Abstract
Acibenzolar-S-methyl (ASM), a plant activator known to induce plant resistance, has been used as foliar sprays to manage several plant diseases including bacterial spot on tomato caused by four distinct Xanthomonas species. This study aimed to investigate the effects of soil application rates of ASM on bacterial spot of tomato and the expression levels of the two pathogenesis-related (PR) genes, PR1a and PR1b, in leaf tissues. Tomato seedlings were leaf-applied with ASM at 18.8 mg/l corresponding to the labeled rate, soil-applied with ASM at 0.84 and 10 mg/l, and sprayed with water served as an untreated control. The soil application of ASM at 10 mg/l consistently reduced the final disease severity and disease progress compared to the untreated control in four growth chamber experiments, whereas the soil application of ASM at 0.84 mg/l and foliar spray of ASM significantly reduced the final disease severity and area under disease progress curve (AUDPC) in three out of the four experiments. The expression levels of PR1a and PR1b in the leaf tissues were significantly induced by both soil and foliar applications of ASM. In addition, field trial results suggested that the soil applications of ASM at 10 mg/l markedly reduced disease progress compared to the control and copper standard. Although the control efficiency of soil applications of ASM depends on rates used, this study suggests that ASM can be used as soil applications to induce tomato resistance against bacterial spot.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Bacterial spot is a destructive disease of tomato (Solanum lycopersicum) caused by at least four taxonomically distinct xanthomonads including Xanthomonas euvesicatoria (T1), X. vesicatoria (T2), X. perforans (T3 and T4), and X. gardneri (T2) (Jones et al. 2004, 2005; Stall et al. 2009). The use of pesticides is a common strategy for controlling this disease. Although copper-based bactericides are heavily utilized (Jones et al. 1991), copper bactericides mixed with an ethylene bisdithiocarbamate (EBDC) compound such as manzeb or mancozeb is necessary to suppress copper-resistant strains of Xanthomonas (Marco and Stall 1983; Pernezny et al. 2008). However, this combination may result in phytotoxicity, reduce yield, and has limited efficacy when environmental conditions are conducive to disease progress (Roberts et al. 2008). The antibiotics streptomycin and kasugamycin have been investigated to control bacterial spot on tomato, but resistance to the antibiotics may develop in Xanthomonas spp. (Stall et al. 2009; Woodcock et al. 1991). Although the use of pesticides is generally effective to control bacterial spot of tomato, consumers may prefer pesticide-free products. It is necessary to explore alternative environment-friendly strategies for managing bacterial spot on tomato.
Acibenzolar-S-methyl (ASM, Actigard® or Bion®, Syngenta Crop Protection, Inc., Greensboro, NC) is a plant activator inducing systemic acquired resistance (SAR) to confer protection against a broad spectrum of plant pathogens (Huang et al. 2012; Meller Harel et al. 2014; Takeshita et al. 2013; Tally et al. 1999). Although ASM has no anti-microbial activity, it induces host plant resistance by activating an up-regulation of pathogenesis-related (PR) gene expression that is thought to contribute to disease resistance (Durrant and Dong 2004; Francis et al. 2009). The expression level of the acidic PR-1 gene (PR1a) has been used as a marker of SAR, whereas ethylene (ET)-mediated responses can be evaluated using the basic PR-1 marker gene (PR1b) (Block et al. 2005; Tornero et al. 1997; van Loon et al. 2006). Previous studies have shed light on the two genes activated by ASM as foliar sprays (Herman et al. 2007, 2008). However, it is not well known whether soil applications of ASM can also induce the expression of the two genes in tomato against bacterial spot.
Previous studies have shown the efficacy of ASM as foliar sprays for bacterial spot control (Huang et al. 2012; Louws et al. 2001; Obradovic et al. 2004, 2005), but it is not well known whether soil applications of ASM is also effective for control of bacterial spot on tomato. In different pathosystems, soil drenches of ASM have been demonstrated to achieve a control efficacy similar to or better than foliar sprays (Barretti et al. 2010; Francis et al. 2009; Johnson and Temple 2016). Moreover, soil applications via irrigation drip tubes may be an easier way to apply ASM while saving application costs for growers. In order to determine whether soil applications of ASM for control of bacterial spot is possible, four growth chamber experiments and two field trials were conducted to investigate the effects of soil application rates of ASM on bacterial spot of tomato and the expression levels of PR1a and PR1b in tomato leaves. Here we report that soil applications of ASM can activate the up-regulation of the two gene expression, and the soil application of ASM at 10 mg/l results in the best control efficacy.
Materials and methods
Inoculum preparation
Inoculum was prepared by growing X. perforans strain T4 (T4) on glucose-nutrient agar plates for 3 d at 28°C, flooded with 10 mM MgSO4, and adjusted to 106 CFU ml−1for inoculations. In order to increase foliar infection, Silwet L-77 (Helena Chemical Co., Collierville, TN) was added to the inoculum to a final concentration of 0.025% (vol/vol).
Effects of soil applied ASM on bacterial spot in growth chamber studies
Tomato seeds of the cultivar ‘Florida 47’ (Seminis Vegetable Seeds, Inc., St. Louis, MO) susceptible to bacterial spot were soaked in 1% (v/v) sodium hypochlorite (NaOCl) for 2 min for surface sterilization, and then washed three times with sterilized water. The sterilized seeds were sowed in seed trays with a commercial growth medium (Fafard Custom Mix, Agawam, MA). Seedlings at the cotyledon stage were removed from the growth medium at two weeks after sowing, their roots were washed, and the seedlings were transplanted in 250 ml-styrene cups each filled with 230 g of quartz sand for experiments 1–3 and of a field soil classified as Myakka fine sand (sandy, siliceous, hyperthermic Aeric Alaquods) for experiment 4. Each styrene cup consisted of one tomato seedling. The tomato plants in the cups were fertigated with 10 ml of the Hoagland’s nutrient solution (Hoagland and Arnon 1950) three times a week after transplanting. The seedlings were placed in a growth chamber held at 28 °C with a 12-h light photoperiod.
After transplanting, the seedlings were treated with or without ASM for three consecutive weeks before inoculation. There were four treatments: (i) non-treated control; (ii) a weekly 20 ml soil application of ASM at a rate of 0.84 mg/l (Actigard 50WG; 50% a.i.; Syngenta Crop Protection) for each plant; (iii) a weekly 20 ml soil application of ASM at 10 mg/l for each plant; and (iv) a weekly foliar application of ASM at 18.8 mg/l corresponding to the labeled rate. Foliar applications were made until runoff using a hand-held sprayer, while soil applications were made directly to the soil surrounding the base of the plant. There were eighteen replicates per treatment arranged in a completely randomized design within the growth chamber. After ASM treatments for the three consecutive weeks, these tomato plants were inoculated with a suspension (106 CFU ml−1) of T4 using a hand-trigger sprayer onto the upper and lower leaf surfaces of each plant, and the inoculated plants were then placed in the growth chamber in which a high relative humidity was maintained using an ultrasonic humidifier.
Effects of soil applied ASM on bacterial spot under field conditions
Two field trials were conducted under field conditions at the Gulf Coast Research and Education Center (GCREC) to evaluate the effect of soil-applied ASM on bacteria spot on tomato cv. SecuriTY 28. Tomato production guidelines established by the University of Florida/IFAS were followed for land preparation, fertilization, irrigation, weed management, and insect control (Olson and Santos 2010). There were five treatments: (i) non-treated control sprayed with water only; (ii) weekly soil application of ASM at a rate of 0.84 mg/l (Actigard 50WG); (iii) weekly soil application of ASM at a rate of 10 mg/l; and (iv) weekly foliar application of ASM at a rate of 17.5 g/ha; and (v) weekly foliar application of a standard copper program including copper hydroxide (815 g a.i./ha; Kocide 3000) plus an EBDC fungicide (420 g a.i./ha; Penncozeb 75 DF). The treatments were arranged in a randomized complete block design with six replicates for each treatment. Each plot was 6.4 m long with 14 plants at 46-cm spacing. A total of 6 and 8 weekly applications starting 4 weeks after transplanting were made per season in field trials 1 and 2, respectively. Foliar applications of treatments (i), (iv), and (v) were made with a CO2-powered backpack sprayer adjusted to deliver 561 to 842 l/ha at 275 kPa depending on plant size. Soil ASM applications used a CO2 injection manifold to deliver each treatment in 2 l of water to each plot using the drip irrigation tape. A valve was inserted at the end of each drip tape to separate plots, and treatment applications were followed by an additional 1.2 l of water to rinse the drip tape. The tomato plants were inoculated with a suspension (106 CFU ml−1) of T4 using a backpack sprayer at 7 weeks after transplanting.
Disease rating
In growth chamber studies, each plant was rated at 0, 5, 7, and 10 days after inoculation using the Horsfall-Barratt scale (Horsfall and Barratt 1945) to evaluate the percentage of foliage infected by bacterial spot. For field trials, the center ten plants within each plot were rated weekly using the Horsfall-Barratt scale to determine the percentage of canopy exhibiting symptoms of bacterial spot. Estimates of disease severity were converted to mid-percentages prior to statistical analysis. The area under disease progress curve (AUDPC) was calculated as previously described (Shaner and Finney 1977).
Tissue collection and RNA extraction
Four leaves were collected from each plant at 0, 5, and 10 days after inoculation in growth chamber experiments 1 and 2. Three plants were sampled for each treatment at each sampling time. The leaves from each plant were pooled, flash-frozen in liquid nitrogen immediately after collection, and stored at −80°C. RNA was extracted using Qiagen RNeasy Mini Kit (Qiagen Inc., Valencia, CA) and further DNase treated with Turbo DNA-free (Ambion Inc., Austin, TX). RNA was quantified with a NanoDrop UV-Vis Spectrophotometer (Thermo Fisher Scientific, Inc., Waltham, MA) and checked for degradation by electrophoresis on a 1.2% agarose formaldehyde gel.
Analysis of gene expression using quantitative real-time PCR
Approximately 1 μg of total RNA was used to generate first-strand cDNA using iScript cDNA synthesis kit (Bio-Rad Laboratories, Hercules, CA) according to the manufacturer’s instructions. RNA samples treated without reverse transcriptase served as controls to check for DNA contamination. Eight candidate reference genes (RGs) were selected to evaluate their expression stability after inoculation with X. perforans (Table 1), whereas PR1a and PR1b were used as respective SA- and ET-mediated response markers. Except those for GAPDH previously described by Mascia et al. (2010), real time PCR primers (Table 1) were designed using Primer Express 3.0 software (Applied Biosystems, Foster City, CA).
Quantitative PCR reactions were performed using the Bio-Rad CFX96 Real-Time PCR Systems (Bio-Rad Laboratories) and utilized SsoFast EvaGreen Supermix (Bio-Rad Laboratories). A total volume of 20 μl of the PCR reaction mixture consisted of following components (final concentration): 1× EvaGreen Supermix (Bio-Rad), 0.5 μM forward and reverse primers, 1 μl of the synthesized cDNA, and PCR-grade water to make up the final volume. Three simultaneous amplifications were performed for each sample to confirm technical reproducibility of the results. A negative control sample consisted of nuclease-free water substituted for the DNA template. The manufacturer’s recommended universal thermal cycle protocol (Bio-Rad Laboratories) was used for PCR amplification: An initial hot start of 30 s at 95°C was followed by 40 cycles of 95°C for 10 s and 60°C for 10 s. Fluorescence emission was measured at 60°C during the annealing and extension phase. Threshold cycles were calculated using Bio-Rad CFX Manager 3.0 software. Immediately after the final PCR cycle, a melting curve analysis was conducted to validate primer specificity.
Appropriate reference genes used for calculating the relative gene expression of PR1a and PR1b were selected using geNorm (Vandesompele et al. 2002). The program determines an expression stability value (M) for each reference gene as the average pairwise variation for a particular gene with all the other reference genes tested in all samples. The expression ratio of two ideal reference genes is identical in all samples, regardless of treatments and experimental conditions. After genes with the lowest expression stability are removed, a new M value is calculated for each of the remaining reference genes until only two genes remain. Normalization factors were derived from the geometric mean of the expression values of the two most stable reference genes. Relative expression levels of PR1a and PR1b were calculated according to the 2-ΔΔCt method (Livak and Schmittgen 2001) normalized by using the most stable reference genes of each experiment. Three biological replicates and three technical replicates were used for measuring the relative expression levels of PR1a and PR1b.
Statistical analysis
Generalized linear mixed models employed in PROC GLIMMIX of SAS (version 9.4; SAS Institute, Gary, NC) were used to analyze the effect of treatments on response variables. The log2 transformation of the relative gene expression level was performed before statistical analysis (Yuan et al. 2006). The least squared means (LSMEANS) statement of the GLIMMIX procedure in SAS was used to compare treatment means at the 5% level of significance according to Fisher’s least significant difference (Fisher’s LSD).
Results
Growth chamber studies
Foliar sprays of ASM did not consistently reduce disease severity of bacterial spot on tomato in all the experiments at 5 days after inoculation, but the two ASM soil application treatments always statistically reduced disease severity compared to the untreated control (data not shown). Compared to the untreated control, soil and foliar ASM applications significantly reduced the final disease severity in experiments 1–3 (P < 0.0001, Table 2). In experiment 4, the weekly soil application of ASM at 10 mg/l significantly reduced the final disease severity in comparison with the untreated control by 14.1%. No significant difference was detected between the other ASM treatments and the untreated control (Table 2). The soil application of ASM at rates as low as 0.84 mg/l was statistically equivalent or even better in reducing the final disease severity compared to the foliar spray standard of ASM in experiments 1–3. However, results of these four experiments suggested that the soil application of ASM at 10 mg/l showed the best control efficacy (65.9%) compared to the untreated control.
Although foliar applications of ASM significantly reduced AUDPC compared to the untreated control in experiments 1 and 3, the soil application of ASM at 0.84 and 10 mg/l significantly decreased AUDPC in three and all of the four experiments, respectively. In general, the two ASM soil treatments were statistically better at reducing disease than the standard foliar applied ASM. Again, the soil application of ASM at 10 mg/l was the most effective treatment to significantly reduce disease progress, showing an average reduction in AUDPC across the four experiments of 65.9% compared to the untreated control.
Field trials
Although in field trial 1 ASM treatments did not significantly reduce the final disease severity, in field trial 2 soil and foliar applied ASM treatments significantly reduced the final disease severity compared to the standard program (Table 3). In comparison with the control and copper standard, weekly soil applications of ASM at a rate of 10 mg/l and weekly foliar applications at a rate of 17.5 g/ha markedly reduced disease progress in both field trials 1 and 2. In contrast, soil applications of ASM at the lower rate (0.84 mg/l) significantly reduced disease progress only in field trial 2 compared to the control and standard.
Expression stability analysis of the reference genes
A ranking of the tested genes analyzed by geNorm was shown on Fig. 1 according to the stability measure M (average pairwise variation of each combination of candidate reference genes), from the most stable (lowest M values) to the least stable (highest M values). All the M values of the tested genes were below the acceptable limit of 1.5 (Vandesompele et al. 2002). UBI and CyP had the lowest M value (0.56) in experiment 1 (Fig. 1a), but EF1α and ACT showed the smallest M of 0.37 in experiment 2 (Fig. 1b). Therefore, the expression values of UBI and CyP were used to calculate normalization factors in experiment 1, and those of EF1α and ACT were used for experiment 2.
Expression stability of the candidate reference genes in tomato plants analyzed by geNorm. Appropriate reference genes are derived from ranking gene pairs that have stable expression patterns relative to each other. Average expression stability values (M) of the eight candidate reference genes are shown for infected leaf tissue samples in experiment 1 (a) and in experiment 2 (b)
Induction of salicylic acid-mediated PR1a
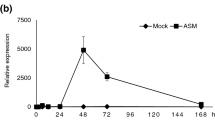
PR1a, a known marker for salicylic acid-mediated responses and SAR in tomato, was induced after the soil and foliar applications of ASM. At the time of inoculation, PR1a expression was significantly greater for ASM applications than the untreated control in both experiments 1 and 2 (Fig. 2). After inoculation, PR1a expression decreased dramatically. In experiment 1, the soil application of ASM at 10 mg/l and the foliar spray of ASM significantly increased the expression of PR1a compared to the untreated control at 10 days after inoculation. In experiment 2, no significant difference in PR1a expression between ASM treatments and the untreated control was detected at 10 days after inoculation.
Expression pattern of the pathogenesis-related (PR)1a gene in the acibenzolar-S-methyl (ASM)-treated and -nontreated leaf tissues of tomato plants following inoculation with Xanthomonas perforans at 0, 5, and 10 days after inoculation. Plants were treated with ASM as soil applications at 0.84 (0.84D) and 10 (10D) mg/l and as foliar sprays at 18.8 mg/l (18.8F). a Experiment 1 and (b) Experiment 2. Relative fold change was calculated by calibrating data to the untreated control. Letters on the bars in the same sampling time are significantly different as denoted by the LSMEANS statement of the GLIMMIX procedure in SAS v9.4 at the 5% level of significance according to Fisher’s least significant difference (Fisher’s LSD)
Induction of ethylene-mediated PR1b
The general trends of activation were similar between PR1a and PR1b. As with PR1a expression, the expression levels of PR1b was significantly induced by both soil and foliar applications of ASM (Fig. 3). There were significant differences between ASM treatments and the untreated control at the time of inoculation. However, PR1b expression decreased remarkably following the inoculation. There were no significant difference in PR1b expression between ASM treatments and the untreated control at 10 days after inoculation in experiments 1 and 2.
Expression pattern of the pathogenesis-related (PR)1b gene in the acibenzolar-S-methyl (ASM)-treated and -nontreated leaf tissues of tomato plants following inoculation with Xanthomonas perforans at 0, 5, and 10 days after inoculation. Plants were treated with ASM as soil applications at 0.84 (0.84D) and 10 (10D) mg/l and as foliar sprays at 18.8 mg/l (18.8F). a Experiment 1 and (b) Experiment 2. Relative fold change was calculated by calibrating data to the untreated control. Letters on the bars in the same sampling time are significantly different as denoted by the LSMEANS statement of the GLIMMIX procedure in SAS v9.4 at the 5% level of significance according to Fisher’s least significant difference (Fisher’s LSD)
Discussion
In this study, we demonstrated that weekly soil application of ASM at 10 mg/l consistently reduced the final disease severity and disease progress compared to the untreated control in all growth chamber studies, whereas the soil application of ASM at 0.84 mg/l and foliar spray of ASM significantly reduced the final disease severity and AUDPC in three out of the four experiments. The control efficiency of soil applied ASM at a rate of 10 mg/l was comparable to that of foliar applied ASM in the field trials, and these two ASM treatments significantly reduced disease progress of bacteria spot on tomato.
The efficacy of ASM as soil applications in reducing bacterial spot on tomato is associated with induction of expression levels of PR1a and PR1b. The most stable reference genes were not identical for determining relative expression levels of PR1a and PR1b, but the expression levels of these two genes were significantly induced by both soil and foliar applications of ASM.
This study confirms that soil applied ASM is as efficacious for control of bacterial diseases as foliar-applied ASM and copper bactericide standards, establishing the use of soil applied ASM for control of bacterial spot on tomato. The efficacy of soil application of ASM was previously established for control of citrus canker (Graham and Myers 2011, 2013) and for control of fire blight in pear and apple (Johnson and Temple 2016). In addition, ASM paints used in conjunction with pruning could be used as a therapy for restoration of health in field-grown pear and apple trees diseased with fire blight (Johnson and Temple 2017). Although the demonstration of efficacy of soil applied ASM is not novel, the use of drip irrigation for ASM application is an advance over soil drench application used in previous studies. Drip application could considerably reduce the labor and cost of application compared to foliar application. Although SAR-inducers such as ASM used as foliar sprays are effective for control of bacterial spot on tomato (Huang et al. 2012), our study indicates that soil applications of ASM not only induce the expression of PR protein genes but also reduce the severity and disease progress of this disease.
While the reference genes used to calculate the expression levels of PR1a and PR1b were different in experiments 1 and 2 probably due to disease severity differences, the outcome was still the same showing that soil applications of ASM remarkably induced the two gene up-regulation. Soil applications of ASM inducing a high and persistent up-regulation of PR gene expression are not uncommon because ASM can be absorbed by roots and then translocated throughout the plant (Francis et al. 2009; Meller Harel et al. 2014). Moreover, the acid metabolite of ASM, benzo [1,2,3] thiadiazole-7-carboxylic acid (CGA210007), can be formed within 2 h in tomato plants after ASM application and degraded to undetectable levels at 72 h to activate induced resistance (Scarponi et al. 2001). The stimulation of induced resistance by ASM, an analogue of salicylic acid, likely results from a high up-regulation of PR gene expression (Durrant and Dong 2004; Francis et al. 2009) and the production of PR proteins (Friedrich et al. 1996; Lawton et al. 1996). In this study, ASM-treated plants showed a systematic induction of both SAR- (PR1a) and ET-regulated (PR1b) genes in agreement with previous studies (Herman et al. 2007; Meller Harel et al. 2014). Significant reduction in disease severity and disease progress of bacterial spot is likely related to increase in the expression levels of PR1a and PR1b induced by ASM as soil and foliar applications before inoculation.
This study also suggests that the efficacy of soil applied ASM in eliciting resistance to bacterial spot depends on the concentration of the product applied. Although soil applications of ASM can activate the expression of PR1a and PR1b, the soil application of ASM at 10 mg/l resulted in the best control efficacy. Francis et al. (2009) demonstrated that a soil application of ASM at 5 mg a.i. per plant is more effective than a foliar spray for controlling citrus canker (Francis et al. 2009). Similarly, our growth chamber studies showed that tomato plants soil-applied with ASM at 0.84 and 10 mg/l before inoculation performed better than the foliar spray treatment with ASM at 18.8 mg/l. The soil application of ASM at 10 mg/l showed a consistent and efficient suppressiveness in all four growth chamber studies and field trial 2, whereas the ASM drench at 0.84 mg/l significantly reduced bacterial spot on tomato in three out of the four growth chamber experiments. This finding suggests that tomato resistance induced by soil applications of ASM at the lower rate may not consistently control bacterial spot. The dissipation rate of ASM in soil presented as half-life values ranges from 0.2 to 14 days depending on soil types and experimental conditions (Myresiotis et al. 2014). Soil applications may provide a prolonged protection since its main metabolite CGA210007, a plant defense activator, can persist in soil up to 10 days after soil applications of ASM (Myresiotis et al. 2014). After foliar applications of ASM, however, CGA210007 persists in tomato plants merely about 3 days (Scarponi et al. 2001). Since our study did not analyze the dissipation rates of ASM and CGA210007 in soil, further investigation is required to determine the relationship between the persistence of the two compounds in soil and disease control efficacy via various soil application rates of ASM.
The effectiveness of soil applied ASM may be related to the application frequency. In growth chamber studies, ASM was not continually soil-applied after inoculation. Apart from induced PR proteins preventing pathogen progress, the expression of PR1a and PR1b decreased markedly at 5 days after inoculation (about 7 days after the last ASM application). Herman et al. (2007) demonstrated that the expression levels of PR1a and PR1b declined to baseline levels by 7 days after ASM application, but reapplication of ASM resulted in the rapid expression of PR-1 within 1–2 days. This gene expression pattern suggests that the soil application frequency of ASM may affect the control efficacy. Therefore, it is necessary to further evaluate whether reapplication of ASM as soil applications after inoculation can achieve more long-lived disease control activity.
A question that may be asked is why soil applications of ASM at 10 mg/l did not outperformed foliar sprays of ASM in reducing the severity and disease progress of bacterial spot on tomato in the field trials in contrast to these results obtained from growth chamber studies. It is possible that disease pressure was higher under field conditions. For example, the final disease severity of the control in the field trials was greater than 89.1%, but that of the control in the growth chamber studies was less than 64.6%. In addition, the rating times after inoculation were different between growth chamber experiments and field trials. Another possibility is that rainfall and irrigation might result in ASM leaching loss in the coarse-textured soil (fine sand) of the two field trials such that soil applications of ASM did not provide sustained resistance to pathogen challenge. Because ASM is relatively insoluble, soil texture and moisture limit its movement in soil and availability for root uptake. Sandy textured soils with low organic matter content are likely to be conducive for soil movement of ASM. The low level of control in the field trial may be accounted for by limitations of soil or delivery with drip irrigation. Therefore, the soil application timing and amount of ASM need to be adjusted according the weather and soil type. Moreover, the environment, genotype, and crop nutrition can influence expression of induced resistance caused by plant activators in the field (Walters et al. 2005). Although experimental conditions vary between our growth chamber and field studies, soil applications of ASM at a higher rate of 10 mg/l is effective for controlling bacterial spot on tomato.
Although the copper-EDBC standard is generally effective in control of bacterial spot on tomato, applications of copper may aggravate disease severity in the field where copper-resistant strains are predominant (Louws et al. 2001). Our field trials suggested that the copper-EDBC standard were statistically ineffective compared to the untreated control. In contrast, soil applications of ASM significantly reduced disease progress of bacterial spot on tomato. Induced resistance by ASM against bacterial spot is generally effective to all races and copper-resistant strains since SAR induced by ASM is considered a broad-spectrum defense (Louws et al. 2001). Taken together, our results suggest that ASM can be used as soil applications to induce tomato resistance against bacterial spot. It is necessary to further determine disease control benefits via combining ASM drenches and other products such as plant growth-promoting rhizobacteria (PGPR) and other plant activators while promoting plant yield. Our study suggests that ASM can be used as soil applications to induce tomato resistance against bacterial spot.
References
Barretti, P. B., Souza, R. M., Pozza, E. A., & Resende, M. L. V. (2010). Effect of application methods and dosages of acibenzolar-S-methyl on protection against bacterial wilt, pathogen populations, and growth of tomato plants. Tropical Plant Pathology, 35, 229–235.
Block, A., Schmelz, E., O'Donnell, P. J., Jones, J. B., & Klee, H. J. (2005). Systemic acquired tolerance to virulent bacterial pathogens in tomato. Plant Physiology, 138, 1481–1490.
Durrant, W. E., & Dong, X. (2004). Systemic acquired resistance. Annual Review of Phytopathology, 42, 185–209.
Francis, M. I., Redondo, A., Burns, J. K., & Graham, J. H. (2009). Soil application of imidacloprid and related SAR-inducing compounds produces effective and persistent control of citrus canker. European Journal of Plant Pathology, 124, 283–292.
Friedrich, L., Lawton, K., Ruess, W., Masner, P., Specker, N., Rella, M. G., Meier, B., Dincher, S., Staub, T., Uknes, S., Metraux, J. P., Kessmann, H., & Ryals, J. (1996). A benzothiadiazole derivative induces systemic acquired resistance in tobacco. The Plant Jounal, 10, 61–70.
Graham, J. H., & Myers, M. E. (2011). Soil application of SAR inducers imidacloprid, thiamethoxam, and acibenzolar-S-methyl for citrus canker control in young grapefruit trees. Plant Disease, 95, 725–728.
Graham, J. H., & Myers, M. E. (2013). Integration of soil applied neonicotinoid insecticides and acibenzolar-S-methyl for systemic acquired resistance (SAR) control of citrus canker on young citrus trees. Crop Protection, 54, 239–243.
Herman, M. A. B., Restrepo, S., & Smart, C. D. (2007). Defense gene expression patterns of three SAR-induced tomato cultivars in the field. Physiological and Molecular Plant Pathology, 71, 192–200.
Herman, M. A. B., Davidson, J. K., & Smart, C. D. (2008). Induction of plant defense gene expression by plant activators and Pseudomonas syringae pv. tomato in greenhouse-grown tomatoes. Phytopathology, 98, 1226–1232.
Hoagland, D. R., & Arnon, D. I. (1950) The water-culture method for growing plants without soil. Circular 347. University of California Agricultural Experiment Station, Berkley.
Horsfall, J. G. & Barratt, R. W. (1945) An improved grading system for measuring plant diseases. (Abstr.) Phytopathology, 35, 655.
Huang, C.-H., Vallad, G. E., Zhang, S., Wen, A., Balogh, B., Figueiredo, J. F. L., Behlau, F., Jones, J. B., Momol, M. T., & Olson, S. M. (2012). Effect of application frequency and reduced rates of acibenzolar-S-methyl on the field efficacy of induced resistance against bacterial spot on tomato. Plant Disease, 96, 221–227.
Johnson, K. B., & Temple, T. N. (2016). Comparison of methods of acibenzolar-S-methyl application for post-infection fire blight suppression in pear and apple. Plant Disease, 100, 1125–1131.
Johnson, K. B., & Temple, T. N. (2017). Induction of systemic acquired resistance aids restoration of tree health in field-grown pear and apple diseased with fire blight. Plant Disease, 101, 1263–1268.
Jones, J. B., Jones, J. P., Stall, J. P., & Zitter, T. A. (1991). Compendium of tomato diseases. St Paul: APS Press.
Jones, J. B., Lacy, G. H., Bouzar, H., Stall, R. E., & Schaad, N. W. (2004). Reclassification of the xanthomonads associated with bacterial spot disease of tomato and pepper. Systematic and Applied Microbiology, 27, 755–762.
Jones, J. B., Momol, M. T., Obradovic, A., Balogh, B., & Olson, S. M. (2005). Bacterial spot management on tomatoes. Acta Horticulturae, 695, 119–124.
Lawton, K. A., Friedrich, L., Hunt, M., Weymann, K., Delaney, T., Kessmann, H., Staub, T., & Ryals, J. (1996). Benzothiadiazole induces disease resistance in Arabidopsis by activation of the systemic acquired resistance signal transduction pathway. The Plant Journal, 10, 71–82.
Livak, K. J., & Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods, 25, 402–408.
Louws, F. J., Wilson, M., Campbell, H. L., Cuppels, D. A., Jones, J. B., Shoemaker, P. B., Sahin, F., & Miller, S. A. (2001). Field control of bacterial spot and bacterial speck of tomato using a plant activator. Plant Disease, 85, 481–488.
Marco, G. M., & Stall, R. E. (1983). Control of bacterial spot of pepper initiated by strains of Xanthomonas campestris pv. vesicatoria that differ in sensitivity to copper. Plant Disease, 67, 779–781.
Mascia, T., Santovito, E., Gallitelli, D., & Cillo, F. (2010). Evaluation of reference genes for quantitative reverse-transcription polymerase chain reaction normalization in infected tomato plants. Molecular Plant Pathology, 11, 805–816.
Meller Harel, Y., Haile Mehari, Z., Rav-David, D., & Elad, Y. (2014). Systemic resistance to gray mold induced in tomato by benzothiadiazole and Trichoderma harzianum T39. Phytopathology, 104, 150–157.
Myresiotis, C. K., Vryzas, Z., & Papadopoulou-Mourkidou, E. (2014). Enhanced root uptake of acibenzolar-S-methyl (ASM) by tomato plants inoculated with selected Bacillus plant growth-promoting rhizobacteria (PGPR). Applied Soil Ecology, 77, 26–33.
Obradovic, A., Jones, J. B., Momol, M. T., Balogh, B., & Olson, S. M. (2004). Management of tomato bacterial spot in the field by foliar applications of bacteriophages and SAR inducers. Plant Disease, 88, 736–740.
Obradovic, A., Jones, J. B., Momol, M. T., Olson, S. M., Jackson, L. E., Balogh, B., Guven, K., & Iriarte, F. B. (2005). Integration of biological control agents and systemic acquired resistance inducers against bacterial spot on tomato. Plant Disease, 89, 712–716.
Olson, S. M., & Santos, B. S. (2010) Vegetable production handbook for Florida 2010–2011. Florida Cooperative Service, University of Florida.
Pernezny, K., Nagata, R., Havranek, N., & Sanchez, J. (2008). Comparison of two culture media for determination of the copper resistance of Xanthomonas strains and their usefulness for prediction of control with copper bactericides. Crop Protection, 27, 256–262.
Roberts, P. D., Momol, M. T., Ritchie, L., Olson, S. M., Jones, J. B., & Balogh, B. (2008). Evaluation of spray programs containing famoxadone plus cymoxanil, acibenzolar-S-methyl, and Bacillus subtilis compared to copper sprays for management of bacterial spot on tomato. Crop Protection, 27, 1519–1526.
Scarponi, L., Buonaurio, R., & Martinetti, L. (2001). Persistence and translocation of a benzothiadiazole derivative in tomato plants in relation to systemic acquired resistance against Pseudomonas syringae pv tomato. Pest Management Science, 57, 262–268.
Shaner, G., & Finney, R. E. (1977). The effect of nitrogen fertilizer on the expression of slow mildewing resistance in Knox wheat. Phytopathology, 67, 1051–1056.
Stall, R. E., Jones, J. B., & Minsavage, G. V. (2009). Durability of resistance in tomato and pepper to xanthomonads causing bacterial spot. Annual Review of Phytopathology, 47, 265–284.
Takeshita, M., Okuda, M., Okuda, S., Hyodo, A., Hamano, K., Furuya, N., & Tsuchiya, K. (2013). Induction of antiviral responses by acibenzolar-S-methyl against cucurbit chlorotic yellows virus in melon. Phytopathology, 103, 960–965.
Tally, A., Oostendorp, M., Lawton, K., Staub, T., & Bassi, B. (1999). Commercial development of elicitors of induced resistance to pathogens. In A. A. Agrawal, S. Tuzun, & E. Bent (Eds.), Induced plant defenses against pathogens and herbivores. Biochemistry, ecology, and agriculture (pp. 357–369). St. Paul: American Phytopathological Society.
Tornero, P., Gadea, J., Conejero, V., & Vera, P. (1997). Two PR-1 genes from tomato are differentially regulated and reveal a novel mode of expression for a pathogenesis-related gene during the hypersensitive response and development. Molecular Plant-Microbe Interactions, 10, 624–634.
van Loon, L. C., Rep, M., & Pieterse, C. M. J. (2006). Significance of inducible defense-related proteins in infected plants. Annual Review of Phytopathology, 44, 135–162.
Vandesompele, J., De Preter, K., Pattyn, F., Poppe, B., Van Roy, N., De Paepe, A., & Speleman, F. (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology, 3, research0034.1–0034.12.
Walters, D., Walsh, D., Newton, A., & Lyon, G. (2005). Induced resistance for plant disease control: Maximizing the efficacy of resistance elicitors. Phytopathology, 95, 1368–1373.
Woodcock, J., Moazed, D., Cannon, M., Davies, J., & Noller, H. F. (1991). Interaction of antibiotics with A- and P-site-specific bases in 16S ribosomal RNA. The EMBO Journal, 10, 3099–3103.
Yuan, J. S., Reed, A., Chen, F., & Stewart, C. N. (2006). Statistical analysis of real-time PCR data. BMC Bioinformatics, 7, 85.
Acknowledgements
This research was supported in part by a grant from the Ministry of Science and Technology, Taiwan, R.O.C. (MOST 104-2311-B-005-003-MY3) to C. -H. Huang. In addition, this study was partially funded through a USDA-AMS Specialty Crop Block Grant (grant number 018015) to the Florida Department of Agriculture and Consumer Services in partnership with the Florida Specialty Crop Foundation. We thank B. C. Hughes, C. M. Dyer, R. C. Willis, and H. M. Adkison for their technical assistance.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Informed consent
All the authors carefully read and approved this version of the manuscript. Authors declare that this manuscript has not been published elsewhere, and that neither the data nor images have been manipulated. This article does not contain any research involving human participants and/or animals.
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Huang, CH., Vallad, G.E. Soil applications of acibenzolar-S-methyl induce defense gene expression in tomato plants against bacterial spot. Eur J Plant Pathol 150, 971–981 (2018). https://doi.org/10.1007/s10658-017-1336-0
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-017-1336-0