Abstract
Ninety five bacterial strains were isolated from the rhizosphere and soils of flower crops, specifically gerbera, carnation and tuberose, were preliminarily screened against Fusarium oxysporum f. sp. gerberae in vitro along with five elite strains Bbv 57, EPCO 16, EPC 5, EPC 8 and Pf 1 obtained from Department of Plant Pathology, TNAU, Coimbatore, India. The results revealed that the strain Bbv 57 had highest inhibition of 50.00% and showed the maximum value for the assays that is siderophore, hydrogen cyanide, IAA, GA3 and salicylic acid production (3.68 O.D. value; 14.05 µg ml-1; 44.40 µg ml-1; 25.28 µg ml-1 and 19.25 µg ml-1 respectively). It also showed resistance to antibiotics namely ampicillin, erythromycin, clindamycin. The highest exopolysaccharides, biofilm production was observed and had lowest protease production clearly indicated that it is non - pathogenic to plants. Further, the polymerase chain reaction using16S rRNA intervening sequencing showed 100% homology to Bacillus subtilis (KF718836) and showed positive for quorum sensing regulator genes of aiiA and comQ. Additionally, the culture filtrate assay also produced significant reduction in egg hatching capacity and juvenile mortality of root-knot nematode Meloidogyne incognita. Further, the HPLC study showed 91.69 µg µl-1 of surfactin with the retention time of 2.304 min and 0.453 µg µl-1 of Iturin with the retention time of 8.739 min at 205 nm. Whereas, GCMS analysis has detected the aliphatic hydrocarbons responsible for antifungal, antibacterial and nematicidal activity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gerbera (Gerbera jamesonii Bolus ex Hooker f.) commonly known as Transvaal daisy, Barberton daisy or African daisy, is a very attractive commercial cut flower crop grown throughout the world under a wide range of climatic conditions (Lhoste, 2002). In India, the flower crops under protected cultivation is gradually increasing in Bangalore, Calcutta, Coimbatore, Darjeeling, Delhi, Kodaikanal, Ludhiana, Nasik, Nilgiris, Palampur, Pune, Shimla, Solan and Srinagar with an area of about 680 ha (Kumar et al., 2014). The area in Tamil Nadu was estimated as 25 ha (Sudhagar, 2013) and it is a potential flower crop in protected cultivation that favors luxuriant growth both for cut flower and plant pathogens. Under these circumstances, major diseases namely foot rot, wilt, root rot complex, blight and grey mold were reported (Padghan and Gade, 2006; Rajendran et al., 2014) and wilt disease is aggravated with root-knot nematode, Meloidogyne incognita (Kofoid et White) Chitw., which is a serious limiting factor. The root-knot nematode infection always increased the severity of Fusarium wilt (Adam et al., 2014; Bonmatin et al., 2003) and results in synergistic flower losses (Mukherjee and Das, 2005). Exotic varieties from Europe suffered 40–60% mortality due to root-knot nematode - Fusarium complex (Nagesh and Parvatha Reddy, 1996). In India, flower yield losses due to M. incognita was estimated as 30% (Kishore, 2007) and controlling one pathogen might not fully solve the problem. Hence, combinations of nematicidal and fungicidal treatments are possible but not always desired due to their negative impact on environment and human health. In recent years, the focus is shifted towards biological control and numerous microbes are antagonistic to soil borne plant pathogens and plant parasitic nematodes. The bacterial genera namely Bacillus, Pseudomonas and Burkholderia are not only promoting plant growth, but also providing resistance against bacterial, fungal and viral diseases (Ramzan et al. 2016). Since, Fusarium and Meloidogyne are soil borne, the ideal biocontrol agent should survive in rhizosphere. Among the biocontrol agents, the strains of Plant Growth Promoting Rhizobacteria (PGPR) are known to survive both in rhizosphere and phyllosphere which are the potential alternative to fungicide and nematicide (Omar et al. 2013). One such plant growth-promoting rhizobacterium is the Gram-positive soil-dwelling Bacillus subtilis Cohn which is found in association with roots of many different plants (Cazorla et al., 2007) and exhibits many beneficial activities for the plant and widely used as a biofertilizer (Lucy et al., 2004). A unique characteristic is the ability to produce endospores when environmental conditions are stressful. Bacillus subtilis with an average of 4–5% of its genome devoted to antibiotic synthesis, has the potential to produce more than two dozen structurally diverse antimicrobial compounds (Stein, 2005; Rajendran et al., 2012). The production of surfactin and other lipopeptides is one of main mechanisms since these molecules can induce systemic resistance (ISR) as well as strongly inhibit Fusarium oxysporum f. sp. gerberae (Ramyabharathi and Raguchander, 2014). Peptide antibiotics or small molecules secreted by Bacillus species contribute their activity against root knot nematodes (Cadena et al., 2008; Kavitha et al., 2012). It naturally colonizes the plant roots by means of biofilm formation which is important for plant root colonization and protection. So far, information on the simultaneous effect of a bio-agent on plant pathogen-nematode complex infecting flower crops grown under protected cultivation is not widely available.
Hence, the objective of the study was to isolate, screen and identify the potential rhizobacteria having an antagonistic, inhibitory effect to both Fusarium and root knot nematode infecting Gerbera under protected cultivation.
Materials and methods
Pathogen and nematode
Fusarium sp. was isolated from infected root sample of gerbera and the roots were washed under tap water, chopped into 2 cm small pieces and surface sterilized in 0.5% sodium hypochlorite for two minutes then rinsed twice with distilled water and placed on Fusarium selective medium (Komada 1975) and for the nematode, Cobb‟s sieving and decanting technique was followed and identified based on key provided by Taylor and Sasser (1978).
Molecular identification
To confirm Fusarium, 18 s rDNA intervening sequence specific ITS FU-F (5‟CAACTCCCAAACCCCTGTGA3‟); ITS FU-R (5‟ GCGACGATTACCAGTAACGA3‟) primers which will yield an amplicon of 389 bp size (Singh and Kumar 2001) were used. A small agarose slice containing the band of interest (observed under long-wavelength [312-nm] UV light) was excised from the gel and purified by using a QIA quick gel extraction kit (Qiagen, Inc., Chatsworth, California) according to the supplier‟s instructions. This purification was performed to remove primer dimers and other residues from the PCR amplification. PCR product was then subjected to sequencing. DNA sequencing was performed at Chromous Biotech Pvt. Ltd, Bangalore, India. The rDNA homology searches were performed using the BLAST program at the National Center for Biotechnology Information (National Institutes of Health, Bethesda, USA). Sequences and accession numbers for compared isolate were retrieved from the Gene bank database. For root knot nematode, perineal patterns were prepared from adult females collected from diseased plants from which nematode species were identified (Taylor et al. 1955). Perineal patterns were identified under a compound microscope with the aid of a pictorial key (Eisenback et al., 1981).
Antagonists and in vitro assay
The bacterial antagonists were isolated by serial dilution technique using King’s B and nutrient agar medium respectively from rhizosphere. Bio-agents namely B. subtilis (Bbv 57, EPCO 16, EPC 5 and EPC8) and P. fluorescens (Pf1), were obtained from the Culture Collection Section, Department of Plant Pathology, TNAU, Coimbatore, India. Bacillus spp. was grown in nutrient broth and the total DNA (including chromosomal and plasmid DNA) was extracted as described by Robertson et al. (1990) with slight modifications. To confirm strains as Bacillus sp., 16 s rRNA intervening sequence specific BCF1 (5‟-CGGGAGGCAGCAGTAGGGAAT-3‟); BCR2 (5‟-CTCCCCAGGCGGAGTGCTTAAT-3‟) primers were used to get an amplicon size of 546 bp (Cano et al. 1994). The amplified samples were sequenced. These bio-agents were tested against F. oxysporum by dual culture technique (Dennis and Webster 1971). Mycelial disc (9 mm) from 7 days old culture was placed at one side of the Petri plate containing 15 ml of PDA medium. The Pseudomonas and Bacillus strains were streaked on the opposite side of the Petri plate by a sterilized inoculation needle. Three replications were maintained for each treatment. The plates were incubated at room temperature (28 ± 2 ºC) for 5 days and the inhibition zone was measured. The radial growth of the pathogen and per cent reduction over control was calculated by using the formula as follows:
Per cent reduction over control = [Mycelial growth of the pathogen in control (cm)-Mycelial growth of the pathogen in dual culture plate (cm)/ Mycelial growth of the pathogen in control (cm)] × 100.
Volatile antibiotics by antagonists
Bacillus strains were evaluated for the production of volatile inhibitory substances in vitro following the technique of Dennis and Webster (1971). Bacillus strains were streaked in Petri plates containing NA medium in triplicates. The test pathogen was inoculated on fresh PDA and the lids of the Petri plates inoculated with the antagonist were replaced by the culture of the pathogen on PDA. The plates were fixed with cellotape and incubated for another seven days. Control plates inoculated with pathogen inverted over dish containing PDA alone. Growth of the pathogen was measured after five days of incubation.
Siderophore, Hydrogen cyanide (HCN) production
The Bacillus strains were assayed for siderophore production by plate assay (Schwyn and Neilands 1987). Ten µl of 48-h-old bacterial cultures were spotted on succinate medium amended with indicator dye. The tertiary complex chrome azural S (CAS) / Fe3+ / hexadecyl trimethyl ammonium bromide served as an indicator. Change of blue colour of the medium surrounding the spotted area to fluorescent yellow indicated the production of siderophore. The area of colour change was measured and compared with control. For HCN, Bacillus culture was streaked onto tryptic soy agar medium (TSA) (HiMedia, Mumbai, India) in a Petri dish. Filter paper disc of 1.5-cm-dia was soaked in picric acid solution (picric acid, 2.5 g; Na2CO3, 12.5 g and distilled water, 1 l) and placed in the lid of each Petri dish (Miller and Higgins 1970). Dishes were sealed with parafilm and incubated for 4 days. HCN production was assessed by the conversion of yellow colour of the filter paper to brown to reddish brown. Reactions were scored as weak (yellow to light brown), moderate (brown) and strong (reddish brown).
Indole Acetic Acid (IAA) and Gibberelic acid production
One ml of the culture at the exponential phase was inoculated in 100 ml Luria-Bertani (LB) medium containing filter sterilized L-tryptophan (0.01% w/v). All the flasks were wrapped with black paper to avoid photo inactivation of the biologically active compounds. The flasks were incubated at room temperature (28 ± 2 °C) for 7 days. The cells were harvested by centrifugation at 10,000 rpm for 5 min and the supernatant was collected and concentrated to 25 ml (Chandramohan and Mahadevan 1968). The estimation has been carried out as per the procedure given by (Gordon and Paleg 1957). For GA3 assay, 1 ml of the culture at the exponential stage was inoculated in Luria-Bertani (LB) broth and incubated for 7 days at room temperature. The cultures were centrifuged for 10 min at 10,000 rpm and the supernatant was collected. The cell pellet was re-extracted with phosphate buffer (pH 8.0) and again centrifuged. Both supernatants were collected and pooled, acidified to pH 2.0 with 5 N HCl and extracted with equal volumes of ethyl acetate thrice. The ethyl acetate phase was evaporated at 32 °C and the residue was redissolved in 2 ml of distilled water containing 0.05% of Tween 80 (Borrow et al. 1955).The estimation has been carried out as per the procedure given by (Mahadevan and Sridhar 1982).
Salicylic acid (SA) estimation
The Bacillus strains were grown in the standard succinate broth at room temperature (28 ± 2 °C) for 48 h (Meyer and Abdullah 1978). The cell-free culture supernatant was used to estimate salicylic acid. The supernatant (4 ml) was acidified with 1 N HCl to pH 2.0 and salicylic acid was extracted in chloroform (2 × 2 ml). Five µl of 2 m ferric chloride and 4 ml of water were added to the pooled chloroform phase. The absorbance of the purple iron-salicylic acid complex, which developed in the aqueous phase was measured at 527 nm (Meyer et al. 1992). Salicylic acid production was measured using salicylic acid as standard and expressed as µg ml− 1 of the culture filtrate.
Intrinsic antibiotic resistance
The Bacillus strains were screened for antibiotics by the disc diffusion method. Pre-impregnated antibiotic discs (Hi-media octa disc) each ring containing eight different antibiotic discs of 0.6 mm diameter was used. Hundred µl of log phase culture containing 2 × 108 cells ml− 1 was spread evenly on solidified Luria-Bertani (LB) medium and the surface was allowed to dry for a few min. The ready-made antibiotic octa disc was placed on the surface of medium and the Petri plates were incubated at 28 ± 2 °C for 48 h. The susceptibility of the bacterial strain was indicated by development of clearing zone around each disc. Control plates were maintained separately without bacterial cultures in the medium. After 2–3 days of incubation at 30 ºC, the inhibition zone was measured.
Exoploysaccharides
For the best effective strain the protein molecular weight of exoploysaccharides was determined. The bacterial cells were lysed by heating at 100 °C in 125 µl of 60 mM Tris HCl (w/v), SDS/1 mM EDTA (pH 6.8) for 5 min and then diluted to 1 ml with the same buffer without SDS. SDS-PAGE was performed following the method of Laemmli (1970) using a separating gel of 15% and a stacking gel of 4% to separate the bacterial protein. The samples containing equal amount of proteins were loaded into the wells of polyacrylamide gels (Sigma-Aldrich Techware system; Sigma). Medium range molecular weight marker (Genei, Bangalore, India) mixed with sample buffer was also loaded in one of the wells. Electrophoresis was carried out at constant voltage of 75 volts. Gels were stained with 0.2% Coomassie brilliant blue R250 solution overnight and then destained. Based on the Rf value of each protein band stained, the molecular weight was calculated.
Biofilm formation
Biofilm formation was assessed by microtitre plate assay of Harvey et al. (2007) with slight modification. Bacterial cultures (125 µl) grown in LB for 18 h were transferred to 5 ml of growth medium (LB) and vortexed for 1 min. Aliquots of 100 µl of the vortexed mixture were transferred to three wells of sterile polystyrene microtitre plates and incubated at 30 °C for 24 or 48 h. Control wells contained sterile growth medium only. Cell cultures were discarded after incubation and the plates were washed thrice with sterile distilled water and air-dried at 30 °C for 30 min. Aqueous 1% crystal violet solution (150 µl) was added to each well and incubated at 30 °C for 45 min. The crystal violet solution was then removed and the plates were washed thrice with sterile distilled water and air-dried. Next, 95% ethanol was added (100 µl) to the wells for destaining the biofilm and the concentration of crystal violet was determined by measuring the optical density (OD) at 595 nm in a microtitre plate reader.
Protease production
Protease was determined on brain heart infusion (BHI) milk medium as described by Sokol et al. (1979) with minor modification. BHI milk medium was prepared by dissolving 18.5 g of BHI broth (Difco, USA) and 7.5 g agar (Difco, USA) in 400 ml of water. One hundred ml of 15% (w/v) solution of skim milk (Guangming, Heilongjiang, China) was prepared, and the solutions were autoclaved separately. These two sterile solutions were mixed at 60 ºC. Ten ml of mixture was dispensed into Petri plates. The bacterial strains were inoculated on BHI agar with a sterile toothpick. Protease activity was determined by measuring the diameter (cm) of zone of clearing around the site of inoculation after incubating the plates for 24 and 48 h at 30 ºC. Each treatment had four replicates, and the experiment was performed twice.
Hatching and mortality test, ESEM study
A single colony of bacterium was cultured in flask containing nutrient broth and incubated at 28ºC on a shaker at 100 rpm for 2–3 days. The culture was subsequently passed through sterilized Whatman filter paper No. 1 and 42, concentrated by centrifugation at 6000 rpm for 10 min and the supernatant was collected and finally passed through a Millipore filter of 0.22 µm which was finally designated as undiluted standard filtrate of 100% concentration (Niknam and Dhawan 2002). About 5 ml cell free culture filtrate of B. subtilis were taken at different concentration of 25, 50 and 100% in a 50 mm Petri dish and five egg mass of M. incognita were placed in each dish and incubated at 28 + 1 ºC. Egg mass placed in NA broth without bacteria served as a standard check along with distilled water as control. The experiment was replicated four times. Observation on number of hatched juveniles was made after 24, 48 and 72 h of exposure. In case of mortality test, five ml of cell free culture filtrate of B. subtilis was taken at different concentrations of 25, 50 and 100% and were poured into separate Petri dishes. The second stage juveniles of M. incognita were introduced at the rate of 100 juveniles in each dish and incubated at 28 + 1 ºC. Juveniles placed in dishes containing nutrient broth without bacteria served as standard check along with distilled water as control. Each treatment was replicated four times. Observations were recorded on the mortality of juveniles after 24, 48 and 72 h of exposure period and % mortality was calculated. The inactive nematodes were transferred separately from each dilution into sterile distilled water and kept overnight to check whether mortality was permanent or temporary. Root knot nematode eggs and female nematode treated with crude antibiotic extract of B. subtilis Bbv 57 were taken and observed under the environmental scanning electron microscope [ESEM (Model: Quanata 250; Manufacturer: FEI, Czech Republic Repulic; Detector; GESD (Gaseous State Electron Detector); Electron source: Tungstan); available in the Department of Nano Science and Technology, TNAU, Coimbatore.
Quorum sensing regulator genes identification
Bacillus subtilis Bbv 57 strain was screened for presence of aiiA homologue gene using PCR. The conditions are initial denaturation at 94 ºC for 10 min, 35 cycles of 94 ºC (30 s), 52 ºC (30 s), 72 ºC (1 min); followed by primer extension at 72 ºC for 5 min. The forward and reverse primers used were aiiA-F (5‟-ATGGGATCCATGACAGTAAAGAAGCTTTAT-3‟) and aiiA-R (5‟-GTCGAATTCCTCAACAAGATACTCCTAATG-3‟) (Gan Chan et al., 2007). The comQ genes of B. subtilis Bbv 57 were amplified by PCR with primers UnicomQ1 (5‟-GGG AGG GGG GAA GTC GTT ATT G-3‟) and UnicomP1 (5‟-AAGAACCGAATCGTGGAGATCGCG-3‟) (Polonca and Mulec 2009) in 20 µl reaction mixture containing 10X buffer (with 2.5 mM MgCl2), 2 µl; 2 mMdNTP mixture, 2 µl; 2 m primer, 5 µl; Taq DNA polymerase, 3 U; H2O, 8 µl and 50 ng of template DNA samples were amplified on DNA thermal cycler (Eppendorf Master Cycler Gradient, Westbury, New York). The PCR profile of the comQXP locus amplification consisted of 30 cycles of denaturation at 94 °C for 30 s, annealing at 55 °C for 45 s, extension at 72 °C for 3 min, and final extension at 72 °C for 5 min. The PCR products were purified with the QIA quick PCR purification kit (Qiagen, Hilden, Germany) prior to sequencing. The resulting amplicons were examined by electrophoresis on a 1.2% agarose gel.
High Performance Liquid Chromatography (HPLC)
Production of iturin and surfactin was verified by using analytical HPLC method described by Huang et al. (1993). B. subtilis Bbv 57 was grown at 28 ± 2 ˚C for 5 days in Pigment Production broth and centrifuged at 11,000 × g for 10 min and the supernatant was acidified to pH 2 with 12 N HCI. Then, the precipitate collected by centrifugation was extracted with 2 ml HPLC methanol for 2 h. After the extracted solution was centrifuged at 10,000 rpm for 2 min, the supernatant was filtered through a 0.2 µm polyvinylidene difluoride (PVDF) membrane (HiMedia laboratories Pvt. Ltd)and injected into a HPLC column (column: Agilent 1100 series TC-C18 (2), 5 µm, 4.6 ID, 25 cm L). The system was operated at a flow rate of 1.0 ml min− 1 with acetonitrile-ammonium acetate - trifluoroacetic acid (10 mM) 3:4 (v/v). The elution pattern was monitored at 205 nm. Authenticated iturin and surfactin were purchased from Sigma-Aldrich (St. Louis, MO, USA) and used as standards.
Gas Chromatography and Mass Spectrometry (GC-MS)
The crude antibiotics of the effective strain Bbv 57 were analyzed for the identification of active bio-molecules responsible for the suppression of M. incognita and F. oxysporum through GC-MS (GC Clarus 500 Perkin Elmer). Volatile components were identified by GC-MS using a coloumn Elite-5MS (100% Dimethyl poly siloxane), 30 × 0.25 mm × 0.25 µm df. The turbo mass-gold-Perkin-Elmer detector was used. The carrier gas flow rate was 1 ml per min, split 10:1 and injected volumes were 3 µl. The column temperature was maintained initially at 110 °C at the rate of 10 °C min− 1 no hold followed by increases up to 280 °C at the rate of 5 °C per min-9 min (hold). The injector temperature was 250 °C and this temperature was held constant for 36 min. The electron impact energy was 70 eV, Juletline temperature was set at 2000 °C and the source temperature was set at 200 °C. Electron impact (EI) mass scan (m/z) was recorded in the 45–450 aMU range. Using computer searches on the NIST Ver.2005 MS data library and comparing the spectrum obtained through GC/MS, the compounds present in the crude sample were identified.
Statistical analysis
The data were statistically analyzed using the IRRISTAT version 92 developed by the International Rice Research Institute Biometrics unit, the Philippines (Gomez and Gomez, 1984). The percentage values of the disease index were arcsine transformed. Data were subjected to analysis of variance (ANOVA) at significant level (P < 0.05) and means were compared by Duncan‟s Multiple Range Test (DMRT).
Results
Pathogen and nematode
The affected gerbera plants exhibited chlorosis of foliage leading to drying of leaves. Infected plants showed typical yellowing of leaves, later leaves turned to straw coloured and wilted. In wilted plants, light brown vascular discolouration was observed which finally led to death of the plants. Plant growth was retarded, bud emergence was suppressed resulting in an absolute loss of flower yield. The root system showed characteristic galls which in severe cases resulted in rotting and decay of suckers. Ten different isolates of F. oxysporum were isolated and the mycelium on PDA was initially white and later turned to light pink. When stained roots were observed under microscope, root knot nematode female population was maximum in root samples collected from Asampur of Salem district (14.00 female nematodes g− 1 of root).
Morphological and molecular Identification
Using the perineal pattern preparations, the prevailing Meloidogyne species was identified as M. incognita. These are characterized by the presence of high, squarish dorsal arch that often continued a distinct whorl in the tail terminal area and the striae were smooth to wavy. The PCR analysis of F. oxysporum resulted in an amplification of 389 bp in all the 10 isolates (Fig. 1). The amplified genomic product was partially sequenced and the rDNA homology searches were performed using the BLAST program though the internet server at the National Center for Biotechnology Information, USA for the virulent isolate FOG 2. The sequence analysis revealed that they had a nucleotide sequence identity of 95–100% with that of F. oxysporum. The partial sequence was submitted to the NCBI, GenBank, New York, USA and assigned with accession number KM523669.
Dual culture plate technique
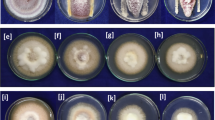
Among the one hundred strains, twenty five strains were found to be effective against F. oxysporum. The Bacillus strain Bbv 57 showed highest inhibition of 50.00% followed by B. subtilis strain EPCO 16 which recorded 46.66% inhibition. Five Bacillus strains RBG 1, RBG 2, RBG 3, RBC 1 and RBC 2 showed more than 44.00% inhibition (Table 1; Fig. 2a). There was no increase or decrease in the inhibition of mycelial growth of Fusarium with increase in the days of incubation even at lower (10 µl) and higher (100 µl) concentration of crude antibiotic (Fig. 2b).
Isolation and characterization of biocontrol agents
In total, 95 strains of bacteria were isolated from rhizosphere soil and plant material of flower crops namely gerbera, carnation and tuberose. Five promising bacterial antagonists, namely Bbv 57 (betelvine rhizobacterium), EPCO 16 (cotton endophyte), EPC 5 (coconut endophyte), EPC 8 (coconut endophyte) and TNAU-Pf 1 (blackgram rhizobacterium), were obtained. The primers amplified a fragment size of 546 bp corresponding to the region of the 16-23 s rRNA intervening sequence for bacteria (Fig. 3). The strains Bbv 57 and RBG 1 were confirmed as Bacillus subtilis and those sequences were submitted to the NCBI, Gene bank, NewYork, USA, with accession numbers KF718836 and KF718837 respectively.
Siderophore production
Among 25 Bacillus strains, nine strains RBG 1, RBG 2, RBG 3, RBG 4, RBC 1, RBC 2, RBC 4, EPCO 16 and Bbv 57 produced yellow colored zone of more than 1.00 cm on blue agar medium. The results presented in Fig. 2c revealed that the intensity of the siderophore production was higher in the above nine strains.
Production of HCN, IAA, GA3 and SA
Among 25 Bacillus strains, only three strains, EPC 5, EPC 8 and Bbv 57 showed medium production of HCN qualitatively (Fig. 2d). Further, four strains namely RBG 1, RBG 2, EPCO 16 and Bbv 57 showed high production of IAA of more than 31.00 µg ml− 1. The lowest production of less than 3.00 µg ml− 1 was recorded in strains RBT 3 and RBT 20. Out of strains tested, six strains, RBG 1, RBG 2, RBG 3, RBG 4, EPCO 16 and Bbv 57 showed high production of Gibberelic acid of more than 20 µg ml− 1. Production of less than 2.00 µg ml− 1 was recorded in strains RBT 3 and RBT 20 (Table 2).Three strains namely RBG1, EPCO 16 and Bbv 57 showed more than 17.00 µg ml− 1 of salicylic acid. The strains RBG 7, RBT 3, RBT 9, RBT 12 and RBT 20 did not show production of salicylic acid quantitatively.
Intrinsic antibiotic resistance
The presence or absence of inhibition zone around the antibiotic discs was observed to validate the susceptibility or resistance of the Bacillus strains to eight different antibiotics. Among the strains, 11 strains RBG 1, RBG 3, RBG 7, RBC 1, RBC 2, RBC 9, RBT 3, RBT 20, EBT 1, EPCO 16 and Bbv 57 showed resistance to ampicillin, erythromycin and clindamycin. The strains RBG 1 and RBG 2 were resistant to cephalothin, chloramphenicol, oxacillin and vancomycin. However, Bbv 57 showed sensitive reaction to gentamicin and vancomycin (Fig. 2e).
Protease production by bacterial strains
Among the strains, one strain Bbv 57 showed the lowest production of protease in BHI milk plates after 24 and 48 h of incubation. It recorded 0.25 and 1.30 cm zone of hydrolysis at 24 and 48 h respectively (Fig. 2f). The next best strain in low protease activity was RBG 1. Hence, the results revealed that Bbv 57 is non - pathogenic to plants.
Production of exopolysaccharides and biofilm formation
Among the strains, the highest yield of EPS was noticed in Bbv 57 (19. 32 mg g− 1) which was followed by strain EPCO 16 with a yield of 17. 53 mg g− 1 of cell dry weight. Five strains, RBC 15, RBG 1, RBG 2, RBG 3 and RBG 4 produced EPS of more than 12.00 mg g− 1 of cell dry weight. The other strains recorded medium to low production of EPS (Table 3). Qualitative estimation through SDS PAGE was performed on 15% (W/V) acrylamide slab gels. The result showed that a clear band of 29 KDa molecular weight which represented the presence of exopolysaccharide (Fig. 4). The Biofilm formation was estimated at 595 nm. Maximum biofilm formation was recorded in Bbv 57 with a O.D. value of 5.90 which was on par with EPCO 16 (O.D. value 5.43). Least production was recorded in strain EBT 1 with an O.D. value of 1.03. The other strains recorded medium to low production of biofilm formation (Fig. 5).
Nematode hatching and mortality test
Significant reduction in egg hatching was observed with B. subtilis strain Bbv 57 (76.67 eggs egg mass− 1) at 25%, (37.33 eggs egg mass− 1) at 50% and (7.00 eggs egg mass− 1) at 100% concentration after 72 h of exposure period compared to control which recoded 202.67 eggs egg mass− 1. The highest juvenile (J2) mortality of 18.67 100 J2− 1 at 25%, 40 100 J2− 1 at 50% and 87 100 J2− 1 at 100% concentration after 72 h of exposure and when observed under microscope the Bbv 57 treated juveniles were found dead (Fig. 6a, b).
Environmental Scanning Electron Microscope (ESEM) study
Nematode eggs and females treated with crude lipopeptide antibiotic were observed under ESEM. The treated egg shell was shriveled whereas the untreated eggs were of oblong shape embedded in a gelatinous matrix.The crude antibiotic treated female nematode wall was shrunken and misshaped whereas untreated control female nematode appeared globose, swollen, pear shaped with a short neck (Fig. 6c, d).
Molecular identification of quorum sensing regulator genes
Primers aiiA-F and aiiA-R were directed against conserved sequences within biosynthetic loci of B. subtilis strain Bbv 57. Primers aiiA-F and aiiA-R amplified the predicted 450-bp fragment from DNA of Bbv 57. Presence of comQ gene in B. subtilis strain Bbv 57 was identified using PCR amplification with gene specific primers UnicomQ1 and UnicomP1 to produce the DNA fragment of approximately 701 bp. The result indicated that the Bbv 57 contained the quorum sensing regulator genes aiiA and comQ (Fig. 7).
HPLC and GCMS study
High Performance Liquid Chromatography (HPLC) analysis for the standard iturin and surfactin at 205 nm recorded retention time of 8.5 and 2.5 min, respectively. B. subtilis strain Bbv 57 showed 91.69 µg µl− 1 of surfactin with the retention time of 2.304 min and 0.453 µg µl− 1 of Iturin with the retention time of 8.739 min. Further, Gas Chromatography and Mass Spectrometry (GCMS) analysis revealed that the crude antibiotics were analyzed through GC/MS to detect the antimicrobial compounds produced by Bbv 57. The compound identity was confirmed through NIST library 2005. The compounds associated included Butanedioic acid, Hexadecanoic acid, Pentanedioic acid 2-oxo-dimethyl ester, Pyrrolo [1,2-a]pyrazine-1,4-dione hexahydro-3-(2-methylpropyl) and Pyrrolo [1,2-a]pyrazine-1,4-dione hexahydro-3-(phenylmethyl) ester with antifungal activity were detected (Fig. 8).
Discussion
In India, cultivation of ornamental crops on commercial scale for domestic and export market is gaining momentum and are being cultivated under controlled environmental conditions. Among the crops, Gerbera is a potential flower crop grown under poly houses (Padghan and Gade, 2006) which was infected by many pathogens namely foot rot, wilt and root rot complexes. In particular, the root-knot nematode M. incognita is a serious limiting factor in commercial polyhouses which always increased the severity of Fusarium wilt incidence. Hence, the management practices by the growers is being followed are fungicide and nematicide application frequently and in excess doses. Due to its negative impact on human, soil health and environment, the focus is towards the use of beneficial microorganisms i.e., biological control, is the suppression of populations of plant pathogens by living organisms (Heimpel and Mills, 2017). The findings of the present study indicated there was a greater association of M. incognita with Fusarium oxysporum causing wilt disease complex in all districts surveyed (data not shown). The causal organism F. oxysporum was isolated by standard tissue isolation method and in total ten isolates were obtained which had amplified at 389 bp in polymerase chain reaction, which were sequenced and identified as F. oxysporum based on the NCBI gene accession. These results are in line with the findings of Mishra et al. (2003) who designed the primer pairs which amplified at 315 bp, 340 bp, 389 and 314 bp for the isolates of F. sambucinum, F. oxysporum, F. equiseti and F. avenaceum respectively. Among the rhizobacterium, B. subtilis Bbv 57 was found to be most effective (50.00% inhibition) and the inhibition suggested that the bacterial antibiotics and other toxic compounds or a direct interaction might be responsible for the inhibition of the pathogen growth. The results are in line with David et al. (2014) who reported that F. oxysporum from soils of cut flower growers was suppressed strongly by native B. subtilis-Beech (45.1%) and B. subtilis-M17 (41.6%). In our study, 10 Bacillus strains namely Bbv 57, RBG 1, RBG 2, RBG 3, RBG 4, RBC 1, RBC 2, RBC 4, RBC 15 and RBT 2 showed highest per cent inhibition, phytohormones production (siderophore, hydrogen cyanide, IAA, GA3 and salicylic acid − 3.68 O.D. value; 14.05 µgml− 1; 44.40 µg ml− 1; 25.28 µg ml− 1 and 19.25 µg ml− 1 respectively) were further selected for molecular characterization. Biocontrol agents may also interact directly with the pathogen by hyperparasitism or antibiosis. Hyperparasites invade and kill mycelium, spores and resting structures of fungal pathogens and cells of bacterial pathogens (Ghorbanpour et al. 2018). Ten strains showed an amplified product of 546 bp corresponding to the region of the 16-23 s rRNA intervening sequence for Bacillus spp. The results are in confirmatory with findings of Rajendran et al. (2014), Ramyabharathi and Raguchander (2014) and Bbv 57 is positively producing Iturin (ItuD gene), Surfactin (srfA gene;sfpgene), Bacilysin (bacABgene;bacDgene), Bacillomycin D (bamDgene), Fengycin (fenBgene), Ericin (eriBgene), Mycosubtilin (mycCgene) and Subtilin (spaBgene) lipopeptides. Among lipopeptides, iturin have strong broad spectrum antifungal and hemolytic activity and the results are commensurate with Mora et al. (2011) who reported the presence of antimicrobial peptide (AMP) biosynthetic genes srfA (surfactin), bacA (bacylisin), fend (fengycin), bmyB (bacyllomicin), spas (subtilin), and ituC (iturin) in 184 isolates of Bacillus spp. Presence of comQ gene in B. subtilis Bbv 57 was identified using PCR amplification (701 bp) with gene specific primers UnicomQ1 and UnicomP1. This results are in line with Stefanic and Mulec (2009); Schneider et al. (2002) who reported the presence of comQ genes in B. subtilis. The bacterial cells secrete polymers such as polysaccharides and proteins that form a hydrated gel-like slime that holds the biofilm together. The microbial populations attached to the roots and the surrounding soil particles may form biofilm communities. Maximum biofilm formation was observed in Bbv57 (O.D. value 5.90) which was on par with EPCO 16 (O.D. value 5.43). This result is in accordance with Ramezani et al. (2014) who reported that B. pumilus (ToIr-MA) and Bacillus sp. (ToIr-10) were found to have significant ability to form maximum biofilm, considerably reducing the number of egg masses and root gall index. The biofilms also provide protection against protozoan predation and are a niche for horizontal gene transfer (Danhorn and Fuqua 2007). Culture filtrate from lip peptide producing.
B. subtilis Bbv 57 were treated with nematode eggs showed decreased egg hatching and increased juvenile mortality. The toxic metabolites or enzymes present in the culture filtrates might have caused the mortality of juveniles and inhibition in hatching of eggs. This result is in agreement with Xiao et al. (2012) who were of the opinion that cell free filtrates of B. cereus X5 on LB significantly decreased egg hatching rates and increased concentration-dependent mortality of J2 larva, suggesting that extracellular nematicidal substances existed in the filtrate. In our study, Bbv 57 showed 91.69 µg µl− 1 of surfactin with the retention time of 2.304 min and 0.453 µg µl− 1 of Iturin with the retention time of 8.739 min at 205 nm. The result is in analogous to Yuan et al. (2011) who quantified Iturin A antifungal lipopeptide by high performance liquid chromatography coupled with aqueous two-phase extraction. The crude antibiotic treated egg shell was shriveled, female nematode wall was shrunken and misshaped in ESEM study. The crude antibiotics were analyzed through GC/MS to detect the antimicrobial compounds produced by Bbv 57. The results revealed that the compounds includes Butanedioic acid, Hexadecanoic acid ethyl ester belonging to fatty acid has antibacterial and antifungal activity. Similarly Pizana et al. (2010) reported that the compound hexatriacontane has antifungal property and suppressed the growth of F. oxysporum f.sp. gladioli. Analysis of the compounds through GC/MS analysis and their properties, clearly indicated that the strain Bbv 57 possess both antifungal and nematicidal properties, which could suppress both Fusarium and nematode in gerbera plant.
To conclude, the bioagent B. subtilis Bbv 57 with diverse antimicrobial peptide, quorum sensing genes, volatile antibiotics was effective in reducing the wilt - root knot nematode complex in gerbera grown under protected cultivation.
References
Adam, M., Heuer, H., & Hallmann, J. (2014). Bacterial Antagonists of fungal pathogens also control root-knot nematodes by induced systemic resistance of tomato plants. PLoS One, 9(2), e90402. https://doi.org/10.1371/journal.pone.0090402
Bonmatin, J. M., Laprevote, O., & Peypoux, F. (2003). Diversity among microbial cyclic lipopeptides: iturins and surfactins. Activity structure relationships to design new bioactive agents. ombinatorial Chemistry & High Throughput Screening, 6, 541–556.
Borrow, A., Brain, P. W., Chester, U. E., Curtis, P. J., Hemming, H. G., Jeffereys, E. C., et al. (1955). Gibberellic acids a metabolic product of the fungus Gibberella fujikuroisome observations on its production and isolation.J. Sci. Food Agric., 6, 340–348.
Cadena, M. B., Burelle, N. K., Kathy, S. L., Santen, E. V., & Kloepper, J. W. (2008). Suppressiveness of root-knot nematodes mediated by rhizobacteria. Biological Control, 47, 55–59.
Cano, R. J., Borucki, M. K., Higby-Schweitzer, M., Poinar, H. N., Poinar, G. O., & Pollard, K. J. (1994). Bacillus DNA in fossil bees: an ancient symbiosis. Appl. Environ. Microbiol., 60(6), 2164–2167.
Cazorla, F. M., Li, X. Z., & Zhang, L. H. (2007). Isolation and characterization of antagonistic Bacillus subtilisstrains from the avocado rhizoplane displaying biocontrol activity. Journal of Applied Microbiology, 103(5), 1950–1959.
Chandramohan, D., & Mahadevan, A. (1968). Indole acetic acid metabolism in soils. Curr. Sci., 37, 112–113.
Danhorn, T., & Fuqua, C. (2007). Biofilm formation by plant associated bacteria. Annual Review of Microbiology, 61, 401–422.
Dennis, C., & Webster, J. (1971). Antagonistic properties of species groups of Trichoderma II. Production of volatile antibiotics. Trans. Br. Mycol. Soc., 57, 41–48.
Eisenback, J. D., Hirschmann, H., Sasser, T. N., & Triantaphyllou, A. C. (1981). A guide to the four most common species of root-knot nematodes (Meloidogynespp.) with a Pictorial Key. A co-operative publication of the Department of Plant Pathology and Genetics (p. 48). Releigh: North Carolina State University and United States Agency for International Development.
Gan Chan, K., Tiew, S.-Z., & Ching-Ching, N. (2007). Rapid isolation method of soil bacilli and screening of their quorum quenching activity. Asia-Pacific Journal of Molecular Biology and Biotechnology, 15(3), 153–156.
Ghorbanpour, M., Omidvari, M., Abbaszadeh-Dahaji, P., Omidvar, R., & Kariman, K. (2018). Mechanisms underlying the protective effects of beneficial fungi against plant diseases. Biological Control, 117, 147–157. https://doi.org/10.1016/j.biocontrol.2017.11.006
Gomez, K. A., & Gomez, A. A. 1984. Statistical Procedure for Agricultural Research.John Wiley and Sons, New York, United States.
Gordon, S. A., & Paleg, L. G. (1957). Quantitative measurement of IAA. Physiol. plantarum, 10, 347–348.
Harvey, J., Keenan, K. P., & Gilmour, A. (2007). Assessing biofilm formation by Listeria monocytogenes strains. Food Microbiology, 24(4), 380–392.
Haseeb, A., Ahmed, V., & Shukla, P. K. 2005.Comparative efficacy of pesticides, biocontrol agents and botanicals against Meloidogyne incognita - Fusariumoxysporumcomplex on Vignamungo. Ann. Plant Protect. Sci., 13(2): 434–437.
Heimpel, G. E., & Mills, N. (2017). Biological Control - Ecology and Applications. Cambridge: Cambridge University Press.
Huang, M. L., van Peer, R., Woestenborghs, R., de Coster, R., Heykants, J., & Jansen, A. A. (1993). Pharmacokinetics of the novel antipsychotic agent risperidone and the prolactin response in healthy subjects. Clin. Pharmacol. Ther., 54(3), 257–268.
Kavitha, P. G., Jonathan, E. I., & Nakkeeran, S. (2012). Effects of crude antibiotic of Bacillus subtilison hatching of eggs and mortality of juveniles of Meloidogyne incognita. Nematologia Mediterranea, 40, 203–206.
Kishore, C. (2007). Studies on diagnosis and management of fungal wilt diseases of carnation and gerbera under protected cultivation. M.Sc. Thesis. University of Agricultural Sciences, Dharwad, India.
Komada, H. (1975). Development of a selective medium for quantitative isolation of F. oxysporum from natural soils. Rev. Plant Protect. Res., 8, 114–125.
Kumar. (2008). Studies on root-knot and wilt complex in Coleus forskohlii(wild.) briq.caused by Meloidogyne incognita (Kofoid and White) Chitwood and Fusariumchlamydosporum(Frag. and Cif.) Booth.M.Sc. Thesis, University of Agricultural Sciences, Dharwad, India.
Kumar, R., Ahmed, N., Lal, S., & Mahendiran, G. (2014). Evaluation of gerbera genotypes for cut flower production under different growing conditions of Kashmir. Indian Journal of Horticulture, 71(1), 138–141.
Laemmli, U. K. (1970). Cleavage of Structural proteins during the assembly of the Head of bacteriophageT4. Nature, 227, 680–685.
Lhoste, A. (2002). Cut gerbera: varietal experiments in Mediterranean climate: PHM -. Revue - Horticole, 435, 24–27.
Lucy, M., Reed, E., & Glick, B. R. (2004). Applications of free living plant growth-promoting rhizobacteria. Antonie van Leeuwenhoek, 86(1), 1–25.
Mahadevan, A., & Sridhar, R. (1982). Methods in physiological plant pathology (p. 316). Madras: Siva Kami Publication.
Meyer, J. M., & Abdallah, M. A. (1978). The fluorescent pigment of Pseudomonas fluorescens: biosynthesis, purification and physicochemical properties. J. Gen. Microbiol., 107, 319–328.
Meyer, J. M., Azelvandre, P., & Georges, C. (1992). Iron metabolism in Pseudomonas: salicylic acid, a siderophore of Pseudomonas fluorescensCHA0. BioFactors, 4, 23–27.
Miller, R. L., & Higgins, V. J. (1970). Association of cyanide with infection of birdsfoot trefoil by Stemphylium loti. Phytopathology, 60, 104–110.
Mora, I., Jordi, C., & Emilio, M. (2011). Antimicrobial peptide genes in Bacillus strains from plant environments. Int. Microbiol., 14, 213–223.
Mukherjee, A. K., & Das, K. (2005). Correlation between diverse cyclic lipopeptides production and regulation of growth and substrate utilization by Bacillus subtilisstrains in a particular habitat. FEMS Microbiology Ecology, 54, 479–489.
Nagesh, M., & Parvatha Reddy, P. (1996). Management of Meloidogyne incognita on carnation and gerbera in commercial polyhouses. In Crop Productivity and Sustainability – Shaping the Future (2, p. 249). New Delhi: International Crop Science Congress, NAAS. nd .
Niknam, G. R., & Dhawan, S. C. (2002). Systemic resistance induced by Pseudomonas fluorescensisolate Pf 1 in tomato against Rotylenchulusreniformis. International Journal of Nematology, 12(2), 203–208.
Omar, A., Almaghrabi, Samia I.M.., & Tamer, S. A. (2013). Influence of inoculation with plant growth promoting rhizobacteria (PGPR) on tomato plant growth and nematode reproduction under greenhouse conditions. Saudi J. Biol. sci., 20(1), 57–61.
Pizana, C.G., Barrera, L.L. & Ma. (2010). Evaluation of the fungicidal activity of leaves powder and extracts of fifteen Mexican plants against Fusariumoxysporumf.sp. gladioli(Massey) synder and Hansen. Plant Pathol. J., 9(3), 103–111.
Polonca, S., & Mulec, I. M. (2009). Social Interactions and Distribution of Bacillus subtilis Pherotypes at Microscale. J. Bacterol., 191(6), 1756–1764.
Ramyabharathi, SA. & Raguchander, T. (2014). Efficacy of secondary metabolites produced by Bacillus subtilis EPCO16 against tomato wilt pathogen Fusarium oxysporum f.sp lycopersici. Journal of Mycology and Plant Patholog, 44(2), 148–153.
Rajendran, L., Raja, P., Jegadeeswari, V., Shanthi, V. P., & Selvaraj, N. (2014). Pseudomonas fluorescens and Trichoderma viride as enriched bioconsortium for the management of Fusarium wilt in Carnation and Gerbera under protected cultivation. Indian Phytopathology, 67(1), 77–81.
Rajendran, L., Ramjegathesh, R., Shanthiyaa, V., Raguchander, T., Karthikeyan, G., & Samiyappan, R. (2012). Biocontrol potential and mode of action of strains EPC5 and EPC8 of endophytic bacterium Bacillus subtilis. Indian Phytopathology, 65(2), 122–127.
Ramarathnam, R. (2007). Phyllosphere bacterial biological contol of Leptosphaeriamaculans, the blackleg pathogen of canola (Brassica napusL.): screening for potential antibiotic producers, investigation of the mechanism of control, biochemical detection of the antifungal compounds and establishment of the role of antibiosis. Ph.D. Thesis, University of Manitoba, Winnipeg, Manitoba.
Ramezani, M. M., Mahdikhani Moghaddam, E., Baghaee Ravari, S., & Rouhani, H. (2014). The nematicidal potential of local Bacillus species against the root-knot nematode infecting greenhouse tomatoes. Biocontrol Science and Technology, 24(3), 279–290.
Ramzan Memoona, B., Tabassum, I. A., Nasir, AnwarKhan, Tariq, M., Awan, M. F., Shahid, Naila, et al. (2016). Identification and application of biocontrol agents against Cotton leaf curl virus disease in Gossypium hirsutum under greenhouse conditions. Biotechnology & Biotechnological Equipment, 30(3), 469–478. https://doi.org/10.1080/13102818.2016.1148634
Schneider, K. B., Tanya, M. P., & Alan, D. (2002). Pheromone in Bacillus subtilis genes required for production of ComX characterization. J. Bacteriol., 184(2), 410–419.
Schwyn, B., & Neilands, J. B. (1987). Universal chemical assay for the detection and determination of siderophore. Anal. Biochem., 169, 47–56.
Singh, V. K., & Kumar, A. (2001). PCR Primer design. Mol. Biol., 2, 27–32.
Sokol, P. A., Ohman, D. E., & Iglewski, B. H. (1979). A more sensitive plate assay for detection of protease production by Pseudomonas aeruginosa. Journal of Clinical Microbiology, 9, 538–540.
Stein, T. (2005). Bacillus subtilisantibiotics: structures, syntheses and specific functions. Molecular Microbiology, 56, 845–857.
Stefanic, P., & Mulec, I. M. (2009). Bacillus subtilis Pherotypes at Microscale. J. Bacteriol., 191(6), 1756.
Sudhagar, S. (2013). Production and marketing of cut flower (Rose and Gerbera) in HosurTaluk. International Journal of Business and Management Invention, 2(5), 15–25.
Taylor, A. L., Dropkin, V. H. and Martin.G.C (1955). Perineal patterns of root-knot nematodes. Phytopathology, 45, 26–34.
Taylor, A. L., & Sasser, J. N. (1978). Biology, Identification and control of root- knot nematodes (Meloidogynespp.) (p.111). Graphics: North Carolina State Univ.
Xiao, T. J., Shi-Yong, T., Qi-Rong, S., & Wei, R. (2012). Bacillus cereus X5 suppresses root-knot nematode of tomato by colonizing in roots and soil. African Journal of Microbiology Research, 6(10), 2321–2327.
Yuan, J., Raza, W., Huang, Q., & Qirong, S. (2011). Quantification of the antifungal lipopeptideiturin A by high performance liquid chromatography coupled with aqueous two-phase extraction. J. Chromatogr. B, 879, 2746–2750.
Acknowledgements
The finding is an outcome of project funded by Department of Biotechnology, Ministry of Science and Technology, New Delhi, India.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics declarations
All authors have participated in the research and manuscript preparation and all have reviewed and approved the manuscript. The manuscript has not been published before and has only been submitted to EJPP for evaluation.
Conflict of interest
Authors declare no conflict of interest.
Human and animal sudies
This article does not contain any studies with human participants or animals performed by any of the authors.
Rights and permissions
About this article
Cite this article
Ramyabharathi, S., Meena, K.S., Rajendran, L. et al. Potential of a rhizobacterium Bacillus subtilis (Bbv 57) on Fusarium oxysporum f. sp. gerberae and Meloidogyne incognita infecting Gerbera grown in protected cultivation. Eur J Plant Pathol 158, 615–632 (2020). https://doi.org/10.1007/s10658-020-02087-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-020-02087-6