fecal coliform. Strains from same sources all the samples exhibit multiple antibiotic-resistant (MAR) patterns, indicating resistance to multiple antibiotic classes, posing significant challenges for effective treatment. Kallayanpur, Uttara, and Lalbag strains have particularly high MAR Indices (0.46), suggesting widespread antibiotic resistance. This emphasizes the urgent need for vigilant antibiotic use and continuous surveillance to curb the escalation of multiple antibiotic-resistant bacterial infections and preserve the effectiveness of available antibiotics [45].
Table 4 outlines the antibiotic resistance patterns of selected E. coli isolates from various wastewater sources. The isolates from different locations exhibited diverse phenotypic resistance profiles, indicating resistance to specific antibiotics. The multiple antibiotic resistance (MAR) Index, a measure of the number of antibiotics an isolate is resistant, ranged from 0.08 to 0.77. Notably, isolates from Kallayanpur, Rampura, Mohammadpur, Mirpur, Dhanmondi, Sayedabad, Lalbag, Sadarghat, and Bangshal were classified as multiple antibiotic-resistant (MAR) based on their resistance to multiple antibiotic classes. In contrast, the Uttara isolate demonstrated a relatively low MAR Index of 0.08, categorizing it as non-multiple antibiotic-resistant (NMAR). This data underscores the prevalence of multiple antibiotic resistance among E. coli isolates in wastewater, emphasizing the need for effective surveillance and control measures to mitigate the spread of antibiotic resistance.
Table 5 presents the antibiotic resistance patterns of Salmonella spp. isolates from various wastewater sources. The phenotypic resistance profiles reveal multiple antibiotic resistances in these isolates. The multiple antibiotic resistance (MAR) Index, calculated based on resistance to different antibiotics, ranges from 0.23 to 0.85. All sources demonstrate a high MAR Index, suggesting exposure to various antibiotics. The Resistance Classification indicates that all isolates from different locations are classified as multiple antibiotic-resistant (MAR), emphasizing the widespread and concerning resistance patterns observed across these environmental samples. The findings underscore the urgency of monitoring and addressing antibiotic resistance in wastewater to mitigate potential public health risks associated with resistant strains of Salmonella spp.
4 Discussion
The findings of this study underscore the concern for the presence of multiple antibiotic-resistant pathogenic bacteria, namely Vibrio spp. and fecal coliform, within the municipal wastewater of Dhaka city. All the collected samples tested positive for Vibrio spp. and fecal coliform, signifying a pervasive contamination across the various locations sampled, including Kallyanpur, Rampura, Mohammadpur, Mirpur, Uttara, Dhanmondi, Sayedabad, Lalbag, Sadarghat, and Bangshal.
The enumeration of bacterial concentrations in these samples revealed varying levels of total viable count (TVC), total Vibrio count, and total fecal coliform count. The highest concentrations of TVC, Vibrio spp., and fecal coliform were notably found in Uttara, Bangshal, and Lalbag, respectively, suggesting diverse contamination levels at these specific locations. Similar findings were found in separate studies investigating antimicrobial resistance in Bangladesh [46, 47]. The high bacterial concentration in Bangshal and Lalbag may be primarily due to their higher population densities and associated increased wastewater production, inadequate sanitation, improper waste disposal, and poorly managed sewage systems in these areas. These factors collectively contribute to elevated bacterial counts in the wastewater [48, 49].
The Antibiotic Susceptibility Testing (AST) results unveiled significant resistance among the tested Vibrio spp. and fecal coliform strains to various antibiotics. Vibrio spp. strains exhibited high resistance to antibiotics such as Cefuroxime, Cefotaxime, Tetracycline, Colistin, Nalidixic Acid, Ampicillin, and Cephradine, highlighting the urgent need for a cautious and precise selection of antibiotics for effective infection management and combating the rise of antibiotic resistance [50, 51]. A study conducted in South Africa found quite similar results to Vibrio isolates from wastewater [52]. The high resistance levels observed in bacteria can be attributed to several factors, including the overuse and misuse of antibiotics in human health, animal treatment, and environmental contamination resulting from the release of antibiotics [53,54,55].
Similarly, fecal coliform strains displayed resistance to antibiotics like Cefuroxime, Chloramphenicol, Colistin, Ciprofloxacin, Erythromycin, Ampicillin, and Cephradine, emphasizing the challenges posed by antibiotic resistance in microbial infections and the critical role of judicious antibiotic use to counteract this resistance effectively [12, 56].
In comparison to the findings of previous studies, our results revealed both similarities and notable differences. Among the E. coli isolates high sensitivity to Meropenem (MEM10) and Cefotaxime (CTX 30) was consistent with some studies [57, 58], emphasizing the efficacy of these antibiotics. However, significant resistance against Cefuroxime (CXM-30) and Ampicillin (AM10) was observed, aligning with reports highlighting the emergence of resistance to commonly used antibiotics [58, 59]. The intermediate resistance observed for Cefotaxime (CTX30) and Cotrimoxazole (COT-25) suggests a moderate level of susceptibility, contrasting with studies reporting lower resistance rates. Notably, the high sensitivity to Erythromycin (E15) and Azithromycin (AZM15) implies potential alternative treatment options [60, 61].
The Salmonella spp. isolates displayed diverse antibiotic resistance patterns, with 100% sensitivity to Meropenem (MEM10) and high resistance rates to Cefotaxime (CTX30) and Ampicillin (AM10), which aligns with previous literature [62, 63]. The varying degrees of resistance observed in this study are consistent with findings from other studies, suggesting a global challenge in combating antibiotic resistance among Salmonella spp. strains [64, 65]. Ciprofloxacin (CIP5) displayed a balanced resistance pattern, with 20% resistance, 40% intermediate susceptibility, and 40% sensitivity. Colistin (CL30) revealed 50% resistance and 50% intermediate susceptibility, emphasizing the need for cautious use of these antibiotics to prevent further development of resistance [66, 67].
The observed multiple antibiotic-resistant (MAR) classification of E. coli isolates from various locations, with MAR Indices ranging from 0.08 to 0.77, aligns with global concerns about the widespread dissemination of multiple antibiotic resistance in environmental samples. The urgency of addressing antibiotic resistance in wastewater is evident, emphasizing the need for ongoing surveillance, prudent antibiotic use, and the development of strategies to mitigate the public health risks associated with resistant strains.
The detection of multiple antibiotic-Resistant (MAR) patterns in several sources signify resistance to multiple antibiotic classes, presenting significant challenges in treating bacterial infections. Particularly, locations like Kallayanpur, Mohammadpur, Lalbag, and Sadarghat exhibited high MAR Indices, indicating widespread resistance to various antibiotics, demanding immediate attention, and stringent surveillance to mitigate the escalation of multiple antibiotic-resistant bacterial infections [46, 47].
Overall, these findings emphasize the urgent need for heightened vigilance in antibiotic use, continual surveillance, and the development of precise treatment strategies to address the widespread antibiotic resistance observed in pathogenic bacteria within Dhaka's municipal wastewater. Taking immediate steps to address this issue is crucial in preserving the effectiveness of available antibiotics and managing the risks associated with multiple antibiotic-resistant bacterial infections.
4.1 Strength and limitations of the study
The study addresses a critical public health concern by investigating the prevalence of antibiotic-resistant pathogenic bacteria in municipal wastewater, focusing on Vibrio spp., fecal coliform, E. coli, and Salmonella spp. Additionally, this is the first study investigating these bacteria simultaneously from wastewater in Bangladesh. While offering valuable insights into Dhaka's municipal wastewater antibiotic resistance, this study has limitations. It focused on specific bacteria, excluding potential contributors to overall resistance. The study's snapshot in August 2023 may not represent evolving resistance patterns. Reliance on phenotypic profiles might miss genetic diversity; genomic studies could enhance understanding. The study concentrated on wastewater, lacking a holistic approach to antibiotic-resistant gene transmission among environmental, human, and animal sources. Specific sources of antibiotic contamination were not explored, hindering targeted interventions for pollution reduction. Additionally, the cross-sectional design hinders the establishment of causal relationships and a nuanced understanding of changes over time. A longitudinal or prospective study would be beneficial in this regard.
5 Conclusion
This study uncovers significant antibiotic-resistant bacteria, Vibrio spp., fecal coliform, E. coli and Salmonella spp. in Dhaka's municipal wastewater. Widespread contamination in neighborhoods like Kallyanpur, Rampura, Mohammadpur, Mirpur, Uttara, Dhanmondi, Sayedabad, Lalbag, Sadarghat, and Bangshal necessitates urgent interventions. Variable bacterial concentrations, particularly in Uttara, Bangshal, and Lalbag, reflect the impact of population density, wastewater production, and sanitation practices. Antibiotic Susceptibility Testing (AST) reveals considerable resistance in Vibrio spp. and fecal coliform, urging cautious antibiotic use. Challenges in E. coli and Salmonella spp. highlight the need for tailored treatments and continuous surveillance. The multiple antibiotic-resistant (MAR) classification, notably in Kallayanpur, Mohammadpur, Lalbag, and Sadarghat, poses a pressing challenge, requiring immediate attention, stringent surveillance, and targeted interventions to address widespread antibiotic resistance. The findings underscore the complexity of factors contributing to resistance, urging comprehensive measures to safeguard public health.
Data availability
Upon reasonable request, the corresponding author will provide access to all the data supporting this article.
Abbreviations
- AST:
-
Antimicrobial susceptibility testing
- CFU/ml:
-
Colony-forming units per milliliter
- CIP:
-
Ciprofloxacin
- CL:
-
Colistin
- COT:
-
Cotrimoxazole
- CTX:
-
Cefotaxime
- CXM:
-
Cefuroxime
- E. coli :
-
Escherichia coli
- MAR:
-
Multiple antibiotic-resistant
- MEM:
-
Meropenem
- NMAR:
-
Non-multiple antibiotic-resistant
- Spp.:
-
Species (plural)
- TVC:
-
Total viable count
References
Yin H, Islam MS, Ju M. Urban river pollution in the densely populated city of Dhaka, Bangladesh: big picture and rehabilitation experience from other developing countries. J Clean Prod. 2021;321:129040.
Cabral, J. P. S. Water microbiology. Bacterial pathogens and water. Int. J. Environ. Res. Public Health 7, 3657–3703 (2010).
Some S, et al. Microbial pollution of water with special reference to coliform bacteria and their nexus with environment. Energy Nexus. 2021;1:100008.
Faruque SM, Nair GB. Vibrio cholera: genomics and molecular biology. Poole: Caister Academic Press; 2008. https://www.caister.com/vib.
Thompson FL, et al. Phylogeny and molecular identification of vibrios on the basis of multilocus sequence analysis. Appl Environ Microbiol. 2005;71:5107–15.
Lee MT, Dinh AQ, Nguyen S, Krucke G, Tran TT. Late-onset Vibrio vulnificus septicemia without cirrhosis. Proceedings (Baylor University. Medical Center). 2019; 32: 286–288. https://doi.org/10.1080/08998280.2019.1580661.
Prakash R, et al. Dielectric barrier discharge based mercury-free plasma UV-lamp for efficient water disinfection. Sci Rep. 2017;7:1–9.
Ogazi AC, Osifo PO. Inhibition of coliforms and escherichia coli bacterial strains in water by 3D printed CS/GO/AgNP filtration membranes. J Polym Environ. 2023;31:4448–67.
Health WSD. of. Coliform Bacteria in Drinking Water. https://doh.wa.gov/community-and-environment/drinking-water/contaminants/coliform#:~:text=They%20appear%20in%20great%20quantities,total%20coliform%20bacteria%20is%20detected.
Turco RF. Coliform bacteria. Methods Soil Anal Part 2 Microbiol Biochem Prop. 1994;5:145–58.
Bhat S, Danek LJ. Comparison of fecal coliform before and after wastewater treatment facility: a case study near a coastal town in the southeastern USA. Water Air Soil Pollut. 2012;223:1923–30.
Łuczkiewicz A, Fudala-Książek S, Jankowska K, Quant B, Olańczuk-Neyman K. Diversity of fecal coliforms and their antimicrobial resistance patterns in wastewater treatment model plant. Water Sci Technol. 2010;61:1383–92.
Cava F. Biology of Vibrio cholera. Editorial overview. Int Microbiol. 2017. https://doi.org/10.2436/20.1501.01.290.
Madigan M, Martinko J. Brock biology of mircoorganisms. London: Pearson Education; 2016. p. 146–8.
Kenneth T. Todar’s Online Text Book of Bacteriology. 2020. https://textbookofbacteriology.net/
Fewtrell L, Kay D. Recreational water and infection: a review of recent findings. Curr Environ Heal Rep. 2015;2:85–94.
Su L-H, Chiu C-H. Salmonella: clinical importance and evolution of nomenclature. Chang Gung Med J. 2007;30:210–9.
Schoch CL, et al. NCBI taxonomy: a comprehensive update on curation, resources and tools. Database: J Biol Databases Curation. 2020. https://doi.org/10.1093/database/baaa062.
Kozak GK, Boerlin P, Janecko N, Reid-Smith RJ, Jardine C. Antimicrobial resistance in escherichia coli isolates from Swine and wild small mammals in the proximity of swine farms and in natural environments in Ontario, Canada. Appl Environ Microbiol. 2009;75:559–66.
Arias Granada Y, Haque SS, Joseph G, Yanez Pagans M. Water and sanitation in Dhaka slums: access, quality, and informality in service provision. World Bank Policy Research Working Paper. 2018.
Sakil AH. Urbanization and housing in developing countries: study of Bangladesh. Acad An Int Multidiscip Res J. 2018;8:28–42.
Haque SS, Yanez-Pagans M, Arias-Granada Y, Joseph G. Water and sanitation in Dhaka slums: access, quality, and informality in service provision. Water Int. 2020;45:791–811.
Chinemerem Nwobodo D, et al. Antibiotic resistance: the challenges and some emerging strategies for tackling a global menace. J Clin Lab Anal. 2022;36:e24655.
Uluseker C, et al. A review on occurrence and spread of antibiotic resistance in wastewaters and in wastewater treatment plants: mechanisms and perspectives. Front Microbiol. 2021;12:717809.
Wengenroth L, et al. Antibiotic resistance in wastewater treatment plants and transmission risks for employees and residents: the concept of the AWARE study. Antibiotics. 2021;10:478.
Nnadozie CF, Odume ON. Freshwater environments as reservoirs of antibiotic resistant bacteria and their role in the dissemination of antibiotic resistance genes. Environ Pollut. 2019;254:113067.
Ng C, Chen H, Tran NH, Haller L, Gin KY-H. Antibiotic resistance in municipal wastewater: a special focus on hospital effluents. In: Manaia C, Donner E, Vaz-Moreira I, Hong P, editors. Antibiotic resistance in the environment. A Worldw Overv. Cham: Springer; 2020. p. 123–46.
WHO. Antimicrobial resistance. 2023. https://www.who.int/campaigns/world-amr-awareness-week/2023.
Catalano A, et al. Multidrug resistance (MDR): a widespread phenomenon in pharmacological therapies. Molecules. 2022;27:616.
Saidi T, Salie F, Douglas TS. Towards understanding the drivers of policy change: a case study of infection control policies for multi-drug resistant tuberculosis in South Africa. Heal Res policy Syst. 2017;15:41.
Cassidy R, et al. Assessments of composite and discrete sampling approaches for water quality monitoring. Water Resour Manag. 2018;32:3103–18.
Lim Y, Totsika M, Mark M, Punyadeera C. The saliva microbiome profiles are minimally affected by collection method or DNA extraction protocols. Sci Rep. 2017. https://doi.org/10.1038/s41598-017-07885-3.
Tavazzi S, et al. Short-term stability of wastewater samples for storage and shipment in the context of the EU Sewage Sentinel System for SARS-CoV-2. J Environ Chem Eng. 2023;11:109623.
Chauhan A, Jindal T, Chauhan A, Jindal T. Biochemical and molecular methods for bacterial identification. Microbiol methods environ food pharm anal. Cham: Springer; 2020. p. 425–68.
Hossain MS, Aktaruzzaman M, Uddin MJ, Rahman SH, Alam MK. Prevalence of multiple drug resistant pathogenic bacteria in cultured black tiger shrimp ( Penaeus monodon Fabricius ). Glob J Environ Res. 2012;6:118–24.
Standardization, international organization for. International Standards in process. 2023. https://www.iso.org/iso-update.html.
Lotz MJ, Tamplin ML, Rodrick GE. Thiosulfate-citrate-bile salts-sucrose agar and its selectivity for clinical and marine vibrio organisms. Ann Clin Lab Sci. 1983;13:45–8.
Mafizur RM, Sangjin L, Chul PY. Prevalence of Salmonella spp. and Escherichia coli in the feces of free-roaming wildlife throughout South Korea. PLoS ONE. 2024;19:e0281006.
Marceddu M, et al. Determination of Salmonella spp., E. coli VTEC, Vibrio spp., and norovirus GI-GII in bivalve molluscs collected from growing natural beds in Sardinia (Italy). Foods. 2017;6:88.
Adesiyan IM, Bisi-Johnson MA, Okoh AI. Incidence of antibiotic resistance genotypes of Vibrio species recovered from selected freshwaters in Southwest Nigeria. Sci Rep. 2022;12:18912.
Bauer AW, Kirby WM, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966;45:493–6.
CLSI. Performance standards for antibiotic susceptibility testing. 2020. https://www.nih.org.pk/wp-content/uploads/2021/02/CLSI-2020.pdf.
Sarita GB. Multiple antibiotic resistances of Vibrio isolates from coastal and brackish water areas. 2005. http://dyuthi.cusat.ac.in/purl/734.
Brand P, Gobeli S, Perreten V. Pathotyping and antibiotic resistance of porcine enterovirulent Escherichia coli strains from Switzerland (2014–2015). Schweiz Arch Tierheilkd. 2017;159:373–80.
Swamy A, et al. Antibiotic stewardship initiative in a Medicine unit of a tertiary care teaching hospital in India: a pilot study. Indian J Med Res. 2019;150:175–85.
Hoque R, et al. Tackling antimicrobial resistance in Bangladesh: a scoping review of policy and practice in human, animal and environment sectors. PLoS ONE. 2020;15:e0227947.
Ahmed I, Rabbi MB, Sultana S. Antibiotic resistance in Bangladesh: a systematic review. Int J Infect Dis. 2019;80:54–61.
Alam M-U, et al. Strategies to connect low-income communities with the proposed sewerage network of the Dhaka sanitation improvement project, Bangladesh: a qualitative assessment of the perspectives of stakeholders. Int J Environ Res Public Health. 2020;17:7201.
Khan TA. Dhaka water supply and sewerage authority: performance and challenges. Web Rep. 2013; 1–13. http://app.dwasa.org.bd/admin/news/Dhaka%20WASA%20Article-for%20BOOK.pdf.
Defoirdt T, Sorgeloos P, Bossier P. Alternatives to antibiotics for the control of bacterial disease in aquaculture. Curr Opin Microbiol. 2011;14:251–8.
Dutta D, Kaushik A, Kumar D, Bag S. Foodborne pathogenic vibrios: antimicrobial resistance. Front Microbiol. 2021;12:638331.
Okoh AI, Igbinosa EO. Antibiotic susceptibility profiles of some Vibrio strains isolated from wastewater final effluents in a rural community of the Eastern Cape Province of South Africa. BMC Microbiol. 2010;10:143.
Serwecińska L. Antimicrobials and antibiotic-resistant bacteria: a risk to the environment and to public health. Water. 2020;12:3313.
Kumar A, Patyal A, Panda AK. Sub-therapeutic use of antibiotics in animal feed and their potential impact on environmental and human health: a comprehensive review. J Anim Feed Sci Technol. 2018;6:25.
Martinez JL. Environmental pollution by antibiotics and by antibiotic resistance determinants. Environ Pollut. 2009;157:2893–902.
Bell JB, Elliott GE, Smith DW. Influence of sewage treatment and urbanization on selection of multiple resistance in fecal coliform populations. Appl Environ Microbiol. 1983;46:227–32.
Iwu CD, Kayode AJ, Igere BE, Okoh AI. High levels of multi drug resistant Escherichia coli pathovars in preharvest environmental samples: a ticking time bomb for fresh produce related disease outbreak. Front Environ Sci. 2022;10:858964.
Yasmin S, Karim A-M, Lee S-H, Zahra R. Temporal variation of meropenem resistance in E. coli Isolated from sewage water in Islamabad, Pakistan. Antibiot. 2022;11:635.
Gekenidis M-T, et al. Antibiotic-resistant indicator bacteria in irrigation water: high prevalence of extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli. PLoS ONE. 2018;13:e0207857.
Mohammadi Gharibani K, et al. High frequency of macrolide-resistant streptococcus pneumoniae colonization in respiratory tract of healthy children in Ardabil, Iran. Tanaffos. 2019;18:118–25.
Jelić D, Antolović R. From erythromycin to azithromycin and new potential ribosome-binding antimicrobials. Antibiotics. 2016;5:29.
Ali Shah SA, et al. Antimicrobial sensitivity pattern of Salmonella Typhi: emergence of resistant strains. Cureus. 2020;12:e11778.
Mumbo MT, et al. Antimicrobial resistance profiles of salmonella spp. and escherichia coli isolated from fresh nile tilapia (oreochromis niloticus) fish marketed for human consumption. BMC Microbiol. 2023;23:306.
Lauteri C, et al. Overcoming multidrug resistance in Salmonella spp. isolates obtained from the swine food chain by using essential oils: an in vitro Study. Front Microbiol. 2021;12:808286.
Qian M, et al. Isolation, antimicrobial resistance and virulence characterization of Salmonella spp from fresh foods in retail markets in Hangzhou, China. PLoS ONE. 2023;18:e0292621.
Fortini D, et al. Colistin resistance mechanisms in human salmonella enterica strains isolated by the national surveillance enter-net Italia (2016–2018). Antibiotics. 2022;11:102.
Vázquez X, et al. Colistin resistance in monophasic isolates of Salmonella enterica ST34 collected from meat-derived products in Spain, with or without CMY-2 Co-production. Front Microbiol. 2021;12:735364.
Acknowledgements
The authors express their sincere appreciation to all the staffs and technicians of microbial laboratory from Department of Public Health and Informatics, Jahangirnagar University, Savar, Dhaka-1342, Bangladesh and Md Rashed Alam for helping to make the study area’s map.
Funding
This research was partially supported by Kurita Water and Environment Foundation Research Grant Program 2022 (Reference number: 22P003-K1).
Author information
Authors and Affiliations
Contributions
Every author listed has thoroughly examined and approved the manuscript, and no individuals who meet the authorship criteria have been omitted. The particular contributions of each author are outlined below: ABS: conceptualization, methodology, microbiological investigation, data curation, data analysis and interpretation, writing—original draft, validation. AM: microbiological investigation, writing—original draft, data curation, validation. MH: microbiological investigation, data curation, data analysis and interpretation, writing—original draft, validation. RR: microbiological investigation, data curation, writing—original draft, validation. MAM: microbiological investigation, data curation, writing—original draft, validation. MTS: conceptualization, methodology, writing—critical review & editing, supervision, validation. TO: conceptualization, methodology, writing—critical review & editing, supervision, validation. AA: conceptualization, methodology, writing—critical review & editing, formal analysis, supervision, validation. MSH: conceptualization, methodology, writing—critical review & editing, supervision, validation.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
No human or animal sample was used in this study.
Consent for publication
Not applicable.
Competing interests
The authors assert that there are no potential competing interests associated with the publication of this paper.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Siddique, A.B., Munni, A., Hasan, M. et al. Investigation and detection of multiple antibiotic-resistant pathogenic bacteria in municipal wastewater of Dhaka city. Discov Water 4, 52 (2024). https://doi.org/10.1007/s43832-024-00114-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s43832-024-00114-9