Abstract
Background
Biocontrol is regarded as a viable alternate technique for managing sugarcane wilt disease caused by Fusarium sacchari. Many fungal antagonists against F. sacchari, have been reported, but the potential of bacterial antagonists was explored to a limited extent, so the present study evaluated the antagonistic potential of rhizoplane Bacillus species and their mode of action.
Results
A total of twenty Bacillus isolates from the rhizoplane of commercially grown sugarcane varieties were isolated. The potential isolate SRB2 had shown inhibition of 52.30, 33.33, & 44.44% and SRB20 of 35.00, 33.15, & 36.85% in direct, indirect, and remote confrontation respectively against F. sacchari. The effective strains were identified as Bacillus inaquosorum strain SRB2 and B. vallismortis strain SRB20, by PCR amplification of 16S-23S intergenic region. The biochemical studies on various direct and indirect biocontrol mechanisms revealed the production of IAA, Protease, Cellulase, Siderophores, and P solubilization. The molecular analysis revealed the presence of antimicrobial peptides biosynthetic genes like fenD (Fengycin), bmyB (Bacyllomicin) ituC (Iturin) and spaS (Subtilin) which provided a competitive edge to these isolates compared to other Bacillus strains. Under greenhouse experiments, the sett bacterization with SRB2, significantly (P < 0.001) reduced the seedling mortality by > 70% followed by SRB20 in F. sacchari inoculated pots.
Conclusion
The study revealed that the isolates B. inaquosorum SRB2 and B. vallismortis SRB20 can be used as potential bioagents against sugarcane Fusarium wilt.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sugarcane (Saccharum officinarum L.) is an ancient valuable cash crop that is the raw material for sugar production. In recent years, it has gained more importance globally due to its associated benefits of biofuel and biogas production [15]. However, there has been a considerable decline in the sugarcane yield due to fungal infections, which has turned many economically significant commercial sugarcane varieties obsolete. Wilt is one of the important fungal diseases caused by Fusarium sacchari (Butler) W. Gams, which causes substantial crop losses. The elimination of many commercial sugarcane varieties was a consequence of wilt epidemics in the last century [1, 20]. Sett germination, productivity, and sugar recovery are negatively impacted by the disease.
Present management strategies involve the juxtaposition of cultural and chemical methods along with resistant varieties. Since F. sacchari is sett borne, the fungicide application is limited to sett treatment. Even fungicidal treatment is insufficient to ward off the infection completely because of its systemic nature and the impervious sett or cane rind makes it hard for the entry of fungicide into the deeper tissues. Moreover, wilt pathogen is also soil-borne but soil treatment with fungicides is not economical and also has several detrimental effects [53]. Among the viable alternative methods, indigenous rhizobacteria play a vital role in growth promotion and disease management in different crop plants [46].
The ability of the rhizobacterial bioagents to colonize the roots and produce hydrolytic enzymes, siderophores, and antibiotics aids the plant to combat the pathogens directly or through inducing systemic resistance [53]. Bacillus species are the most extensively studied among the various genera reported to possess plant growth promotion and antagonistic activity [24].
Any biocontrol investment would be appropriately compensated by increased crop yield as sugarcane is a commercial commodity with significant economic value. The sugar industry manages the crop effectively through large-scale cultivation across continuous areas that make it simple to deliver biocontrol formulations. Ratooning and monocropping make the biocontrol agents self-sustainable without interruption. Additionally, the by-product of the sugar industry known as "pressmud" makes a perfect carrier in the mass production of biocontrol agents [30]. Hence there is a specific advantage in adopting biocontrol measures for the control of sugarcane diseases. Therefore, the present investigation was undertaken to isolate and screen antagonistic Bacillus species from rhizoplane of various sugarcane genotypes against F. sacchari under in-vitro and in-vivo conditions and to reveal the biocontrol mechanisms of potential candidates through biochemical and molecular studies.
Methods
Isolation of Bacillus species from rhizoplane
The rhizoplane samples were collected by uprooting the sugarcane plants along with root portion from various sugarcane genotypes grown in Anakapalle district of Andhra Pradesh, India (Table 1), and 1 g of the sample was briefly vortexed in 10 ml sterilized distilled water. The bacterial population was brought down to 10–6 concentration by serial dilution, and 1 ml of the final dilution was inoculated on the Bacillus Agar media containing Bacillus selective supplement (8 ml per 1000 ml of media) and incubated at 28 ℃. Distinct colony types were purified and maintained on nutrient agar. The gram reaction of isolated bacteria was evaluated using 3% KOH, and the non-stickiness of the KOH-treated colony indicated the Gram-positive reaction of bacteria.
Cultural characterization
Twenty bacterial colonies were characterized by observing culture plates under a compound microscope (40X). Colony characters like shape, margin, texture, opacity, and pigmentation were studied and morphologically characterized as described by Dasgupta [6].
Phytopathogen
A single virulent isolate of F. sacchari was obtained from the Plant Pathology Division, Regional Agricultural Research Station, Anakapalle, Andhra Pradesh, India.
Screening of the isolates for antagonistic potential
Twenty isolates of bacteria obtained from sugarcane rhizoplane were evaluated for their antagonism against F. sacchari at three different levels, i.e., direct, indirect, and remote confrontation.
Direct confrontation
Dual culture assay was followed using the protocol given by Dennis and Webster [8]. A 5 mm culture disc of F. sacchari was centrally placed on a PDA containing Petri plates. A loopful of 24-h-old culture of test bacterial antagonist was streak inoculated on either side of the pathogen disc at a distance of 2 cm and incubated at 28 °C for 9 days. The pathogen-inoculated PDA plate without bacterial streak, served as control. Each treatment was replicated thrice in a completely randomized design (CRD). The radial growth of the pathogen was recorded at an interval of 3 days, and per cent inhibition was calculated using the formula given by Vincent [47].
- Xi:
-
colony diameter (cm) at the ith observation,
- ti:
-
time (days after inoculation) at the ith observation and
- n:
-
total number of observations.
Indirect confrontation
The selected three bacterial isolates inoculated in 100 ml of Nutrient broth, were incubated on a rotary shaker at 100 rpm for 48 h. The cultures were centrifuged at 10,000 rpm for 5 min. The supernatant was collected and filtered through a 0.25 \(\mathrm{\mu m}\) membrane filter under sterile conditions, and the obtained filtrates were amended to PDA at various concentrations (10, 20, and 30%). A 5 mm mycelial disc of F. sacchari was placed centrally on each plate. Three replications were maintained for each treatment. Non-amended PDA was used as the control. The per cent inhibition of pathogen was calculated using the above-mentioned formula.
Remote confrontation
The selected three isolates were evaluated for their antifungal property by the sealed plate method. The compartment inoculated with a 5 mm mycelial disc of F. sacchari was placed in an inverted position over the compartment streaked with potential antagonistic bacteria. Control plates were maintained with fungal inoculation alone. The tightly sealed plates were incubated at 28 ºC in an incubator and were observed for pathogen inhibition. The per cent inhibition was calculated using the above-mentioned formula.
Molecular characterization of potential antagonistic isolates
Genomic DNA was extracted from all the twenty isolates grown in Nutrient broth for 24 h using a modified CTAB method [39]. The integrity and concentration of purified DNA was determined using a Nanodrop Spectrophotometer.
Initially, the extracted DNA of all the twenty isolates was subjected to PCR amplification using Bacillus genus-specific primers BCF1 (5'- CGG GAG GCA GCA GTA GGG AAT -3') & BCF2 (5'- CTC CCC AGG CGG AGT GCT TT -3') for their identification. The species-level identification of two potential isolates was done using 16S-23S intergenic region primers i.e. 16F945 (5'- GGG CCC GCA CAA GCG TGG -3') and 23R458 (5'- CTT TCC CTC ACG GTA C -3'). The reaction mixture (25 μl) contained 2.5 μl of 10X PCR buffer (Thermo Scientific), 2 μl of 25 mM MgCl2, 1 μl of 0.01 mM dNTPs, 0.3 μl of 1U Taq polymerase (Thermo Scientific), 1 μl of 0.001 M forward and reverse primer each, 15.2 μl of molecular grade water and 2 μl of sample DNA. Steps in PCR include initial denaturation at 95 ℃ for 5 min and 35 cycles of denaturation at 94 ℃ for 1 min, annealing at 53 ℃ for 1 min, primer extension at 72 ℃ for 2 min, and final extension at 72 ℃ for 10 min. Samples (5 μl) of the PCR products were separated on a 1.2% agarose gel in 1X TBE buffer (90 mM Tris borate, 2 mM EDTA [pH 8.0]) in a horizontal electrophoresis unit (SCIE-PLAS) at 60 V and 60 mA for 1 h and the PCR products were visualized in gel documentation unit.
Amplified PCR products of 16S-23S intergenic region of the two potential isolates were sequenced by Bioserve Biotechnologies (India) Pvt. Ltd., Hyderabad. The obtained gene sequences were analysed using BLASTn in NCBI. The sequences were aligned in Clustal W and a phylogenetic tree was constructed using the maximum likelihood method with 1000 bootstrap values in MEGA 7 software.
Biochemical assays to screen Bacillus isolates for their biocontrol potential activity
All the twenty bacterial isolates were tested for their plant growth promoting and disease supressing traits through the following qualitative biochemical tests.
Phosphate Solubilisation
The phosphate solubilisation ability of bacterial isolates was detected by inoculating them on Pikovaskaya’s agar plates [33]. The inoculated plates were observed for clear zone around the colonies after incubation for three days at 28 °C.
IAA Production
A modified agar plate assay was used to qualitatively estimate IAA production by Bacillus isolates [41]. Luria Bertani Agar plate amened with 100 µg ml−1 of tryptophan was used. A cavity of 5 mm diameter and 0.2 cm depth was made using a sterile cork borer. Each cavity was filled with 50 µl of overnight-grown culture and incubated at 30 oC for 24 h. After the incubation, two drops of Salkowski reagent was added. The development of pink colour after the addition of Salkowski reagent was considered positive for IAA production.
Protease Activity
All bacterial isolates were screened for protease activity by inoculating on skim milk agar plates. The inoculated plates were observed for clear zone around the colonies after incubation for three days at 28 °C [43].
Cellulase Production
The Czapek-mineral salt Agar medium supplemented with Carboxy methyl cellulose was used for qualitative estimation of cellulase production by bacterial bioagents [27]. A 5 mm agar disc of one-day-old bacterial culture was placed on the media and incubated at 28 °C in darkness for 4 days. The plates were washed with aqueous Congo red (2% w/v) solution for 15 min. Further, it was washed with NaCl (1 M) for 1.5 min. The yellow opaque area around the colonies indicates cellulase production.
Siderophore Production
Modified CAS medium was used for qualitative estimation of siderophore production by bacterial isolates [40]. The plates were inoculated with bacterial discs and were incubated in the dark at 28 °C for 5 days. The orange zones around the wells were considered siderophore-positive.
Molecular analysis to screen Bacillus isolates for their biocontrol potential activity
The gene-specific primers (Table 2) were used to decipher the distribution of antimicrobial peptide genes fenD (Fengycin), bmyB (Bacyllomicin) ituC (Iturin) and spaS (Subtilin) in the isolates [31]. Steps in PCR include initial denaturation at 94 ℃ for 5 min and 40 cycles of denaturation at 94 ℃ for 1 min, annealing temperature (Table 2) for 1 min, primer extension at 72 ℃ for 1 min, and final extension at 72 ℃ for 5 min. Samples (5 μl) of the PCR products were separated on a 1.2% agarose gel in 1X TBE buffer in a horizontal electrophoresis unit at 100 V and 60 mA for 90 min. The PCR products were visualized in a gel documentation unit for desired bands that confirm the antibiotic production by the isolates.
Greenhouse experiment
Single node setts of wilt susceptible cultivar, 2009A 107 were first soaked in a conidial suspension of pathogen (F. sacchari) overnight and then treated with a cell suspension of selected three bacterial bioagents (109 CFU ml−1) for 30 min. Five single-budded setts were sown in each pot. For healthy check, setts were soaked in autoclaved distilled water and for inoculated check, setts were treated with only pathogen (F. sacchari). The experiment was performed in a completely randomized block design with five replications per treatment. The setts were watered regularly. Data on germination percentage, plant stand, and mortality were recorded till 60 DAS.
Statistical analysis
Data analysis was performed based on standard procedures followed by Gomez and Gomez [12] and also analysed in R statistical programming environment. The following R packages were used in data analysis: datasets, agricolae, tidyverse (a collection of R packages designed for common data science tasks such as data cleaning and visualization), and readx1 (data import into R from Excel). The figures were plotted in Excel sheet and Prism software.
Results
Isolation of Bacillus Species
A total of twenty bacterial isolates were obtained on Bacillus specific media, and all the isolates were classified as Gram-positive bacteria as they remain unaffected in the 3% KOH test. The isolated Bacillus strains were preserved and available in the Division of Plant Pathology, Regional Agricultural Research Station, Anakapalle, India.
Cultural Characterization of Rhizoplane Bacteria
Sixteen out of 20 isolates were round in shape and the other isolates SRB1, SRB7, SRB10, and SRB20 were irregular in shape. Many isolates had flat colonies (13) with even margins (14) with some exceptions, viz., SRB5, SRB6, SRB8, SRB9, SRB15, SRB18, and SRB19 had raised colonies and SRB1, SRB4, SRB11, SRB15, SRB16, and SRB20 had wavy margin. There was no much variation in the texture, opacity, and pigmentation of the colonies, most of the isolates were opaque with smooth texture and creamy to milky whitish colonies except SRB2 and SRB20 which were rough textured and SRB2 and SRB11 were transparent in nature.
Direct Confrontation
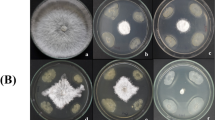
In dual culture assay, the direct confront between F. sacchari and the isolates SRB2, SRB4, and SRB20 had shown inhibition of 52.30, 17.22 and 35.00%, respectively on the ninth DAI (Table 1) and were considered antagonistic to F. sacchari. A clear zone of inhibition was seen in SRB2 (2.5 mm) (Fig. 1) and minimum AUMGC was observed with isolates SRB2 (21.34) and SRB20 (27.12) indicating their better antagonistic activity in comparison to other isolates and reduction of AUMGC with respect to control by 48.73 and 34.85%, respectively (Graph 1). The three potential isolates (SRB2, SRB4, and SRB20) were selected for further tests.
Antagonistic efficacy of Bacillus isolates against F. sacchari at Direct (a, b, c & d) Indirect (e, f, g & h) 30% concentration and Remote (i, j, k & l) confrontation. Inhibition of mycelial growth was observed with SRB2 (d, h & l), SRB20 (c, g & k) and SRB4 (b, f & j) when compared to control (a, e & i) inoculated with only F. sacchari
Indirect Confrontation
The three potential isolates were evaluated for their antifungal nature in the absence of active colonies. Here, instead of bacterial colonies, the culture filtrate of the selected isolates at three different concentrations (10, 20, and 30%) was tested for their antifungal property (Table 3). The increase in concentration of culture filtrate from 10 to 30% resulted in an increased inhibition of the pathogen from 11.94 to 26.54% (Graph 2). The pathogen was highly inhibited by the culture filtrates obtained from SRB2 (33.33%) and SRB20 (33.15%) at 30% concentration (Fig. 1). The lowest inhibition was seen in SRB4 (10.93%) at 10% concentration.
Remote confrontation
The paired plate assay of three potential isolates with F. sacchari showed varied levels of antagonism (Fig. 1). F. sacchari was significantly (P < 0.001) inhibited by 26.85 (SRB4), 36.85 (SRB20) and 44.44% (SRB2) at ninth DAI (Graph 3) demonstrating their antagonistic nature.
Based on the three in vitro confrontation studies, SRB2 and SRB20, were found to be the most potential antagonistic rhizobacteria against F. sacchari.
Molecular characterization of potential antagonistic Bacillus species
Initially, all the isolates were confirmed as Bacillus using genus-specific primers (Fig. 2). Partial 16S-23S rRNA gene sequencing of the most potential isolates SRB2 and SRB20 (Fig. 3) and its analysis using BLASTn in NCBI database showed that SRB2 had 99.62% similarity with B. inaquosorum (CP096592), and SRB20 had 99.70% similarity with B. vallismortis (CP026362). The sequences were submitted to the NCBI database and accession numbers for SRB2 (ON964473) and SRB20 (ON954849) were obtained. Further, the phylogenetic tree construction confirmed that SRB2 and SRB20 were clustered with B. inaquosorum and B. vallismortis respectively (Fig. 4).
Phylogenetic tree showing position of the biocontrol agents (▲) used in this study. The 16S-23S rDNA sequences were aligned with the ClustalW and the tree was constructed with the maximum likelihood method based on the Tamura-Nei model integrated in the MEGA7 software. The GenBank accession numbers of the DNA sequences are shown in parentheses. Numbers at branches represent bootstrap values > 70% from 1000 replicates
Biochemical analysis for potential biocontrol mechanisms
P Solubilization
Seven isolates SRB1, SRB5, SRB6, SRB8, SRB13, SRB14 and SRB16 showed phosphate solubilization by producing a halo zone around the colonies on Pikovskaya’s agar plates after 3 DAI (Table 4; Fig. 5).
Biochemical tests for potential biocontrol mechanisms. a – IAA production (development of pink colour on tryptophan amended media). b—Phosphate solubilization (halo zone around the colony on Pikovskaya’s agar media). c—Protease activity (halo zone around the colonies on Skim milk agar). d—Cellulase activity (the yellow-opaque area around the colonies on Czapek-mineral salt Agar medium). e—Siderophore production (orange-coloured zone on CAS media)
IAA Production
Only four isolates i.e., SRB9, SRB11, SRB18 and SRB19, were detected to produce IAA as indicated by the development of pink colour after the addition of Salkowski reagent at 2 DAI (Table 4; Fig. 5).
Protease activity
Nearly 90% (18) of the isolates tested positive for protease activity as they produced colorless halo zones around the bacterial colonies on Skim milk Agar plates. SRB4, SRB7, SRB12 and SRB20 had shown higher protease activity (> 50 mm dia.), SRB2, SRB6, SRB9, SRB10, SRB11, SRB13, SRB14, SRB15 and SRB18 had medium (30–50 mm dia.) protease activity and low protease activity (< 30 mm dia.) was shown by SRB1, SRB5, SRB8, SRB17 and SRB19 (Table 4; Fig. 5).
Cellulase activity
Five among all isolates i.e., SRB5 (31 mm), SRB8 (10 mm), SRB11 (30.5 mm), SRB19 (13 mm) and SRB20 (45 mm) were qualitatively detected to produce cellulase by the formation of the yellow-opaque area around the colonies when the plates were washed with aqueous Congo red (2% w/v) solution (Table 4; Fig. 5).
Siderophore Production
Among 20 isolates, six isolates i.e., SRB4, SRB7, SRB12, SRB16, SRB17, and SRB20 formed orange coloured zone due to chelation of Fe3+ from CAS medium thereby confirming siderophores production (Table 4; Fig. 5).
Molecular analysis for potential biocontrol mechanisms
Four gene-specific primers were used to detect Antimicrobial Peptide (AMP) genes viz. fenD, bmyB, ituC and spaS that are involved in the production of fengycin, bacillomycin, iturin, and subtilin respectively (Table 4). The desired bands confirmed the antibiotic production by the Bacillus isolates (Figs. 6, 7, 8 & 9 respectively). Among the tested isolates, 60% of the isolates had fenD gene (270 bp), 50% of the isolates had bmyB gene (370 bp), 30% of the isolates had spaS gene (375 bp) and 30% of the isolates had ituC gene (423 bp).
The study on the frequency of AMP genes revealed that all four AMP genes were present in only 10% of the tested isolates (SRB2 and SRB20), 20% of the isolates had three AMP genes (SRB5, SRB8, SRB9, and SRB11), 20% of the isolates had two AMP genes (SRB12, SRB13, SRB14, and SRB18), 30% of the isolates had a single AMP gene (SRB1, SRB3, SRB4, SRB6, SRB10, and SRB15) and none of the AMP genes were detected in remaining 20% of the isolates (SRB7, SRB16, SRB17, and SRB19). The unique combination of different antibiotic genes provided a competitive edge to these isolates over other Bacillus strains for survival and biocontrol efficacy in different ecological niches.
Greenhouse experiment
Higher germination of F. sacchari inoculated setts treated with bioagents was observed than in inoculated check ranging from 73.33% (SRB4) to 80.00% (SRB2). The highest plant stand was reported in SRB2-treated setts (73.33% @ 30DAS; 66.67% @ 60DAS) and was found significantly (P < 0.001) superior to other bioagents (Table 5; Graph 4). The lowest pre-emergence mortality (PEM) and post-emergence seedling mortality (PESM) were observed in the SRB2-treated setts (20.00% and 15.00% respectively) in comparison to inoculated check (Fig. 10). Though SRB4 showed on par PEM with SRB2 and SRB20, it showed much higher PESM proving that SRB2 and SRB20 performed very well under greenhouse conditions (Table 6; Graph 5).
Discussion
The diverse rhizoplane microbiome serves as an infinite reservoir for biological control that can be a game changer in a zero-chemical plant disease management strategy [11]. Among the diverse soil microbiota, Bacillus species are the best fit for this purpose because they are in perpetual competition with other pathogenic microbes to colonize the plant roots [42]. Innate traits like multilayered cell wall, stress-resistant endospore production, and secretion of antibiotic peptides, extracellular enzymes and peptide signal molecules promote their survival under dynamic environmental conditions [24]. Hence, this study attempted to explore the potentiality of Bacillus species isolated from the rhizoplane of different sugarcane cultivars at different growth stages for biocontrol of Fusarium wilt of sugarcane.
A total of 20 Bacillus isolates were screened for their inhibitory effect on the growth of F. sacchari at three different levels (Direct, Indirect and Remote confrontation). Among them the three isolates SRB2, SRB4 and SRB20 were found antagonistic in dual culture assay that brings pathogen and bioagent in direct confront [9, 35, 50, 52]. In the next level, the culture filtrate (Indirect confrontation) from SRB2 and SRB20 significantly inhibited the pathogen [14]. This antagonistic nature of culture filtrate is attributed to the non-volatile compounds and secondary metabolites that inhibited the pathogen by antibiosis. Further, these two isolates were also found antagonistic in remote confrontation as revealed by paired plate assay could be due to the antifungal nature of the various volatile compounds released by the isolates [13].
The variable regions of the 16S rRNA gene are generally used to identify microorganisms. However, closely related microorganisms have minimal sequence variation in their 16S rRNA genes [3]. So, the isolates were initially identified as Bacillus by Bacillus-specific probes BCF1 and BCF2 (16S rRNA intervening sequence) [10, 34] but the polymorphisms were sought in the 16S-23S intergenic spacer region of the rRNA genes using the primer set 16F945 and 23R458 that differentiated potential bacteria upto species level as B. inaquosorum SRB2 and B. vallismortis SRB20 [19, 23, 26, 51].
Plant growth promotion is an additional benefit offered by the bioagents. The higher vigour of plants equips them against pathogen invasion. The plant growth-promoting activity of different root-colonizing bacteria could be attributed to biofertilization [24]. Rhizobacteria stimulates plant growth directly through nutrient solubilization thereby enhancing the availability of inaccessible nutrients to the plants via organic acids production, viz., citric acid, oxalic acid, gluconic acid, etc. [38]. Likewise, Thongponkaew et al. [44] reported that isolates from sugarcane rhizospheric soils were capable of solubilizing tricalcium phosphate, and also reported the increased sugarcane yield with the application of phosphate solubilizing Bacillus species [5].
Phytostimulation through the production of plant growth regulators by root colonizing Bacillus species is a notable phenomenon [16, 22]. Among the PGRs produced, IAA plays a multifaceted role in plant growth and usage of bacterial IAA by plants leads to cell division, elongation, and differentiation [28] and is a vital component in shaping the architecture of plant roots such as initiation of lateral roots, differentiation of vascular tissues, and root gravitropism [24].
The biocontrol activity of rhizoplane isolates B. inaquosorum SRB2 and B. vallismortis SRB20 against Fusarium could be due to several mechanisms viz., protease, siderophore and antimicrobial peptides (fengycin, bacillomycin, iturin and subtilin) production by SRB2 and protease, cellulase, siderophore and antimicrobial peptides (fengycin, bacillomycin, iturin and subtilin) production by SRB20 as confirmed by in vitro qualitative tests. Secretion of extracellular lytic enzymes (protease, cellulase, and other hydrolytic enzymes) will degrade the fungal mycelium through lysis of the cell wall and thereby retards the pathogen growth [2, 29]. Siderophores produced by isolates have a high affinity for ferric ions thereby forming ferric siderophore complexes. This makes iron unavailable to pathogens that require it for their pathogenesis [21, 32].
Bacillus species produce different secondary metabolites, including various antibiotic compounds, which have been scrutinized mainly by genetic techniques for biocontrol activity. Identifying antimicrobial genes from the rhizobacterial isolates enabled us to understand the molecular mechanism involved in biological control. Antibiotics produced by Bacillus species are deleterious to other microorganism's growth or metabolic activities. Antifungal compounds include Fengycin, a plipastatin-like lipopeptide that is helpful in the biological control of plant pathogens, degrading their cell structure and permeability [4, 7]. They are also reported to induce systemic resistance in plants [36] and degrade polycyclic aromatic hydrocarbons . Iturins are a large family of antibiotic compounds, which includes bacillopeptin, ituin A and C, and mycosubtilin, that have strong antibiotic effect with moderate surfactant activity and can enhance swarming motility [18]. Furthermore, the antifungal activity of bacillomycin and subtilin is reported by [25, 37, 45] and [10] in various Bacillus species. The dominance of these particular genes in Bacillus strains SRB2 and SRB20 associated with plants reinforces the competitive role of subtilin, bacyllomycin, fengycin, and iturin in the fitness of strains in the natural environment.
Pathogen-inoculated pots treated with SRB2 and SRB20 displayed a reduction in mortality. This could be due to their higher root colonizing property, saprophytic ability, and increased fitness of the isolate against abiotic and biotic factors [17]. The reduced germination could be attributed to the pre-emergence effect of the pathogen [48]. The variation in antagonistic efficacy in in-vitro and greenhouse conditions could be due to microbe–microbe interactions that are very sensitive to several abiotic and biotic factors that affect the extent of microbial species interactions and composition in the rhizoplane [49].
Conclusion
Among the indigenous soil-borne antagonists, B. inaquosorum SRB2 and B. vallismortis SRB20 were found to have better efficacy in controlling F. sacchari, reducing the severity of wilt disease in sugarcane under greenhouse conditions and can be used for further studies under field conditions. Biochemical and molecular analysis of the 20 rhizobacteria isolated from the rhizoplane has unveiled the hidden biocontrol mechanisms like production of IAA, Protease, Cellulase, Siderophores, P solubilization and the presence of antimicrobial peptide biosynthetic genes involved in the production of Fengycin, Bacyllomicin, Iturin, and Subtilin. These genes can be further used in the improvement of the biocontrol potential of the isolates through molecular techniques.
Future line of work
The other plant growth-promoting traits and inhibitory principles of the potential Bacillus isolates, which were not determined in this research, have to be studied in detail for complete knowledge of unknown mechanisms by which bioagents inhibit the pathogens and further studies on different formulations and compatibility for practical implementation of such propositions would pave the way for better establishment of biological control.
Abbreviations
- DNA:
-
Deoxyribose Nucleic Acid
- CTAB:
-
Cetyl Trimethyl Ammonium Bromide,
- PCR:
-
Polymerase Chain Reaction,
- TBE:
-
Tris Borate EDTA buffer
- EDTA:
-
Ethylene Diamine Tetra Acetic acid
- CRD:
-
Completely Randomized Design
- PDA:
-
Potato Dextrose Agar
- AUMGC:
-
Area Under Mycelial Growth Curve
- F. sacchari :
-
Fusarium sacchari
- IAA:
-
Indole-3-Acetic Acid
- KOH:
-
Potassium Hydroxide
- CAS:
-
Chrome Azurol S
- UV:
-
Ultra-Violet light
- CFU:
-
Colony Forming Unit
- PEM:
-
Pre-Emergence Mortality
- PESM:
-
Post Emergence Seedling Mortality
- SRB:
-
Sugarcane Rhizoplane Bacillus species
- DAI:
-
Days After Inoculation
- AMP:
-
Anti Microbial Peptide
References
Agnihotri VP, Rao GP (2002) A century status of sugarcane wilt in India. In: Singh SB, Rao GP, Eswaramoorthy S (eds) Sugarcane crop management. Sci Tech Publishers, Houston, Texas, USA, pp 145–160
Ali S, Hameed S, Shahid M, Iqbal M, Lazarovits G, Imran A (2020) Functional characterization of potential PGPR exhibiting broad-spectrum antifungal activity. Microbiol Res 232:126389
Barry T, Colleran G, Glennon M, Dunican LK, Gannon F (1991) The 16S–23S ribosomal spacer region as a target for DNA probes to identify eubacteria. Genome Res 1:51–56
Chan Y, Savard M, Reid L, Cyr T, McCormick W, Seguin C (2009) Identification of lipopeptide antibiotics of a Bacillus subtilis isolate and their control of Fusarium graminearum diseases in maize and wheat. Biocontrol 54:567–574
Chungopast S, Thongjoo C, Islam AKM, Yeasmin S (2021) Efficiency of phosphate-solubilizing bacteria to address phosphorus fixation in Takhli soil series: a case of sugarcane cultivation, Thailand. Plant Soil 460:347–357
Dasgupta MK (1988) Principles of plant pathology. Allied Publishers.
Deleu M, Paquot M, Nylander T (2005) Fengycin interaction with lipid monolayers at the air-aqueous interface-implications for the effect of fengycin on biological membranes. J colloid interface sci 283(2):358–365
Dennis C, Webster J (1971) Antagonistic properties of species groups of Trichoderma. I. Production of non-volatile antibiotics. Trans Br Mycol Soc 57:25–39
Desai I, Patel N (2020) Characterization and in vitro efficacy of Bacillus subtilis and Pseudomonas aeruginosa as biological control against wilt causing Fusarium species of sugarcane. South Asian J Exp Biol 10:363–373
Devi NO, Devi RK, Debbarma M, Hajong M, Thokchom S (2022) Effect of endophytic Bacillus and arbuscular mycorrhiza fungi (AMF) against Fusarium wilt of tomato caused by Fusarium oxysporum f. sp. lycopersici. Egypt J Biol Pest Control 32:1–14
Gardener BB, Driks A (2004) Overview of the nature and applications of biocontrol microbes: Bacillus spp. Phytopathol 94:1244
Gomez KA, Gomez AA (1984) Statistical procedures for agricultural research. John Wiley & Sons. Ltd., Singapore
Guevara-Avendano E, Bejarano-Bolivar AA, Kiel-Martinez AL, Ramirez-Vazquez M, Mendez-Bravo A, Wobeser EA, Sanchez-Rangel D, Guerrero-Analco JA, Eskalen A, Reverchon F (2019) Avocado rhizobacteria emit volatile organic compounds with antifungal activity against Fusarium solani, Fusarium sp. associated with Kuroshio shot hole borer, and Colletotrichum gloeosporioides. Microbiol res 219:74–83
Hassan MN, Afghan S, Hassan Z, Hafeez FY (2014) Biopesticide activity of sugarcane associated rhizobacteria: Ochrobactrum intermedium strain NH-5 and Stenotrophomonas maltophilia strain NH-300 against red rot under field conditions. Phytopathologia Mediterranea 229–239.
Hoang NV, Furtado A, Botha FC, Simmons BA, Henry RJ (2015) Potential for genetic improvement of sugarcane as a source of biomass for biofuels. Front Bioeng Biotechnol 3:182
Idris EES, Iglesias DJ, Talon M, Borriss R (2007) Tryptophan-dependent production of Indole-3-Acetic Acid (IAA) affects level of plant growth promotion by Bacillus amyloliquefaciens FZB42. Mol Plant Microbe Interact 20:619–626
Islam A, Kabir MS, Khair A (2019) Molecular identification and evaluation of indigenous bacterial isolates for their plant growth promoting and biological control activities against Fusarium wilt pathogen of tomato. Plant Pathol J 35:137
Jaques P (2011) Surfactin and other lipopeptides from Bacillus spp. In G. Soberon-Chavez (eds.) Biosurfactants from genes to applications. Allied publishers. 216.
Kamali M, Guo D, Naeimi S, Ahmadi J (2022) Perception of biocontrol potential of Bacillus inaquosorum KR2-7 against tomato Fusarium wilt through merging genome mining with chemical analysis. Biol 11:137
Kirtikar GP, Singh SB, Shukla R (1972) Role of seed material in carry over of wilt disease of sugarcane. Indian Sugar 22:89–90
Kloepper JW, Leong J, Teintze M, Schroth MN (1980) Enhanced plant growth by siderophores produced by plant growth-promoting rhizobacteria. Nature 286:885–886
Kloepper JW, Ryu CM, Zhang S (2004) Induced systemic resistance and promotion of plant growth by Bacillus spp. Phytopathol 94(11):1259–1266
Korir H, Mungai NW, Thuita M, Hamba Y, Masso C (2017) Co-inoculation effect of rhizobia and plant growth promoting rhizobacteria on common bean growth in a low phosphorus soil. Front Plant Sci 8:141
Kumar A, Prakash A, Johri BN (2011) PGPR in crop ecosystem. In: Maheshwari DK (ed) Bacteria in agrobiology: crop ecosystems. Springer-Verlag, Berlin, Heidelberg, pp 37–59
Kumar PR, Adhipathi P, Nakkeeran S (2014) Antimicrobial peptide genes of PGPR for the management of Fusarium wilt of carnation under protected cultivation. Plant Pathol 44:54–61
Li Y, Wang R, Liu J, Xu L, Ji P, Sun L, Pan H, Jiang B, Li L (2019) Identification of a biocontrol agent Bacillus vallismortis BV23 and assessment of effects of its metabolites on Fusarium graminearum causing corn stalk rot. Biocontrol Sci Technol 29:263–275
Liang YL, Zhang Z, Wu M, Wu Y, Feng JX (2014) Isolation, screening and identification of cellulolytic bacteria from natural reserves in the subtropical region of China and optimization of cellulase production by Paenibacillus terrae ME27-1. BioMed res int 48:83–89
Mirza SM, Ahmad W, Latif F, Haurat J, Bally R, Normand P, Malik KA (2001) Isolation, partial characterization, and the effect of plant growth-promoting bacteria (PGPB) on micro-propagated sugarcane in vitro. Plant Soil 237:47–54
Modi K, Patel P (2017) Isolation and characterization of plant growth promoting rhizobacteria associated with Saccharum officinarum L. Curr Synth Sys Biol 5:132
Mohanraj D, Kumaresan S, Sreenivasan TV (2002) Molecular characterization of isolates of the sugarcane red rot pathogen Colletotrichum falcatum Went. Indian Phytopathol 55:147–151
Mora I, Cabrefiga J, Montesinos E (2011) Antimicrobial peptide genes in Bacillus strains from plant environments. Int Microbiol 14:213–223
Pahari A, Mishra BB (2017) Characterization of siderophore producing Rhizobacteria and Its effect on growth performance of different vegetables. Int J Curr Microbiol App Sci 6(5):1398–1405
Pikovskaya RI (1948) Mobilization of phosphorus in soil in connection with vital activity of some microbial species. Mikrobiologiya 17:362–370
Prabhukarthikeyan SR, Karthikeyan G, Jeyarani S, Raguchander T (2014) PCR based identification and characterization of lipopeptides producing Bacillus against Fusarium oxysporum f. sp. lycopersici. Biochem Cell Arch 14:133–140
Raton TMO, Yano R, Gamez OR, Floh EIS, Diaz MJS, Barbosa HR (2012) Isolation and characterization of aerobic endospore forming Bacilli from sugarcane rhizosphere for the selection of strains with agriculture potentialities. World J Microbiol Biotechnol 28:1593–1603
Romero D, Vicente A, Rakotoaly RH, Dufour SE, Veening JW, Arrebola E, Cazorla FM, Kuipers OP, Paquot M, Perez-Garcia A (2007) The iturin and fengycin families of lipopeptides are key factors in antagonism of Bacillus subtilis toward Podosphaera fusca. Mol Plant Microbiol 20:430–440
Roongsawang N, Thaniyavarn J, Thaniyavarn S, Kameyama T, Haruki M, Imanaka T, Morikawa M, Kanaya S (2002) Isolation and characterization of a halotolerant Bacillus subtilis BBK-1 which produces three kinds of lipopeptides: bacillomycin L, plipastatin, and surfactin. Extremophiles 6:499–506
Sadia A, Khalil S, Ayub N, Rashid M (2002) In vitro solubilization of inorganic phosphate by phosphate solubilizing microorganisms (PSM) from maize rhizosphere. Int J Agric Biol 4:454–458
Sambrook J, Russell DW (2001) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, New York, pp 631–632
Schwyn B, Neilands JB (1986) Universal Chemical Assay for the detection and determination of siderophores. Anal Biochem 140:47–56
Shrivastava UP, Kumar A (2011) A simple and rapid plate assay for the screening of Indole-3-acetic acid (IAA) producing microorganisms. Int J Appl Biolo Pharm Tech 2:120–123
Sivasakthi S, Usharani G, Saranraj P (2014) Biocontrol potentiality of plant growth promoting bacteria (PGPR)-Pseudomonas fluorescens and Bacillus subtilis: A review. Afr J Agric Res 9:1265–1277
Sneha S, Das MP, Rebecca LJ (2014) Isolation and screening of protease producing bacteria from marine waste. J Chem Pharm Res 6:1157–1159
Thongponkaew C, Chittamart N, Tawornpruek S, Pinjai P (2016) Screening of phosphate-solubilizing bacteria from sugarcane rhizospheric soil and their abilities to improve growth and yield of sugarcane. Proc Int Soc Sugar Cane Technol 29:1329–1337
Varma PK, Kumar KVK, Sekhar VC, Adilakshmi D, Suresh M, Kumar NR, Jayachandra K, Anitha R (2017) Evaluation of endophytic bacteria for plant growth promotion and pathogen suppression traits in Saccharum officinarum. J Agric Sci Technol 7:537–545
Vejan P, Abdullah R, Khadiran T, Ismail S, Nasrulhaq BA (2016) Role of Plant Growth Promoting Rhizobacteria in Agricultural Sustainability-A Review. Molecules 21:573
Vincent JM (1947) Distortion of fungal hyphae in the presence of certain inhibitors. Nature 159:850
Viswanathan R (2013) Status of sugarcane wilt: 100 years after its occurrence in India. J Sugarcane Res 3:86–106
Wei Z, Huang J, Yang T, Jousset A, Xu Y, Shen Q, Friman V (2017) Seasonal variation in the biocontrol efficiency of bacterial wilt is driven by temperature-mediated changes in bacterial competitive interactions. J Appl Ecol 54:1440–1448
Xia Y, Farooq MA, Javed MT, Kamran MA, Mukhtar T, Ali J, Tabassum T, Rehman S, Munis MFH, Sultan T, Chaudhary HJ (2020) Multi-stress tolerant PGPR Bacillus xiamenensis PM14 activating sugarcane (Saccharum officinarum L.) red rot disease resistance. Plant physiol biochem 151:640–649
Xu D, Cote JC (2003) Phylogenetic relationships between Bacillus species and related genera inferred from comparison of 3’ end 16S rDNA and 5’ end 16S–23S its nucleotide sequences. Int J Syst Evol Microbiol 53:695–704
Zain M, Yasmin S, Hafeez FY (2019) Isolation and characterization of plant growth promoting antagonistic Bacteria from Cotton and Sugarcane plants for suppression of phytopathogenic Fusarium species. Iran J Biotechnol 17:61–70
Zia MA, Yasmin H, Shair F, Jabeen Z, Mumtaz S, Hayat Z, Shah H, Afghan S, Hafeez FY, Hassan MN (2019) Glucanolytic rhizobacteria produce antifungal metabolites and elicit ROS scavenging system in Sugarcane. Sugar Tech 21:244–255
Acknowledgements
We would like to express our gratitude to the Regional Agricultural Research Station, Anakapalle, and Acharya NG Ranga Agricultural University, Guntur, India, for providing good laboratory facilities for the better conduct of work. Indian Council of Agricultural Research for providing Junior Research Fellowship.
Funding
Indian Council of Agriculture Research (ICAR) provided a fellowship to the researcher throughout the research period and Acharya NG Ranga Agriculture University provided funds for laboratory needs.
Author information
Authors and Affiliations
Contributions
Author Chetan K.K. conducted all the experiments, performed the statistical analysis, tabulated and interpreted the data, and wrote the first draft of the manuscript. Authors Kishore Varma. P, Chandrasekhar. V, and Anil Kumar. P designed the experiments, provided the ideas, and revised the manuscript. Authors Vasanthi. V and Vamshi Krishna. G conducted part of the experiment and helped during the experimental period and data collection.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This article does not contain any studies with human participants or animals performed by any of the authors.
Consent for publication
Not applicable.
Data availability
The data sets used and/or analyzed during the study can be obtained on a reasonable request.
Competing interests
We have no known competing interests that could have appeared to influence the work reported in this paper.
Additional information
Responsible Editor: Jerri Zilli
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chetan, K.K., Kishore Varma, P., Chandrasekhar, V. et al. Unveiling the Biocontrol Potential of Rhizoplane Bacillus Species against Sugarcane Fusarium Wilt through Biochemical and Molecular Analysis. Braz J Microbiol 55, 1883–1896 (2024). https://doi.org/10.1007/s42770-024-01307-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42770-024-01307-z