Abstract
In recent years, several applications have emerged which require access to consolidated information that has to be computed and provided in near real-time. Traditional record linkage algorithms are unable to support such time-critical applications, as they perform the linkage offline and provide the result set only when the entire process has completed. To address this need, in this paper we propose the first summarization algorithms that operate in the blocking and matching steps of record linkage to speed up online linkage tasks. Our first method, called SkipBloom, efficiently summarizes the participating data sets, using their blocking keys, to allow for very fast comparisons among them. The second method, called BlockSketch, summarizes a block to achieve a constant number of comparisons for a submitted query record, during the matching phase. Our third method, SBlockSketch, operates on data streams, where the entire data set is unknown a-priori but, instead, there is a potentially unbounded stream of incoming data records. Finally, we introduce PBlockSketch, which adapts BlockSketch to privacy-preserving settings. Through extensive experimental evaluation, using real-world data sets, we show that our methods outperform the state-of-the-art algorithms for online record linkage in terms of the time needed, the memory used, and the recall and precision rates that are achieved during the linkage process. Following the evaluation of our approaches, we introduce SFEMRL, a novel framework that uses them to enable the linkage of electronic health records at large scale, while respecting patients’ privacy. Under this framework, patient records first undergo a data masking process that perturbs sensitive information in data fields of the records to protect it. Subsequently, they participate in a parallel and distributed ecosystem, whose goal is to persist these records in order to be queried efficiently and accurately. We demonstrate that the integration of our framework with Map/Reduce offers robust distributed solutions for performing on-demand large-scale privacy-preserving record linkage tasks in the health domain.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Record linkage, also known as entity resolution or data matching, is the process of identifying records that match, i.e., refer to the same real-world entity, in absence of common unique identifiers for records that belong to disparate data sources. Additional obstacles, such as the existence of writing variations (lack of standardization), errors, misspellings, and typos in various data fields, are commonly met in record linkage tasks and constitute record linkage a very challenging task. Traditional approaches to record linkage perform the linkage process offline and provide the result set only when the entire linkage process has completed. The process itself typically consists of two main steps, namely blocking and matching. In the blocking step, records that potentially match are grouped into the same block. Subsequently, in the matching step, records that have been blocked together are examined to identify those that actually match. Matching is implemented using either a distance function, which compares the respective field values of a record pair against specified distance thresholds, or a rule-based approach, e.g., “if the surnames and the zip codes match, then classify the record pair as matching”.
Several blocking approaches have been developed with the aim to boost the scalability of record linkage to Big data sets, without sacrificing accuracy [3, 9, 18, 39]. Given the massive size of modern data sets and the costly operations that have to be performed for record linkage, offline methods can take a significant amount of time—typically measured in hours or even days—to produce the matchings. This can be highly problematic in many real-world use cases where the linkage process must return a fast response in order to allow for emergency actions to be triggered. Let us assume, for example, a central crime detection system that collects data from several sources, such as crime and immigration records, central citizens’ repositories, and airline transactions. Query data about a suspect could be submitted to this system in order to be matched with any possible similar records found therein. The results of this process have to be reported as fast as possible or, at least, within an acceptably low time period, in order to trigger police enforcement actions.
As a second example, consider the recent series of bank and insurance company failures, which triggered a financial crisis of unprecedented severity. In order for these institutions to recover and return to normal business operation, they had to engage in merger talks. One of the driving forces of such mergers is the appreciation of the extent to which the customer databases of the constituent institutions are shared, so that the benefits of the merger can be proactively assessed in a timely manner. A very fast estimation of the extent of the overlap of the customer databases is thus a decisive factor in the merger process. To achieve this, the data custodians could use summaries of their databases in order to quickly estimate the overlap of their customers, instead of engaging in a tedious record linkage task. Although our motivation comes from the summarization of the blocking structure of a database, we believe that database summarization is an area of great interest with applications beyond record linkage.
As can be easily observed from the use cases discussed above, there exist many real-world scenarios in which the data records that have to be linked from the disparate data sources represent sensitive personal information about individuals. Such information must be sufficiently protected during the linkage task. Specifically, in this paper, we consider the summarization and linkage of patients electronic health records as a major use case in which record linkage has to be performed with high accuracy, at large scale, and with privacy provisioning. Performing record linkage on patients’ electronic health records enables hospitals to gain a comprehensive view of a patient’s medical history, as patients’ information is usually distributed across multiple disparate health providers. Besides the benefit to the patients, who can receive better treatment when doctors are aware of their complete medical history, a holistic view of patients’ medical information enables performing accurate medical research studies. Given the non-existence of a universal patient identifier across health care organizations, such an integration is not currently possible without the use of sophisticated, state-of-the-art record linkage techniques. Moreover, due to the high sensitivity of patients’ medical information, the integration of such data cannot be performed without the use of Privacy-Preserving Record Linkage (PPRL) [17] techniques. Such techniques are expected to not only comply with existing privacy legislation, but to offer privacy guarantees that go beyond legislative requirements, effectively offering protection to patients’ records from re-identification and sensitive information disclosure attacks. Even more, record linkage techniques are expected to deliver a high level of linkage quality, by performing accurate record matching and by significantly reducing the number of record pairs that require human intervention to be classified as matching or non-matching pairs. PPRL is the first important step towards the collective mining of data, coming from various health care providers, in order to facilitate the discovery of valuable insights. For instance, linked data from different medical providers can be used to support the discovery of new drugs, aid researchers in identifying novel drug targets, as well as new indications for existing drugs. Personalized care plans and automated care management workflows, that have resulted from a data analysis, allow to create informed action plans. Moreover, medical tests can be interpreted faster and with greater accuracy by analyzing and drawing inferences from large volumes of medical data.
To support real-world applications where record linkage (and PPRL) has to be performed in near real-time, several online record linkage approaches have been proposed in the research literature [7, 14, 30, 38]. All these approaches require the availability of large amounts of main memory, which is necessary in order to store their corresponding data structures. For instance, [7] utilizes large inverted indexes, while [14, 30, 38] sort the records to form blocks by leveraging large matrices or huge graphs. Despite several efforts to utilize small amounts of memory, e.g., [30], the results in terms of performance clearly indicate the inability of these algorithms to handle an increasingly large volume (or a continuous stream) of records in a real-time fashion. Given that main memory is always bounded and the number of records may in several real-world applications be unbounded, the performance of these data structures quickly degrades significantly. Furthermore, in order to deal with this plethora of records and detect the matching pairs, the proposed methods usually resort to conducting an excessive number of distance computations. This strategy, however, is not efficient, since it incurs significant delays to the record linkage process.
In the first part of the paper, we introduce four methods for efficiently managing large volumes of records in the context of online record linkage. Our first method, called SkipBloom, performs a summarization (synopsis) of the blocking structure of a data set using a small footprint of main memory, whose size is logarithmic in the number of distinct processed blocking keys. This synopsis can be easily transferred to another site (or used remotely) to estimate the common number of blocking keys. Such a preliminary estimation may bring to surface important insights, which can be further analyzed by the data custodians. The outcome of such analyses may encourage (or discourage) the data custodians to conduct a full-scale record linkage task. Our second method, called BlockSketch, tackles the problem of blocks that are overwhelmed with records, which should be compared against a query record to detect matching pairs. BlockSketch instead of implementing the naïve linear approach, compares the query record with a constant number of records in the target block, which entails a bounded matching time. In order to achieve this optimization, BlockSketch compiles, for each block, a number of sub-blocks, which reflect the distances of the underlying blocked records from the blocking key. The algorithm places a query record to the sub-block whose records exhibit the smallest distances from the query record. Our third method, called SBlockSketch, operates on data streams, where the entire data set is not known a-priori but, instead, there is an unbounded stream of incoming data records. SBlockSketch maintains a constant number of blocks in main memory at the cost of a time overhead during their replacement with blocks that reside in secondary storage. In this scheme, we propose a selection algorithm to effectively select the blocks that should be replaced, by taking into account their selectivity (by the incoming records) and age. Finally, we introduce PBlockSketch, which adapts BlockSketch to privacy-preserving settings. To the best of our knowledge, SkipBloom is the first algorithm for creating an appropriate synopsis of a blocking structure, while BlockSketch and SBlockSketch are the first methods for sufficiently summarizing a block for the needs of the matching phase of a record linkage task. SkipBloom, BlockSketch, and SBlockSketch were first introduced in [25].
In the second part, we propose a framework for linking and summarizing patients’ electronic records in a privacy-preserving manner. Our framework, called SFEMRL (Summarization Framework for Electronic Medical Record Linkage), incorporates PBlockSketch and a privacy-preserving algorithm for online operations called FPS [22]. SFEMRL allows for approximate matching in an embedding privacy-protected space by preserving the original distances. These methods can identify, with high probability, similar patients’ records, by applying an efficient blocking/matching mechanism. In the heart of SFEMRL resides LSHDB [23], which uses the Locality-Sensitive Hashing [15] technique to efficiently block the masked records, and store the produced blocking structures on disk for further use. LSHDB achieves very fast response times, which makes it ideal for online settings, thanks to the utilization of efficient algorithms and the employment of flexible and robust noSQL systems for storing the data. Utilizing a ring of LSHDB instances, we establish distributed data stores than can be easily queried, and we integrate them with Map/Reduce [11] for effectively processing big volumes of records. We also propose a new algorithm for the online operation of SFEMRL, which relies on the median trick and FPS, to accelerate its response time.
The rest of this paper is structured as follows: Sect. 2 presents the related work, while Sect. 3 outlines the building blocks utilized by our algorithms and provides the formal problem definition. Section 4 describes our proposed algorithms from both a practical and a theoretical point of view. Section 5 presents SFEMRL, used as shorthand for Summarization Framework for Electronic Medical Record Linkage, which is a complete framework for identifying electronic health records corresponding to the same patient that appear in medical data sets held by disparate health providers. The results of our experimental evaluation, including a detailed comparison with baseline methods, are reported in Sect. 6, while Sect. 7 concludes this work.
2 Related work
A significant body of research work has been conducted in record linkage during the last four decades. This work has been nicely summarized in a number of survey articles [6, 8, 13, 37]. However, only a very limited amount of work has targeted the area of near real-time record linkage, such as [1, 2, 7, 12, 19].
In [7], Christen et al. present an approach that involves a preprocessing phase, where the authors compute the similarities between commonly blocked values, using a set of inverted indexes. The authors use the double metaphone [5] method to encode the string values, which are then inserted into the inverted indexes. This scheme is extended in [34], where a heuristic method is presented to index the most frequent values of data fields. This method, though, requires a-priori knowledge of the values in certain fields and is not well-suited for settings where highly accurate results are needed. Ramadan and Christen in [33] utilize a tree structure where a sorting order is maintained according to a chosen field(s). A query record scans not only the node that is inserted, but also its neighboring nodes where similar records may also reside. Nevertheless, the distance computations that should be performed may degrade considerably the performance of this method in online settings.
Dey et al. in [12] develop a matching tree to speed up the decision about the matching status for a pair of records, so that it can be made without the need to compare all field values. However, the performance of this method depends heavily on the training of the matching tree, which requires a large number of record pairs. Moreover, the authors do not draw any attention to the acute problem of reducing the record pairs comparisons. Ioannou et al. in [19] resolve queries under data uncertainty, using a probabilistic database. The effectiveness of their method heavily depends on the potential of the underlying blocking mechanism, which is used implicitly, to produce blocks of high-quality. In [1], Altwaijry et al. propose a set of semantics to avoid resolving certain record pairs. Their scheme, however, focuses on how to resolve generic selection queries (e.g., range queries), rather than on minimizing the query time.
There is also another body of related literature that deals with progressive record linkage (e.g., [14, 30, 38]). These techniques report a large number of matching pairs early during the linkage process and are quite useful in the event of an early termination of the linkage process, or when there is limited time available for the generation of the complete result set.
The solutions proposed by Whang et al. in [38] and Papenbrock et al. in [30] are empirical and rely heavily on lexicographically sorting the input records to formulate clusters of similar records. Although the sorting technique is quite effective in finding similar values in certain cases, it cannot guarantee identification of matching record pairs. Consider, for example, the similar strings ‘Jones’ and ‘Kones’, where the first letter has been mistyped; using [30, 38], the corresponding records would definitely reside in different clusters (assuming a large number of records). Consequently, the corresponding pair of records would never be considered as matching.
More recently, Firmani et al. [14] introduced two progressive strategies that provide formal guarantees of maximizing recall, focusing though only on minimizing the number of queries to an oracle (which is an entity that replies correctly about the linkage status of a pair) and not on minimizing the running time. Both strategies implicitly assume an underlying blocking mechanism that has been applied on the data sets, and heavily rely on the effectiveness of that blocking mechanism. Their most serious shortcoming is the excessive amount of time-consuming similarity computations, which need to be performed between the formulated pairs in the blocks, without achieving any increase in recall. For example, in a data set of 3 million records (including the query set), more than 1.3 billion similarity computations should be performed without reporting any results!
There is also another body of work, termed as meta-blocking [28, 29], which investigates how to restructure the generated blocks with the aim of discarding redundant comparisons. Meta-blocking techniques, however, conduct a cumbersome transformation of a blocking structure into a graph, which renders these techniques not applicable to online settings.
In Sect. 6, we elaborate further on the approaches of Christen et al. and Firmani et al., which are the state-of-the-art methods with which we compare our proposed techniques.
3 Background and problem formulation
In this section, we first introduce the necessary background and terminology for the understanding of our proposed schemes, and subsequently derive the problem statement.
3.1 Skip list
A skip list [31] is a probabilistic data structure that is designed to provide fast access to an ordered set of items. It is actually a sequence of lists, or levels, where the first list, termed as the base level, contains all the items inserted so far in sorted order. Each successive list is a copy of the previous with some elements skipped, until the empty list is reached. Its randomization lies in the number of levels an item will join, determined by tossing a fair coin.Footnote 1 Each item of each list is linked to the same item in the previous list, as well as to the next item at the same level. The query operation for an item starts at the top-level, by horizontally scanning the items therein until it encounters either the target item or a larger item. In the case of a larger item, the same process is repeated at the lower level until the base level is reached. The running time to insert an item, as well as to report the existence of an item, is \({\mathcal {O}}(\log (n))\), where n is the number of inserted items.
3.2 Bloom filter
A Bloom filter [4] is a probabilistic data structure for representing a large number of items using a small number of bits, which are initialized to 0, to efficiently support membership queries. Each item is hashed by a set of universal hash functions that map it to certain positions, chosen randomly and uniformly, in the Bloom filter. Accordingly, these positions are set from 0 to 1. Upon querying for an item, the same process is followed, where:
-
one can definitely infer that this item has not appeared, if all retrieved positions are set to 0.
-
one can conjecture that this item has appeared with certain probability, if all retrieved positions are set to 1. The probabilistic nature of the reply is due to the fact that these positions may have been set to 1 by other items and not the query item.
3.3 Locality-sensitive hashing
For the blocking step, we employ the well-established randomized Locality-Sensitive Hashing (LSH) technique, which has been studied in detail as a blocking scheme in [21, 24]. There have been devised several LSH families, which work in certain metric spaces, e.g., the Hamming [15], Min-Hash [32], or the Euclidean [10] space. The hash functions of each such family are locality-sensitive to the corresponding distance metric.
LSH guarantees that each similar record pair is identified with high probability using a strong theoretical foundation. The similarity between a pair of records is defined by specifying an appropriate distance threshold \(\theta \) in the metric space that is used.
LSH is implemented by building a redundant structure T which consists of L independent hash tables, each denoted by \(T_l\) (where \(l=1,\ldots ,L\)). We assign to each hash table a composite hash function \(g_{l}\), which consists of a fixed number k of base hash functions. Depending on the used LSH family, a composite hash function applied to a masked record returns a result that is used as a hash key to the corresponding hash table. Figure 1 conveys this multi-hash operation. The masked records which exhibit the same hash key for some \(T_l\) share a common bucket. Intuitively, the smaller the distance of a pair of masked records is, the higher the probability of a \(g_l\) to produce the same hash result.
The optimal number L of hash tables used depends on the chosen values of k and \(\theta \). For the optimal values of these parameters, we refer the interested reader to relevant discussions in [20, 21].
During the matching step, those sanitized records that have been inserted into a common bucket, formulate pairs which are then compared and classified as similar or dissimilar pairs according to the specified distance threshold.
3.4 Overview of LSHDB
In order to perform PPRL tasks, we use LSHDB [23], a distributed engine that we developed, which leverages the power of LSH and parallelism to perform record linkage and similarity search tasks. LSHDB creates data stores for persisting records using the key-value primitive as its fundamental data model. The key part is the hash result of an LSH hash function, while the value is the Id of the record being hashed. It is important to note that hashing the records and maintaining them to a ready-for-linkage state may save working hours, since PPRL is an ongoing process that may be executed several times by changing the configuration parameters to obtain the most accurate result set.
Upon the creation of a data store, the developer needs to specify only two parameters: (i) the LSH method that will be employed, e.g., Hamming, Min-Hash, or Euclidean LSH, and (ii) the underlying noSQL data engine that will be used to host the data. Support of any noSQL data store and/or any LSH technique can be provided by extending/implementing the respective abstract classes/interfaces. The concreteFootnote 2 classes include the definition of the locality-sensitive hash functions and the implementation of the generic get/set concepts of the key/value primitive. By default, LSHDB supports the Hamming and Min-Hash LSH methods and two open-source Java-embedded noSQL engines: LevelDBFootnote 3 and BerkeleyDB.Footnote 4
LSHDB resolves each submitted query in parallel, by invoking a pool of threads, to efficiently scan large volumes of data. Moreover, a query can be forwarded to multiple instances of LSHDB to support data stores that have been horizontally partitioned into multiple compute nodes. To the best of our knowledge, LSHDB is the first record linkage and similarity search system in which parallel execution of queries across distributed data stores is inherently crafted to achieve fast response times.
3.5 Problem statement
Consider two data custodians who own data sets A and B, respectively. For each record r of A (or B), the data custodians use a function \(k=\textit{block}(r)\) that generates the blocking key k of r. This key is used to locate a target block in the blocking structure to either insert r into the target block (blocking), or iterate all the records already found therein and compare them with r (matching). We use \(D_A\) and \(D_B\) to denote the set of blocking keys of each of these data sets. Moreover, we refer to the fraction \({\mathcal {D}}=\frac{|D_A \cap D_B|}{|D_B|}\), as the overlap coefficient between A and B.
In this work we introduce three algorithms, namely SkipBloom, BlockSketch, and SBlockSketch, for addressing the following problemsFootnote 5:
Problem Statement 1
Calculate the overlap coefficient for A and B, by accurately summarizing \(D_A\) and \(D_B\) using sublinear memory requirements and sublinear running time in the number of inserted blocking keys.
Problem Statement 2
For each query record of A (or, equivalently, B), find the set of its matching records from B (or, equivalently, A) in constant running time.
Problem Statement 3
For each query record of A (or, equivalently, B), find the set of its matching records from B (or, equivalently, A) in constant time, using also a constant amount of main memory.
4 Algorithms and data structures
In this section, we present our proposed algorithms for efficiently managing large volumes of records in the context of online record linkage.
4.1 The operation of SkipBloom
SkipBloom is an efficient blocking data structure that reports membership queries of blocking keys (derived from a large data set) to the blocking structure, using only a small footprint of main memory. It implements the following generic operations:
-
query(k): Reports the membership (true or false) of key k to SkipBloom.
-
insert(k): Inserts key k into SkipBloom.
The operation of SkipBloom is based on a skip list that implements a mechanism to locate efficiently a blocking key, as well as on a series of Bloom filters, which are used as fast memory-bounded buffers.
SkipBloom maintains, in expectation, \(\sqrt{n}\) blocks in main memory, stored in the base level of the skip list. Each such block, which is represented by its key,Footnote 6 includes a list of Bloom filters in order to store keys that have been driven by the mechanism of the skip list to this block. This actually means that the keys stored in the Bloom filters of a block are greater than the value of the corresponding key.
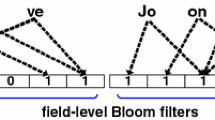
The operation of SkipBloom is illustrated in Fig. 2. In this figure, a skip list is shown that contains five keys in the base level. Upon receiving a query record, which is first filtered by a blocking function to generate its key (e.g., \(k={`John'}\)), SkipBloom locates the block \({`Jack'}\) very fast, using the logarithmic runtime property of the underlying skip list. According to the operation of skip lists, this block is alphabetically the nearest key to k from the left. The next step is a simple insertion of k into a Bloom filter of \({`Jack'}\). Each block has an active Bloom filter, termed as current, and a number of inactive Bloom filters, which are used only during the query process, as we will shortly explain.
To answer a query on whether a certain key k exists or not, SkipBloom follows almost the same process as described above. Assume, for example, that SkipBloom receives the query \(k={`Jonathan'}\). First, the skip list will be scanned to eventually locate ‘Larry’. Subsequently, each Bloom filter of this block will be iteratively queried until k is found, or the Bloom filters of ‘Larry’ are exhausted.
In what follows, we provide details that will justify certain design choices, such as the reason for maintaining a series of small (in length) Bloom filters in each block, instead of having a larger one. In order to populate the skip list with keys, we apply a simple Bernoulli random sampling algorithm that chooses each key with probability equal to \(n^{-1/2}\). This sampling process ensures the uniform reflection of the distribution of keys from the data set to the skip list. This is an appealing feature, since SkipBloom easily tackles distribution anomalies, such as skews of certain ranges of keys, by choosing these keys and inserting them into the skip list to effectively reduce the bottleneck of certain keys and maintain uniformity (in expectation). Any uniform sampling method can be applied; we refer the interested readers to a comprehensive survey in [16].
If a large number of similar keys are generated, then the sampling routine will choose randomly similar keys to create the corresponding blocks. For example, consider the case of blocking a large number of surnames from the US census data. Then, possible blocks might be ‘Johns’, ‘Johnson’, and ‘Johnston’, which will be created in this particular chronological order. Consequently, there will be keys other than ‘Johns’, e.g., ‘Jordan’ or ‘Jolly’, that will be inserted into the Bloom filters of ‘Johns’. These Bloom filters should be now transferred to (or referenced by) ‘Johnson’, and then to (by) ‘Johnston’. For this reason, we keep the number of keys that can be inserted into each Bloom filter small; this number will be accurately specified later. Moreover, we annotate each Bloom filter with its smallest and its greatest key, in terms of alphabetical order. By doing so, upon inserting ‘Johnson’, SkipBloom scans iteratively the Bloom filters of ‘Johns’ to locate Bloom filters that might contain ‘Johnson’, or any greater values. If such Bloom filters exist, a simple reference is established between the block of ‘Johnson’ and the corresponding Bloom filters. Fig. 3 illustrates the reference of a block to a Bloom filter that belongs to the previous block.
Eventually, a record is stored into a key/value database system, maintaining its original blocking key, regardless of the block that was used in SkipBloom.
4.2 Algorithms
Algorithm 1 illustrates the query operation of SkipBloom. First, the skip list SL is queried to locate the nearest key p to the query k (line 1). Then, the Bloom filters that are both maintained and referenced by pFootnote 7 (line 2) are scanned iteratively to find k using the min and max values of each Bloom filter (line 4). If k is found, then the algorithm terminates (line 6). In case of composite keys, we perform a conjunction using the individual keys.

Algorithm 2 outlines the insertion of a key in SkipBloom. For each key k derived from each record, we determine with probability \(\frac{1}{\sqrt{n}}\) whether k will be inserted into the skip list or not (line 1). In more detail, we generate a random value in (0, 1) and then pick k if this value is less than \(\frac{1}{\sqrt{n}}\). Since the generation of a random value is an expensive operation, we exploit the fact that the number of keys skipped between successive inclusions follow a geometric distribution [16]; accordingly, each time we pick a key, we generate the position of the next key, in the stream of records, that will be picked.

If a key k will be inserted into the skip list as a base level key, then a block is created after the nearest key to k (line 2). Then, SkipBloom has to locate each Bloom filter of p that may contain keys that should be now transferred to the newly created block of k (lines 4–8). In order to easily locate these Bloom filters, we annotate each Bloom filter used with the min and max keys it contains (line 5). The inclusion of a Bloom filter with a valid range of keys is achieved through a reference from p to k.
If a key will not be stored in the skip list, then the nearest key p to k is located in order to insert k in the current Bloom filter of p (lines 10–12). Algorithm 2 eventually updates the min and max annotations of the current Bloom filter of p (lines 13–18).
4.3 Accuracy and complexity analysis
As we expect \(\sqrt{n}\) blocks in the base level of the skip list, where the sampling process ensures a uniform distribution of the corresponding blocking keys, the expected number c of keys in each block is:
By setting \(u=\sqrt{n}/m\) to be the maximum number of keys that will be stored in each Bloom filter, where m is a constant value (e.g., \(m=10\)), the number of Bloom filters in each block will be (in expectation) equal to m. Furthermore, the number \(m_{bt}\) of Bloom filters contained in block b at time t, specifies the upper and lower bound of the number \(n_{bt}\) of the distinct keys inserted, which is:
The accuracy of SkipBloom to report the existence of a key depends on the false positive probability parameter \(\textit{fp}\) of the Bloom filters. First, consider the event where a query key does not exist in any Bloom filter of the resulting block. The probability of reporting correctly this event, using one such Bloom filter, is \(1-\textit{fp}\). Hence, the same probability by using collectively all the m Bloom filters is:
since the content of a Bloom filter is independent from that of another filter.
In the case that a query key does exist in anyFootnote 8 Bloom filter of the resulting block, the probability of reporting this event is 1. Therefore, we bound from below the error probability of SkipBloom by \(1 - (1 - \textit{fp})^{m}\).
Computational complexity Based on Algorithm 1, the running time of querying SkipBloom is \({\mathcal {O}}(\log (\sqrt{n})+m+m\sqrt{n})\), where the first term denotes the time of scanning the skip list to locate the appropriate block, the second term denotes the time of scanning the Bloom filters found therein, and the last term is the time of scanning the Bloom filters referenced directly or indirectly by the chosen block.
Algorithm 2 suggests that the running time of an insertion of a key into SkipBloom is \({\mathcal {O}}(\log (\sqrt{n})+m)\), where the two terms are the time of inserting a key into the skip list and the time of scanning the Bloom filters of the nearest key, respectively.
Memory complexity The memory requirements of SkipBloom are \({\mathcal {O}}(2\sqrt{n}+\sqrt{n}m) = {\mathcal {O}}(\sqrt{n}(2+m)\), because the skip list contains \({\mathcal {O}}(2\sqrt{n})\) keys and each key in the base level of the skip list consists of \({\mathcal {O}}(m)\) Bloom filters.
4.4 Using SkipBloom as a synopsis of the universe of blocking keys
SkipBloom can be used as a synopsis, termed also as summarization, of the universe of the blocking keys of a database, in order to facilitate an accurate pre-blocking process. During the execution of this process, the data custodians will resolve very fast the common blocks, which will be of great assistance in estimating the running time, in terms of the number of comparisons that will be needed (by exchanging the number of records in each common block). In turn, the data custodians will determine whether they will perform the linkage process or not, by considering several factors based on these preliminary results. For instance, if the number of common blocks is very small, then (a) the chances of identifying similar, or matching, record pairs are rather slim, and (b) the record linkage process itself may not be cost-effective.
Let us now consider the following scenario. Data custodian A generates a SkipBloom from database A, which is submitted to data custodian B. Subsequently, data custodian B iterates her blocking keys and queries the SkipBloom, which reports positive or negative answers for the existence of the query keys. This entails \({\mathcal {O}}(n(\log (\sqrt{n})+m+m\sqrt{n}))={\mathcal {O}}(n(\log (\sqrt{n})+\sqrt{n}))\) running time,Footnote 9 since each key of B is queried against the SkipBloom of A.
To further accelerate this process, data custodian B also generates a SkipBloom, to compile a uniform sample of keys and to use this SkipBloom to report membership queries. The keys found in the base level of the skip list are now queried against the SkipBloom of A, as illustrated in Fig. 4.
Since, the keys of B constitute a randomly and uniformly chosen sample, they can be used as input to a Monte Carlo simulation [27], which will estimate the proportion (or the number) of identical blocking keys between the data sets of the two data custodians. Using only the synopses, the data custodians will acquire a clear picture about the overlapping keys with certain approximation guarantees. Monte Carlo simulation requires \((\epsilon ^2\vartheta )^{-1}\) (ignoring a small constant factor) keys from B in order to exhibit relative error \(\epsilon \) with high probability. Since the proportion of identical keys is unknown, we bound it from below with a reasonable value, e.g., \(\vartheta =0.05\), to approach the number \(\sqrt{n}\) of the sampled keys that are contained in the SkipBloom of B. Even for a relatively small \(n=10^8\), the Monte Carlo simulation will provide its guarantees, since \(\sqrt{n}\) is greater than the required number of sampled keys for \(\epsilon \ge 0.05\). The fraction of the overlapping keys found in the sample is used as an estimate for the overlap coefficient of the keys between the two databases. By comparing the synopses, we eventually achieve the much faster \({\mathcal {O}}(\sqrt{n}(\log (\sqrt{n})+\sqrt{n}))\) running time, compared to using only the synopsis of data custodian A.
4.5 The operation of BlockSketch
The existence of blocks that contain a large number of records makes the matching phase (i.e., the comparison of query records against every record found in a target block) prohibitively expensive in highly demanding environments. The situation becomes even more challenging in environments where the matching record pairs have to be reported in near real-time.
To address this shortcoming, in this work we opt for a different strategy: we compare the query record with a constant number of records of the target block, which entails a bounded matching time. This optimization requires maintaining \(\lambda \) sub-blocks \((S_1, S_2,\ldots ,S_\lambda )\) in each block, whose aim is to represent sufficiently the records inserted so far. In our proposed representation, a number of records play the key role of representatives for each sub-block. This allows to formulate groups of records inside each block that are more likely to match. We term our proposed algorithm as BlockSketch, because a small number of records comprise a sketch that represents sufficiently the records of an entire block. The concept of sufficient representation boils down to choosing representatives that exhibit certain distances from the corresponding blocking key. We note that BlockSketch can operate either autonomously or in conjunction with SkipBloom, where the latter will be used as a fast bounded memory to report whether a certain blocking key has appeared or not.
The fact that certain records are inserted into a block, using a blocking function, implies that all these records share some degree of similarity. Therefore, it is reasonable to assume that the distance between a key and a recordFootnote 10 will be upper bounded by \(\lambda \theta \). Hence, BlockSketch formulates \(\lambda \) sub-blocks, each of which represents records with distances \( \le \theta , \le 2\theta , \cdots , \le \lambda \theta \) from the key, where \(\theta \) is the distance threshold of the keys of a pair of matching records. Upon receiving a key, BlockSketch aims to insert this record into the sub-block of the target block, where it is more likely to formulate matching record pairs. For this reason, each key is compared against all representatives found in a block, in order to locate the sub-block whose representative exhibits the smallest distance from the newly arrived key.
As an example, assume that we use edit distance as the similarity metric, \(\theta =2\) and \(\lambda =3\), and a blocking key is used that consists of the first three letters and the whole value of the surname and given name attributes, respectively. As Fig. 5 shows, record \(<{`John'}, {`Jones'}, 1970>\), whose key values exhibit a total distance of \(2 \le \theta \) from \(<{`John', `Jon'}>\), is inserted into the 1-st sub-block, because of the representative \(<{`John'}, {`Jon'}>\). Similarly, \(<{`John'}, {`Jonker'}, 1975>\), whose distance is \(3 \le 2\theta \) from \(<{`John'}, {`Jon'}>\), is inserted into the 2-nd sub-block, due to the comparison with the representative \(<{`John'}, {`Jonkers'}>\).
It is important to note that for threshold \(\theta \) any metric that is used in record linkage processes can be supported, whether satisfying the triangle inequality or not. For example, a very commonly used metric in record linkage is the Jaro-Winkler similarity function [5], which takes on values in [0, 1]. Hence, one by setting the similarity threshold to \(\theta '\), and then by choosing \(\theta =1-\theta '\), produces very reasonable sub-blocks.
The probability for a record to fall into a certain sub-block that holds its matching record, depends on the representatives of the target sub-block, as well as on the left and right neighboring sub-blocks. For instance, assume two neighboring sub-blocks with representatives ‘Jacks’ and ‘Jackson’, respectively. The keys of these representatives comprise the values of the ‘surname’ attribute. Key ‘Jackson’ arrives, whose record is inserted into the identical sub-block of ‘Jackson’. At a later time, ‘Jacksn’ arrives, that suffers from a typo, whose record is inserted into the sub-block of ‘Jacks’. We have thus missed the formulation of one matching record pair. BlockSketch tackles this deficiency by using more than one representatives for each sub-block,Footnote 11 so as to give more chances for grouping together matching record pairs. By doing so, if record a has been inserted into a sub-block, BlockSketch compares the key of its matching record b with more similar representatives to record a. To keep the number of representatives of a sub-block constant, whenever a key is chosen for inclusion in a sub-block, the algorithm tosses a coin to determine if this newly inserted key would be a representative as well. If it is chosen, a randomly picked old representative is evicted from the set of representatives.
Illustration of a block with \(\lambda =2\) sub-blocks, whose key is \(<{`John', `Jon'}>\). BlockSketch inserts records into the sub-blocks based on the distance of the key values of these records from the chosen representative(s). The sub-block for which one of its representatives exhibits the smallest distance from the key values of a record, is chosen as the target sub-block
As a last step, the query record is inserted into that sub-block which is maintained by a key/value database. The pairs formulated in this sub-block constitute the final result set.
4.6 Algorithm
Algorithm 3 outlines the basic operation of BlockSketch. For a query record q, the algorithm first retrieves an object S that contains the corresponding sub-blocks, either from a key/value database or from a cache structure in main memory (line 2). BlockSketch then iterates over the representatives of each sub-block and performs the distance computations between the key values of q and these representatives,Footnote 12 whose results are stored in array u (line 5). The representative that exhibits the smallest distance from the key values of q specifies the sub-block (line 12) into which q is finally inserted (line 17). For ease of presentation, we omit from Algorithm 3 the details regarding the random choice and eviction of a representative from a sub-block.

4.7 Accuracy and complexity analysis
The probability of a record to fall into the correct sub-block is \(1/\lambda \), since it completely relies on the distance from the corresponding representative. Hence, the inverse probability of a record not falling into the correct sub-block, and therefore not formulating a record pair, is \(\le 1 - 1/\lambda \). In order to amplify the probability of formulating a matching record pair, we give more chances for grouping together the two constituent records, by comparing each key with a number \(\rho \) of representatives, chosen randomly and uniformly from the underlying stream. We rigorously specify the required number of representatives that each sub-block should maintain, as the following lemma suggests.
Lemma 1
If a pair of records, which constitute a matching pair, has been brought in a certain block, then by maintaining \(\rho = \lambda \ln (\frac{1}{\delta })\) representatives in each sub-block, this matching pair is detected with probability at least \(1- \delta \).
Proof
The probability of not detecting a matching pair that exists in a certain block is \((1 - \frac{1}{\lambda })^\rho \). We bound this probability above by \(\delta \) and solve for \(\rho \) in the following:
since \(\ln (1-\frac{1}{\lambda }) \le -\frac{1}{\lambda }\). \(\square \)
We subsequently apply the ceiling function on the value of \(\rho \) \((\lceil \cdot \rceil )\), in order to select the smallest integer following \(\rho \) for the sake of optimality.
Computational complexity The running time of BlockSketch is \({\mathcal {O}}(\log {n}+\lambda \rho )\), which consists of the time to retrieve a block from the database (which is logarithmicFootnote 13), and the execution of the subsequent \(\lambda \times \rho \) distance computations (\(\rho \) representatives for each of the \(\lambda \) sub-blocks).
Memory complexity The storage requirements of BlockSketch are \({\mathcal {O}}(\lambda n)\), where n is the number of blocking keys.
4.8 The operation of SBlockSketch
Let us now suppose that the number of records, which are initiated from multiple sources, e.g., from different hospitals, is unbounded (or endless). This literally turns the record linkage scenario of a large number of records, into the record linkage of a stream of records. Therefore, BlockSketch will grow in both directions; it will not grow only in terms of sub-blocks, but also its number of blocks might unexpectedly grow considerably. Since our main memory is bounded, BlockSketch adapts its operation to record linkage tasks that involve streams of records.
In this version of BlockSketch, called SBlockSketch, we bound the number of blocks, that are maintained in main memory, by an integer value \(\mu \) which depends on the available main memory. Since the number of blocks is bounded, SBlockSketch applies an eviction strategy, so as to insert a newly arrived blocking key from the stream, when there is not an empty slot to accommodate the corresponding block. We annotate each liveFootnote 14 block with (a) the number of incoming records that generated its key, i.e., the number \(\xi \) of times this block has been chosen as the target block, and with (b) its age \(\alpha \), in terms of the number of times that this block has survived eviction, since its admission into main memory. We derive the eviction status of each block as follows:
where factor w adjusts the weight of successes \(\xi \) of a block to its es. The intuition behind this scheme is that we promote (a) newer blocks against older ones, and (b) blocks that exhibit higher eligibility. The status of old blocks, that are additionally not chosen by the incoming records, will exponentially decay, which will result in their eviction from the main memory. SBlockSketch is materialized by a hash table, which holds the live blocks, and the corresponding sub-blocks, and a priority queue, that is used to indicate which of these live blocks should be evicted in case of a newly arrived block (key).
In this example, SBlockSketch uses a hash table T with \(\mu =4\) blocks, \(\lambda =3\) sub-blocks, and the weight of successes set to \(w=1.5\). On the arrival of an incoming new key, the block with key \(k_4\) is evicted because of its low eviction status. The priority queue pq stores the eviction status (on a logarithmic scale) of each live block
Figure 6 illustrates the components of SBlockSketch, namely the hash table T and the priority queue pq. T exists in main memory and contains a specified number \(\mu \) of rows, each of which holds a block, as a function of the available main memory. Each row of T contains the sub-blocks of the corresponding block. The priority queue pq stores the eviction status of each live block in ascending order, so as to return the key of the block that holds the minimum eviction status. In the example shown in Fig. 6, we observe that the block with key \(k_4\) has survived \(\alpha =4\) evictions and has not been chosen as target block since its admission into T. These two events lead inevitably to its eviction, despite the existence of block \(k_2\), which has \(\alpha =10\) survivals, but it additionally exhibits \(\xi =6\) successes.
4.9 Algorithm
Algorithm 4 illustrates the operation of SBlockSketch, using a stream of data records. Upon receiving a record from the stream, the algorithm first derives its key, and then queries T (line 2). Only if this query is fruitless, SBlockSketch resorts to the structures of secondary storage (line 4). If the block that corresponds to the incoming record exists neither in T nor in secondary storage, then SBlockSketch initiates the eviction of the block from T that exhibits the minimum eviction status, as indicated by pq (line 7). Eventually, SBlockSketch computes the eviction status of each live block and rebuilds pq.

4.10 Accuracy and complexity analysis
The accuracy of SBlockSketch is not affected by the use of T, since the block in question may exist either in main memory or in secondary storage. However, T, whose operations are of \({\mathcal {O}}(1)\) time, affects running time and space.
Computational complexity The running time depends on two mutually exclusive possibilities. The first one is when a block exists in T, where the running time is \({\mathcal {O}}(\lambda )\) (see Sect. 4.7), while the other possibility is when a block should be evicted from T. The eviction requires accessing the priority queue, which is of \({\mathcal {O}}(\sqrt{\mu })\) time, and then transferring the incoming block into T. The latter step consumes, as we discussed in Sect. 4.7, \({\mathcal {O}}(\log (n))\) time in the number n of available blocks found in the secondary storage. Finally, we have to add the time to build the priority queue, which is \({\mathcal {O}}(\mu \log (\sqrt{\mu }))\). Hence, the total running time for replacing a block is \({\mathcal {O}}(\sqrt{\mu }+\log (n)+\mu \log (\sqrt{\mu }))\).
Memory complexity The space occupied in main memory is exactly \({\mathcal {O}}(\mu \lambda )\), where \(\mu \) corresponds to the rows and \(\lambda \) to the cells of T (by assuming T as a two-dimensional array).
4.11 The operation of PBlockSketch
In this section, we present PBlockSketch, used as shorthand for Private BlockSketch. This version adapts BlockSketch to the LSH scheme using Bloom filters to perform PPRL.
An LSH base hash function (see Sect. 3.3), when applied to a Bloom filter with length \(\gamma \), returns the value of its i-th position, where \(i \in \{0,\ldots ,\gamma -1\}\), chosen uniformly at random. Those Bloom filters which exhibit the same hash value for some \(T_l\) share a common bucket. Intuitively, the smaller the Hamming distanceFootnote 15 is, the higher the probability of a \(g_l\) to produce the same result. During the matching step, those Bloom filters which have been inserted into a common bucket, formulate pairs which are compared and classified as similar or dissimilar, according to the used Hamming distance threshold.
Applying the above-mentioned straightforward technique using voluminous data sets of Bloom filters will inevitably result in overpopulated blocks in each \(T_{l}\). Consequently, the matching phase would fail to generate results in a timely manner. PBlockSketch unloads the matching phase by rehashing each query Bloom filter to obtain a fresh key value using another pre-generated \(g_l\) for each \(T_l\). The newly created blocking key will be compared with each representative of the target block in a \(T_l\).
For example using Hamming distance, assume that we set \(\theta =1\) and \(\lambda =3\). Bloom filter B1 is inserted into the 1-st sub-block because its rehashed key exhibits distance equal to 1 from the respective representative. Similarly, Bloom filter B3 occupies the first slot of the 2-nd sub-block due to its distance from the representative ‘1101001011’ (Fig. 7).
5 SFEMRL: a summarization framework for electronic medical record linkage
In this section, we present SFEMRL, used as shorthand for Summarization Framework for Electronic Medical Record Linkage. SFEMRL is a complete framework for identifying electronic health records corresponding to the same patient, which appear in medical data sets held by disparate health providers, thereby enabling the integration of these records in order to produce a holistic view of the patient’s medical history.
The most common model for offering PPRL involves a Trusted Third Party (TTP), e.g., a ministry of health, that receives the records, which have undergone a maskingFootnote 16 process, from the data custodians through a secure channel and performs their linkage. The TTP is assumed to hold the records in a secure environment that is trusted by the data custodians. This model is known as the three-party model and is typically enforced by established specialized linkage units. A different approach to offering PPRL is via the two-partyFootnote 17 model. In this model, the only participants are the data custodians. The adversary PPRL models are extensively discussed in [37].
In the first step of PPRL, the healthcare providers mask their collection of electronic patient records in order to protect certain (common) direct identifiers, such as patients’ names and home addresses, that are useful for enabling record linkage [37]. This masking process is specially crafted to both protect the identity of the patients represented, and simultaneously enable the blocking and matching phases of PPRL in an approximate manner. Other direct identifiers, such as patients’ medical record numbers, are removed from the data as they are both sensitive and not useful for PPRL (due to not being universal). Last, non direct identifiers, such as symptoms or medication, remain unmasked to facilitate data analysis based on these dimensions. The processed data are securely transmitted to a TTP and stored in a secure environment (following legal requirements). The TTP performs the PPRL process using the masked data to detect those records that describe the same patients. The integrated records, deprived of patient identifiers, are subsequently stored into a data lake from where they can be queried and retrieved for research and data analysis purposes. Figure 8 highlights this process.
Certain legislation has also been enacted that currently governs the collection and release of private medical data on both sides of the Atlantic. In the United States, the Health Insurance Portability and Accountability Act (HIPAA),Footnote 18 establishes national standards for electronic health care transactions and national identifiers for providers, health insurance plans, and employers. The HIPAA Privacy Rule requires that certain field values, which might uniquely identify an individual, such as names or biometric information, must be sanitized in patients’ records, before these records are released or shared with another health provider. Similarly in the European Union, the GDPR (General Data Protection Regulation)Footnote 19 protects the privacy of all personal data related to EU citizens. Moreover, it is important to note that there are established specialized linkage units,Footnote 20 which play the role of the TTP by respecting the above-mentioned regulations.
The main requirements of a framework that aims to perform an effective and efficient linkage of electronic health records are:
-
Protect the privacy of the patients Confidentiality is central to the preservation of trust between health providers and their patients. Additionally, when breaching patient confidentiality, it is very hard to provide adequate justifications on a legal basis.
-
Achieve a high level of matching accuracy Accurately matching health electronic records has a great impact on business analysis and intelligence. Correctly identifying records held by different health care providers that refer to the same patient can have a tremendous effect in the quality and the accuracy of the performed analysis on the collected data.
-
Support many masking methods The diversity of masking methods is expanding rapidly in the literature, each of which exhibits different features and characteristics. Supporting state-of-the-art masking methods, or being capable of easily incorporating them, will give more options of choosing the suitable blocking method that will be applied on the masked records.
-
Handle large data volumes Nowadays, health care organizations are struggling to manage large volumes of data that exist within and outside of their systems and infrastructures. PPRL solutions should be able to scale well to big data, by offering effective blocking algorithms and efficient matching techniques.
The TTP maintains a number of sites, as shown in Fig. 9, each of which holds a horizontal partition of the masked records that have been previously submitted by the data custodians. PBlockSketch along with an LSHDB instance should be up and running in each site, and be populated with the corresponding Bloom filters. A query record, in terms of a Bloom filter, is submitted to a central site, which has been specified to receive the query records, forward them to the remaining sites, and eventually collect the results asynchronously.
This mode has two main advantages: (a) there is no mass release and maintenance of records at a single site, and (b) SFEMRL may easily scale, e.g., geographic of administrative scalability, to deal with significantly increased loads.
In the following, we delve into the details of SFEMRL, which first masks, then blocks, and finally matches the records at hand using appropriate masking methods and LSH techniques. For the sake of simplicity, assume two data custodians who might be two health care institutes that own databases A and B, respectively, and have a-priori agreed to use a common schema. Configuration parameters, such as the the hash functions used for the generation of the sanitized records, are securely communicated between the data custodians. We also demonstrate the integration of our framework with Map/Reduce to provide robust solutions using very large volumes of records.
5.1 Integration with map/reduce
SFEMRL utilizes the Map/Reduce [11] programming paradigm to scale up its performance to large volumes of records. The map phase of Map/Reduce pertains to the blocking step of PPRL, while the reduce phase to the matching step. In the following, we summarize the functionality of a generalized PPRL Map/Reduce pipeline:
-
Map phase. Each map task builds the L hash keys of each Bloom filter at hand and emits them to the partitioner task along with the Id of the corresponding record. A pair of a hash key and an Id is termed as a tuple.
-
Distribution of tuples. Each partitioner task, which is always bound to a map task, controls the distribution of the formulated tuples to the reduce tasks. Tuples with the same hash key will be forwarded to a specific reduce task. An efficient partitioning scheme distributes the computational load equally to the available reduce tasks, which has as a result the least possible idle time for each reduce task.
-
Reduce phase. Each reduce task processes the load of the received tuples forwarded by the partitioner tasks. This processing usually includes the retrieval of the corresponding masked records from a data store to perform the distance computations.
This pipeline constitutes a PPRL Map/Reduce job. More complex settings may require additional jobs to execute a PPRL task, where the output of some job is used as the input to the next in-line job.
We modified slightly the above basic scenario to cover our domain-dependent requirements by including the services of PBlockSketch. First, we will present some details regarding the topology of data. Specifically, there is a big cluster of compute nodes, each of which holds very large amounts of historical records of patients that comprise their medical history. Each such record is of the form:
where BF denotes the Bloom filter that corresponds to a patient. The values of the symptom, medication, year, and state attributes are securely transmitted and stored in plain text, because they will be used as dimensions to build certain data views depending on business requirements. The goal is to perform a global PPRL task using this large volume of masked patient records. By linking all these islands of records based on the Bloom filters, which include their given name and surname, one can then build a data lake to perform mining and analysis.
First, we discuss the topology of the data. There is a big cluster of compute nodes, each of which holds very large amounts of electronic health records transformed into Bloom filters. The goal is to perform a global PPRL task, by linking all these islands of Bloom filters. Then, the TTP can deposit the integrated records to the data lake to perform mining and analysis.
Initially, each Bloom filter of each data island is hashed and the L key values are forwarded to the appropriate reduce tasks by the underlying partitioner tasks. Subsequently, each reduce task deploys a PBlockSketch whose aim is to perform the comparisons between the blocked Bloom filters in a bounded matching time. This setting is contrasted to the naive matching of all the Bloom filters in a certain block aggregated by a reduce task. Figure 10 shows the pipeline of actions to populate the corresponding LSHDB stores.
5.2 Online SFEMRL
SFEMRL can be also applied to settings where the PPRL task has to return a fast response in order to allow for emergency actions to be triggered. An illustrative example is a public health surveillance system, which analyzes data on a continuous basis to uncover correlations between symptoms of patients and administered drugs. By detecting at an early stage such correlations, we can prevent the outbreak of diseases or epidemics, by triggering certain measures when a specified number of symptoms has occurred. The successful realization of such system mandates the unique representation of each patient. To this end, data from several sources need to be integrated, such as electronic health records from public hospitals, private medical offices, and records from pharmacy stores. In such cases, we require a very fast response that includes a result set of high accuracy. SFEMRL meets these requirements, which is attributed to the high-quality blocking of LSH and the performance of the underlying noSQL systems; LSH scores highly accurate results, while the noSQL systems retrieve the hash keys and the masked records from disk using efficient data structures, which exhibit logarithmic search times.
To accelerate the query response time, we used an efficient algorithm, namely the FPS (Frequent Pairs Scheme), which was introduced in [22]. FPS is based on Hamming LSH and instead of performing the distance computations for all the formulated pairs, it scans the hash tables to count the number of collisionsFootnote 21 of all pairs. This number, for a single pair, is binomially distributed and is expected to be above a specified collision threshold if this pair is a matching one. For this set of pairs, which are called the frequent pairs, FPS performs the distance computations. Correspondingly, FPS discards the record pairs that do not achieve the required number of collisions. The weakness of FPS is the larger number of hash tables that are required than baseline Hamming LSH blocking uses. In the following, we give a short description of FPS and also propose a technique, that is theoretically justified, to apply FPS without increasing the number of the hash tables used.
5.2.1 Applying the median trick on FPS
Hamming LSH blocking [21] embeds the records into the Hamming space using Bloom filters. The probability of collision in a hash table for two Bloom filters is \(p^k\), where \(p=1 - \frac{t}{s}\), s is the total number of components of each Bloom filter, t is the specified distance threshold, and k is the number of components chosen randomly and uniformly of the Bloom filters to formulate each hash key. The value of k should be large enough \(k > 25\) so as not to overpopulate the buckets of the hash tables [21, 22]. The set of indexes of the chosen components of the Bloom filters is identical for each hash table.
However, FPS requires more hash tables than the standard Hamming LSH mechanism. By applying the median trick, we can easily locate highly similar Bloom filter pairs, without increasing the number of hash tables. The number of hash tablesFootnote 22 used is \(L={\mathcal {O}}(\ln (\frac{1}{\delta })\) [21], where \(\delta \) is the probability of failure for a similar Bloom filter pair to collide in a hash table. Applying the median trick in this iterative mechanism, we expect that two similar Bloom filters will collide in more than half of these hash tables, namely:
when the probability of collision is fairly high:
Making some algebraic manipulations, we arrive at:
where \(t'\) is the upper distance bound, which is an integer value, that Bloom filter pairs should meet in order to be safely characterized as frequent pairs, and then be classified as similar pairs with probability at least \(1-\delta \).
5.2.2 Ubiquitous FPS
As we have described previously, the theoretical premise behind FPS is the binomial distribution of the number of collisions that record pairs achieve in the hash tables [22]. Although, it is quite trivial to count these collisions in a single LSHDB instance, applying the same counting mechanism on multiple LSHDB instances universally, i.e., in each such instance, may yield considerable time delays. Nevertheless, the query records that are submitted may assist in this counting process acting as bridge records.
Consider the following scenario. Two LSHDB instances maintain Bloom filters \(r_1\) and \(r_2\), respectively, in their corresponding hash tables. Query record q is submitted and, in turn, this record collides with \(r_1\) and \(r_2\) more times than the collision thresholdFootnote 23 specifies in each respective LSHDB instance. Finding the number of common hash keys between \(r_1\) and \(r_2\), we essentially apply FPS between the records of two data stores maintained in two distinct LSHDB instances.
The distance between \(r_1\) and \(r_2\) is also implied by the triangle inequality, however the corresponding distance bound is not tight enough to make reasonable inferences about the distance between the record pair involved. A necessary precondition for the operation of this ubiquitous mechanism, illustrated in Fig. 11, among different LSHDB instances is the sharing of the LSH hash functions.
5.3 Interoperability
SFEMRL, using the interoperability layer of LSHDB, produces the results using either Java or JSON objects. In an LSHDB ecosystem, it is preferable for the instances involved to communicate using Java objects, because it is much faster and less prone to errors. In a diverse environment though, where potentially any third-party piece of software might consume services from an LSHDB instance, SFEMRL exposes its services using JSON objects. Therefore, a trusted user can invoke a set of simple commands to query a data store and receive the results using a web browser. For instance, upon completion of a linkage process, SFEMRL may forward the results, in terms of JSON objects, to a trusted stub which can initiate next in-line actions, such as clerical review to disambiguate any record pairs whose linking status is not clear and imposes human intervention.
In this example, the data custodians utilize FPS and have set the collision threshold to 2. We observe that q collides with \(r_1\) and \(r_2\) in \(T_1\) and \(T_3\), and in \(T_1\) and \(T_4\), respectively. We also observe that \(r_1\) and \(r_2\), which have been brought together by q, exhibit identical hash keys in \(T_1\), \(T_2\), and \(T_4\), which exceeds the specified collision threshold
6 Experimental svaluation
For the experimental evaluation, we used three real-world data sets, namely (a) DBLP,Footnote 24 which includes bibliographic data records, (b) NCVR,Footnote 25 which comprises a registry of voters, and (c) LAB,Footnote 26 which includes biological assays (e.g., albumin, hepatitis, or creatinine) and their corresponding results. The technical characteristics of these data sets are summarized in Table 1. For each record of each data set, denoted by Q, we generated 1,000 perturbed records, which were placed in a separate data set symbolized by A. We perturbed all the available fields using at most four edit, delete, insert, or transpose operations, chosen at random.
We also used the DBLP data set to generate arbitrary clusters of matching records by perturbing (including the missing values operations) the names of authors and titles of certain publications. This dirty data setFootnote 27 includes matching records with the same (or almost the same) set of authors especially in conference/journal extensions scenarios.
We ran each experiment 20 times and plotted the average values in the figures. The software components were developed using the Java programming language (ver. 1.8) and the experiments were conducted in a virtual machine utilizing 4 cores of a Xeon CPU and \(32\hbox {GB}\) of main memory.
6.1 Evaluating the Algorithms
The blocking methods that were used for the needs of the evaluation were standard [6] and LSH blocking [21], which relies on the Locality-Sensitive-Hashing [15] technique. LSH blocking generates from a single record a certain number of blocking keys that are placed in multiple hash tables. This number of blocking keys is a function of several parameters [22] of LSH blocking, such as the distance threshold. The LSH technique is commonly used in the domain of record linkage [21, 24, 26, 36] because of its efficiency and accuracy guarantees. We used Hamming LSH blocking [21], in which records are embedded into the Hamming space using record-level Bloom filters [35]. LSH blocking implements redundant blocking, because a record is inserted into multiple independent blocks, which are accommodated into independent hash tables. In contrast, standard blocking inserts records that exhibit identical values, in an appropriately chosen blocking field(s), into the same block.
For performing the standard and the LSH blocking, we utilized LevelDBFootnote 28 and LSHDB [23], respectively. The length of each Bloom filter, utilized by SkipBloom, was set to 32, 000 bits for storing 5000 keys, with false positive probability set to \(\textit{fp}=0.05\).
We evaluated our schemes and their competitors according to the time needed, and the memory that was consumed to perform the record linkage process, as well as the recall and precision rates that were achieved.
6.1.1 Baseline methods
We compared our schemes with three state-of-the-art methods for online record linkage. The first method, termed as INV [7], uses inverted indexes as its basic blocking structure. The main idea behind this method is the pre-computation of similarities between field values that have been inserted into the same block. An inverted index is used for this purpose, which stores the blocking keys encoded by the double metaphone method.Footnote 29 A weakness of this structure regards the storage of all field values of a record into the same set of indexes. As a result, one cannot be certain for a value encountered therein, to which field this value belongs. This ambiguity affects negatively the recall rates of INV.
The second method we compared against is the Edge Ordering strategy, termed as EO, which was introduced in [14]. EO utilizes an oracle, which is aware of the ground-truth, to resolve the matching status of a record pair. A graph is constructed by assuming each record pair, which materializes an edge connecting two vertices/records, formulated in each block. The algorithm performs all similarity computations in the target block in order to assign a probability estimate to each edge (pair) based on its similarity. In turn, EO selects those edges that are expected to maximize the recall, and submits them to the oracle that returns their matching status.
Finally, we compare against method PDB [19] which resolves queries under data uncertainty. Records are pairwise linked using a record linkage algorithm and then according to the pairs formulated, the corresponding records should be divided into disjoint sets, called factors. PDB creates these factors by applying standard blocking and restructures the generated blocks for all blocking keys in order to generate the universally disjoint factors. Each pair in a block is then annotated by its similarity result. For a query record submitted , PDB considers only these records found in the same factor. The final step is to compute the probability of each possible world which is the product of the annotated similarity results of each formulated pair.
EO, INV, and PDB utilize only key/value pairs, materialized by hash tables that map a key to list of record Id’s. These methods do not offer any component to report efficiently the membership of a certain key, or to adequately summarize the data set. Thus, in order to be fair in our comparison with these methods, we maintained the key/Id’s mappings, as well as the entire records, in secondary storage. All the baseline methods and our proposed schemes used the Jaro-Winkler [5] function as the similarity measure, where the corresponding threshold was set to \(\theta =0.75\).
6.1.2 Experimental results
In our first set of experiments, we evaluated the running time and memory performance, as well as the ability of the SkipBloom algorithm to provide accurate estimates in the pre-processing step of record linkage.
Figure 12a shows the total time needed to build the SkipBloom, by scaling the number of the streaming records using the NCVR data set. It is quite obvious that the time increases by a constant factor, depending on the number of records that are processed. The consumption of main memory is illustrated in Fig. 12b, where SkipBloom exhibits almost linear performance. Specifically, although the number of records increases by 10 and 50 times, SkipBloom utilizes \(0.6\hbox {GB}\), \(0.8\hbox {GB}\), and \(1.4\hbox {GB}\) of main memory, respectively. In contrast, a map data structure, symbolized by MAP, e.g., a HashMap in the Java programming language, exhibits a steep linear performance. In both scenarios, MAP throws fatal errors and terminates when it reaches the processing of \(500\hbox {M}\) records.
Table 2 illustrates the time consumed by SkipBloom to report the existence of a key. We remind to the reader the probabilistic nature of SkipBloom, whose performance depends on the number of comparisons that will take place until the target block is located (which is \({\mathcal {O}}(\log (\sqrt{n}))\)). For this reason, we observe that SkipBloom almost consumes the same amount of time when it has to process either \(100\,\hbox {M}\) or \(500\,\hbox {M}\) records.
The accuracy of SkipBloom is evaluated by the fraction of overlapping keys it estimates using the above-mentioned data sets. Table 3 clearly shows that SkipBloom approximates the overlap coefficient of A and Q for each data set, where in the worst case it exhibits an error nearly equal to 0.06 (which is within its approximation guarantees specified by \(\epsilon \)).
In the next set of experiments, we compared our schemes against EO, INV, and PDB. Figures 13a and b display the recall rates achieved by all methods using standard and LSH blocking, respectively. We observe in Fig. 13a that EO exhibits slightly better recall rates than BlockSketch, by using all data sets, although the differences lie in the small range [0.01, 0.04]. The scores of PDB are also high, slightly lower than BlockSketch. PDB formulates all possible pairs from a merge of all the factors, which contributes to the increase of the recall rates. In NCVR, it was impossible to obtain measurements due to excessively high memory requirements. Specifically, the inverted indexes maintained for storing the factors and the records pushed the utilization of main memory to its maximum extent, which resulted in a large disk swap activity. Finally, INV falls short in formulating those matching pairs that exhibit a high degree of perturbation, which is due to the weakness of the double metaphone scheme to group together such pairs into the same blocks.
The recall rates of DBLP and NCVR are also higher than LAB, which is due to the longer (in characters) blocking keys, which render them more tolerant to the perturbation errors. BlockSketch achieves to maintain high recall rates, although we have to stress that the underlying blocking method drives the whole linkage process. As Fig. 13b suggests, LSH blocking, which leverages redundancy, scores much better rates than standard blocking. Only BlockSketch and EO can use LSH blocking, because they essentially run on top of the blocking mechanism. On average, BlockSketch and EO achieve \(10\%\) and \(8\%\) higher recall rates, respectively, using LSH blocking.
Figures 13c and d show the precision rates using the two different blocking approaches described before. As one can observe, BlockSketch outperforms EO, INV, and PDB by a large margin, due to the effective categorization of records into the sub-blocks of each block. This minimizes significantly the required number of comparisons. Specifically, the precision rates of the competitors fall by \(18\%\), \(21\%\), and \(23\%\), respectively, compared with the rates of BlockSketch. The reasons for this recession vary between the three methods. EO starts to produce meaningful recall rates after performing a large number of comparisons to derive the probability estimates for each pair. These comparisons, however, considerably reduce the precision rates. On the other hand, the double metaphone scheme of INV groups a large number of non-matching pairs into the same block, whose comparisons also result in low precision rates. The merge of all factors in PDB generates many irrelevant pairs, which in turn has negative impact on the precision rates. The redundancy of LSH blocking accounts for the reduced precision rates of both BlockSketch and EO, as shown in Fig. 13d, since both methods perform a larger number of comparisons for the pairs formulated in the blocks of each hash table. We observe though that BlockSketch retains its superiority over EO by scoring, on average, rates that are very close to 0.75.
Figure 14a and b display the performance of BlockSketch and its competitors using the dirty DBLP data set. We observe that the recall rates have dropped for all methods mainly due to the missing values in this version of the data set. BlockSketch exhibits the smallest losses which are on average 4.5% lower that the rates in Fig. 13a. This superiority of BlockSketch is justified by its smart blocking functionality that accounts both for its accuracy and speed. The precision rates in Fig. 14b do not exhibit any noticeable fluctuation.
The time needed to perform the blocking step is illustrated in Fig. 15a and b. EO and PDB perform a simple blocking for each record, which results in a faster time for building their accompanying structures than the combination of SkipBloom and BlockSketch. Specifically, BlockSketch, through a single get operation, retrieves the representatives of a block from the database, as well as replaces them, through a single set operation, when needed. INV utilizes three hash tables to store the precomputed similarities, the encoded, and the original field values, which leads to certain delays.
In Fig. 15c and d, we present the time performance of BlockSketch and its competitors for resolving the query data sets, symbolized by Q (see Table 1), after having populated the blocking structures with the records of A. For each query record of Q, BlockSketch performs a constant number of comparisons in each target block, which results in superior performance. As Fig. 15c suggests, BlockSketch is \(2\times \) faster than EO and \(1.5\times \) faster than INV and PDB, respectively, which struggle to compare all records found in a block. Moreover, EO should build the graph to locate these record pairs that are expected to maximize the recall. Finally for PDB, the computation of all possible worlds increases noticeably the response time.
Using LSH blocking, which is shown in Fig. 15d, both BlockSketch and EO exhibit longer time rates, which are nearly \(3\times \) slower than before, due to the inherent redundancy of LSH. Since, a record pair might appear several times during the matching phase, for each record of Q, we utilize a map data structureFootnote 30 to discard the comparisons of duplicate record pairs. Table 4 illustrates the time for resolving a single query record of Q during the matching phase. The constant number of distance computations for a single record accounts for the stable time performance of BlockSketch regardless of the size of the corresponding data set. In contrast, EO and INV consume running times which apart from the fact that in most cases they are almost the double of those of BlockSketch, they also depend on the number of records found in each block.
In SBlockSketch, we initially set \(\mu \) to a moderate size (\(\mu =1\)MFootnote 31). In Fig. 16a and b, we observe an average of \(10\%\) increase in time consumption than BlockSketch, only in NCVR and DBLP. The large number (over 60M) of distinct blocking keys that are generated in these data sets, resulted in relatively frequent evictions and disk seeks for the replacement of blocks in T. Nevertheless, the eviction status of highly selective (high \(\xi \)) but old (high \(\alpha \)) blocks remained high during the blocking phase, which prevented their eviction from T. The running time of LAB remained almost intact due to the small number of blocking keys (about 10M) and the corresponding replacements. Since, SBlockSketch utilizes a single hash table T, LSH keys were formulated in a composite format HashTableNo_Key to accommodate all of them in T.
We next varied the values of \(\mu \) and initiated the streaming of records of the NCVR data set. Figures 17a and b illustrate the time performance of SBlockSketch, where we observe that by doubling \(\mu \), we achieve significantly lower running time. For instance, by setting \(\mu =1\)M, the corresponding time value is 43 minutes, which is almost \(4\times \) faster than the previous value (156 minutes) on the y-axis. In LSH blocking, the number of incoming records increases by a constant factor, which is the number of the LSH keys that are generated for each record. Since a large number of these keys are identical, the running time increases by \(156\%\) on average, as Fig. 17b suggests, compared to the use of standard blocking.
6.2 Evaluating SFEMRL
Initially, we evaluated the memory consumption of SFEMRL using LSHDB and LevelDB as the underlying noSQL engine. LevelDB uses an in-memory highly efficient multi-level data structure, which enables logarithmic disk seeks in the number of stored keys. We used a data store, whose total size was nearly \(60\hbox {GB}\) holding \(10\hbox {M}\) of records, which generated almost \(1\hbox {B}\) hash keys. We varied the amount of main memory that was available to the host node from \(5\hbox {GB}\) to \(50\hbox {GB}\) and the average time response was surprisingly constant. Figure 18 illustrates these results, where we observe that the amount of main memory utilized did not affect the response time of SFEMRL, which maintained an almost stable performance. Setting the main memory to \(50\hbox {GB}\), we achieved the highest response time, because we essentially eliminated the need for disk seeks.
In the next series of experiments, we evaluated the integration of SFEMRL with Map/Reduce. We employed HadoopFootnote 32 utilizing a cluster of 64 compute nodes, each of which may run up to 2 reduce tasks. The map tasks of each node had to process 1M of records reproducing the NCVR data set and appending randomly to each record a value for the state attribute. The goal was twofold; we had to achieve both the PPRL task, and the aggregation of these records based on the state attribute. The reduce tasks retrieved the whole masked records from a CassandraFootnote 33 database and then inserted each such record into the appropriate LSHDB instance. This robust infrastructure resulted in 25 LSHDB instances, since each instance was populated with records from a pair of U.S. states. Fig. 19 shows the total time that was consumed to complete the whole task, where parallel processing of records resulted in setting up a distributed LSHDB store in a fast and reliable manner.
Finally, Fig. 20a presents the results by employing FPS as the mechanism for detecting the matching pairs of the NCVR data set by scanning the hash tables and counting the number of times (collisions) each record pair was formulated in the hash tables. We achieved to prune a large number of distance computations and accelerate the matching phase by discarding pairs that did not collect the required number of collisions at the expense of a tiny accuracy loss. We observe that the difference of the PC rates between FPS and the baseline LSH is negligible, while the time savings, shown in Fig. 20b are important in settings that require real-time responses. Moreover, M-FPS, which is the FPS using the median trick for identifying Bloom filter pairs with small distances, exhibits the fastest response times, while it maintains the PC rates of FPS.
7 Conclusions
In recent years, several applications have emerged which require access to consolidated information that has to be computed and presented in near real-time, through the linkage of records residing in voluminous disparate data sources. To address this need, we proposed the first summarization algorithms that operate in the blocking and matching steps of online record linkage to boost their performance. SkipBloom compiles a synopsis of the blocking structure of a data set using a small footprint of main memory, while BlockSketch compares each query record with a constant number of records in the target block, which results in a bounded matching time. Our experimental findings indicate that SkipBloom and BlockSketch (and PBlockSketch for privacy-preserving settings) outperform the state-of-the-art algorithms, in terms of the time needed, the memory used, and the recall and precision rates that are achieved during the linkage process. SBlockSketch utilizes a constant memory footprint to perform the linkage in settings that use streaming data.
We then introduced SFEMRL, a complete privacy-preserving framework for masking records and performing record linkage to identify matching records. The core component of SFEMRL is LSHDB, a parallel and distributed data engine that imports the masked data and leverages LSH to perform the blocking and matching step of privacy-preserving record linkage. The integration of LSHDB with Map/Reduce resulted in building a distributed data store for performing on-demand PPRL tasks. Our thorough experimental evaluation, using four synthetically perturbed data sets, reveals the applicability of SFEMRL to the linkage of patients’ records in a privacy-preserving fashion.
Notes
As long as tails come up, we add the item to each successive list. We terminate this process when we encounter heads.
A concrete class implements the inherited methods of an abstract class and/or interfaces in the Java programming language.
SkipBloom aims to address Problem 1, BlockSketch targets Problem 2, while SBlockSketch tackles Problem 3.
Henceforth, key and blocking key will be used interchangeably.
SkipBloom locates these Bloom filters performing a recursive process.
Since, we expect to have duplicate keys, it is quite natural that the same key may be stored into multiple Bloom filters of a block.
We assume that the number of distinct blocking keys is n in both A and B.
The distance either between a pair of records, or between a blocking key and a record, is determined by the distances of the certain field values, part of which usually make up the blocking key.
The exact number of representatives will be specified later.
A representative, being essentially a blocking key, has only key values.
For instance, LevelDB (see https://github.com/google/leveldb) uses an in-memory highly efficient multi-level data structure, which enables logarithmic disk seeks in the number of stored blocking keys.
A live block is a block that is stored in main memory.
The Hamming distance between two Bloom filters is equal to the number of components in which these filters have different bits.
We use the term masking to refer to data obfuscation and perturbations operations that are applied to protect the plain-text (original) values.
In general, in a multi-party model, there are more than two data custodians involved.
A collision is a record pair formulation in a certain hash table.
Each hash table can be assumed as an independent Bernoulli trial for the collision of a Bloom filter pair.
The collision threshold is essentially the number of collisions that should be counted between a pair of records, so as to be considered as a potential matching pair.
in terms of the predefined clusters of matching records.
Using the double metaphone encoding method, ‘SMITH’ and ‘SMYTH’ are both encoded as ‘SM0’.
The map structure is initialized for each record of Q.
We had 32GB of main memory available.
References
Altwaijry, H., Kalashnikov, D., Mehrotra, S.: Query-driven approach to entity resolution. In: International Conference on Very Large Data Bases (PVLDB), vol. 6, pp. 1846–1857 (2013)
Bhattacharya, I., Getoor, L., Licamele, L.: Query-time entity resolution. In: International Conference on Knowledge Discovery and Data Mining (KDD), pp. 529–534 (2006)
Bilenko, M., Kamath, B., Mooney, R. J.: Adaptive blocking: learning to scale up record linkage. In: International Conference on Data Mining (ICDM), 87–96 (2006)
Broder, A., Mitzenmacher, M.: Network applications of Bloom filters: a survey. In: Internet Mathematics, pp. 636–646 (2002)
Christen, P.: Data matching—concepts and techniques for record linkage, entity resolution, and duplicate detection. Springer, Data-Centric Sys. and Appl. (2012)
Christen, P.: A survey of indexing techniques for scalable record linkage and deduplication. Trans. Knowl. Data Eng. (TKDE) 12(9), 1537–1555 (2012)
Christen, P., Gayler, R., Hawking, D.: Similarity–aware indexing for real-time entity resolution. In: International Conference on Information and Knowledge Management (CIKM), pp. 1565–1568 (2009)
Christophides, V., Efthymiou, V., Stefanidis, K.: Entity Resolution in the Web of Data. Morgan and Claypool Publishers, San Rafael (2015)
Cohen, W.W., Richman, J.: Learning to match and cluster large high-dimensional data sets for data integration. In: International Conference on Knowledge Discovery and Data Mining (SIGKDD), pp. 475–480 (2002)
Datar, M., Immorlica, N., Indyk, P., Mirrokni, V.: Locality-sensitive hashing scheme based on p-stable distributions. In: Symp. on Comp. Geom., pp. 253–262 (2004)
Dean, J., Ghemawat, S.: Mapreduce: simplifed data processing on large clusters. CACM 51(1), 107–113 (2008)
Dey, D., Mookerjee, V., Liu, D.: Efficient techniques for online record linkage. Trans. Knowl. Data Eng. (TKDE) 23(3), 373–387 (2011)
Elmagarmid, A., Ipeirotis, P., Verykios, V.: Duplicate record detection: a survey. Trans. Knowl. Data Eng. (TKDE) 19(1), 1–16 (2007)
Firmani, D., Saha, B., Srivastava, D.: Online entity resolution using an oracle. In: International Conference on Very Large Data Bases (PVLDB), vol. 9, pp. 384–395 (2016)
Gionis, A., Indyk, P., Motwani, R.: Similarity search in high dimensions via hashing. In: VLDB, pp. 518–529 (1999)
Haas, P.J.: Data-stream sampling: basic techniques and results. In: Garofalakis, M., Gehrke, J., Rastogi, R. (eds.) Data Stream Management: Processing High-Speed Data Streams, pp. 13–44. Springer, Berlin (2016)
Hall, R., Fienberg, S.: Privacy-preserving record linkage. In: PSD, pp. 269–283 (2010)
Hernandez, M., Stolfo, S.: The merge/purge problem for large databases. In: International Conference on Management of Data (SIGMOD), pp. 127–138 (1995)
Ioannou, E., Nejdl, W., Niederee, C., Velegrakis, Y.: On-the-fly entity-aware query processing in the presence of linkage. International Conference on Very Large Data Bases (PVLDB), vol. 3(1), pp. 429–438 (2010)
Karapiperis, D., Verykios, V.: A distributed near-optimal LSH-based framework for privacy-preserving record linkage. COMSIS 11(2), 745–763 (2014)
Karapiperis, D., Verykios, V.: An LSH-based blocking approach with a homomorphic matching technique for privacy-preserving record linkage. Trans. Knowl. Data Eng. (TKDE) 27(4), 909–921 (2015)
Karapiperis, D., Verykios, V.: A fast and efficient Hamming LSH-based scheme for accurate linkage. Knowl. Inf. Syst. (KAIS) 49(3), 861–884 (2016)
Karapiperis, D., Gkoulalas-Divanis, A., Verykios, V.: LSHDB: a parallel and distributed engine for record linkage and similarity search. In: International Conference on Data Mining (ICDM) demos, pp. 1–4 (2016)
Karapiperis, D., Vatsalan, D., Verykios, V., Christen, P.: Efficient record linakge using a compact Hamming space. In: International Conference on Extending Database Technology (EDBT), pp. 209–220 (2016)
Karapiperis, D., Gkoulalas-Divanis, A., Verykios, V.S.: Summarization algorithms for record linkage. In: EDBT, pp. 73–84 (2018)
Kim, H., Lee, D.: Fast iterative hashed record linkage for large-scale data collections. In: International Conference on Extending Database Technology (EDBT), pp. 525–536 (2010)
Motwani, R., Raghavan, P.: Randomized Algorithms. Cambridge Univ. Press, Cambridge (1995)
Papadakis, G., Koutrika, G., Palpanas, T., Nejdl, W.: Meta-blocking: Taking entity resolution to the next level. TKDE 26(8), 1946–1960 (2014)
Papadakis, G., Papastefanatos, G., Koutrika, G.: Supervised meta-blocking. In: PVLDB, pp. 1929–1940 (2014)
Papenbrock, T., Heise, A., Naumann, F.: Progressive duplicate detection. Trans. Knowl. Data Eng. (TKDE) 27(5), 1316–1329 (2015)
Pugh, W.: Skip lists: a probabilistic alternative to balanced trees. CACM 33(6), 668–676 (1990)
Rajaraman, A., Ullman, J.: Mining of Massive Datasets, Chapter Finding Similar Items. Cambridge Univ. Press, Cambridge (2010)
Ramadan, B., Christen, P.: Forest-based dynamic sorted neighborhood indexing for real-time entity resolution. In: CIKM, pp. 1787–1790 (2014)
Ramadan, B., Christen, P., Liang, H., Gayler, R., Hawking, D.: Dynamic similarity-aware inverted indexing for real-time entity resolution. In: PAKDD Workshops, pp. 47–58 (2013)
Schnell, R., Bachteler, T., Reiher, J.: Privacy-preserving record linkage using Bloom filters. Med. Inform. Decis. Making (BMC) 9, 41 (2009)
Steorts, R., Ventura, S., Sadinle, M., Fienberg, S.: A comparison of blocking methods for record linkage. In: Privacy in Statistical Databases (PSD), pp. 253–268 (2014)
Vatsalan, D., Christen, P., Verykios, V.: A taxonomy of privacy-preserving record linkage techniques. Inf. Sys. 38(6), 946–969 (2013)
Whang, S.E., Marmaros, D., Garcia-Molina, H.: Pay-as-you-go entity resolution. Trans. Knowl. Data Eng. (TKDE) 25(5), 1111–1124 (2013)
Whang, S.E., Menestrina, D., Koutrika, G., Theobald, M., Garcia-Molina, H.: Entity resolution with iterative blocking. In: SIGMOD, pp. 219–232 (2009)
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Karapiperis, D., Gkoulalas-Divanis, A. & Verykios, V.S. Summarizing and linking electronic health records. Distrib Parallel Databases 39, 321–360 (2021). https://doi.org/10.1007/s10619-019-07263-0
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10619-019-07263-0