Abstract
Nanotechnology is increasingly developing area including more than 700 commercial products such as clothing, food preparation, cosmetics, mechanics, electronics and also health industry. People generally contact with nanoparticles by inhaling from air. Thus, it is becoming important issue to understand harmful effects of nanoparticles on human health and prepare risk reports for common nano-sized materials. In this paper, synthesis, characterization and cytotoxicity evaluation of boron nitride (BN) nanoparticles were performed on human primary alveolar epithelial cells (HPAEpiC) since, main exposure to nanoparticles would generally happen through lung via inhalation. Chemically synthetized BN nanoparticles were characterized by using X-ray crystallography, transmission electron microscope, scanning electron microscope and energy-dispersive X-ray spectroscopy techniques. 3-(4,5-dimethyl-thiazol-2-yl) 2,5-diphenyltetrazolium bromide, neutral red and lactate dehydrogenase release assays were used to analyze cytotoxicity after nanoparticles exposure. Whole genome microarray analysis was used to find out the effects of BN NPs on gene expressions of HPAEpiC cells. Finally, the database for annotation, visualization and integrated discovery analysis was used to reveal relationships between different cellular pathways and nanoparticle exposure. According to cytotoxicity analysis LC20 value for BN nanoparticles was 125.051 mg/L. Microarray results showed that 2159 genes expression change (FC ≥ 2) significantly over 40,000 genes analysis. When the gene pathways were analyzed, it was seemed that BN nanoparticles mostly affect cell cycle, cell–cell interactions, cancer affecting genes and signal transduction. In a conclusion, our results supported for the first time that BN nanoparticles could be used as a safe nanomaterial in both pharmacological and medical applications.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The rapid development of nanotechnologies results in application of nanomaterials in wide spectrum areas. When nanomaterials used in every aspects of consumers lives, enormous concerns arise about potential toxic effects of nanoparticles on human health (Singh and Nalwa 2007; Oberdörster et al. 2009). Previous researches reveal that different nanoparticles can cause detrimental health effects which may depend on NPs chemical features, physical components and functional group on their surface (Hardman 2006). Because of NPs wide surface area with very small particle size, they may generate harmful oxyradicals (ROS) which attack DNA and other organic structure of cells (Brown et al. 2001). Also, most possible toxicity of NPs is resulted from oxidative stress but, exact mechanisms of nanotoxicity for specific nanomaterials is still unknown (Lewinski et al. 2008).
BN becomes very popular compound because of its features, which relate closely to carbon. Extreme hardness and thermal conductivity of cubic form boron nitride is also similar to diamond properties (Karim et al. 1991). It is reported that boron nitride nanotubes (BNNTs) have similar properties with carbon nanotubes (CNTs) and boron nitride nanotubes with unique physical properties are nontoxic like carbon nanotubes. On the other hand, BNNTs can direct protein, cell binding and deliver DNA oligomers inside of a cell without any toxic effect. These properties make BNNTs superior to CNTs in aspect of biomaterial and probe production (Chen et al. 2009). It is known that BNNTs do not have cytotoxic features but, so many properties of BNNTs are still unknown that biomedical and cell interaction properties have not been investigated yet (Ciofani et al. 2008a, b; Horváth et al. 2011).
Toxicogenomics is fast growing area, which compares genomic data with toxic molecule effects. By investigating whole genome gene expression in respect to toxic compound exposure, different pathways can be related with toxicity and health risk reports for various compounds can be assessed. Furthermore, different molecules with similar toxicity shows also similar gene expression pattern. This information prove that toxic molecule data bases can be constituted with toxicogenomics and health risks of unknown molecules can be estimated with these databases (Bulera et al. 2001). Comparable information can give us insights about unknown compounds and their effects on gene expression and toxicity. But, there must be very large data pool to conclude this type of comparisons and this type of information can only be used as preliminary information (Reilly et al. 2001). Gene expression profiles of toxic molecules can be used to find parallel pathways and similar cellular mechanisms. Generally closely related drug interactions and toxicity analysis can be investigated and also irrelevant or unlikely pathways relevance can be established via toxicogenomics (Waring et al. 2002).
In this study we aimed to reveal BN NPs safe and effective uses in nanomedicine and nanopharmaceutics for the first time. Therefore, BN nanoparticles were synthesized chemically and characterized by using XRD, TEM, SEM and EDX techniques, and BN nanoparticles were applied on the human lung alveolar epithelial cell (HPAEpiC) culture. To evaluate cytotoxicity of BN nanoparticles different viability assays (MTT, LDH and NR) were carried out after incubation of 72 h. BN NPs were applied to the cell culture again according to IC20 concentration and total RNAs were isolated from cultures, and analyzed in Illumina microarray technologies. Finally, microarray data were investigated with the database for annotation, visualization and integrated discovery (DAVID) analysis and functional categories for these genes revealed to understand effects of BN NPs on biological pathways. The main aim of this article is to reveal characteristics, living cell interactions and any cytotoxic effect of BN NPs for the first time.
Materials and methods
Synthesis of boron nitride nanoparticles
To synthesis BN nanoparticles boric acid (H3BO3), sodium azide (NaN3) and hydrazine monohydrate (N2H4·H2O) was used. At first, 2, 5 g H3BO3 and 8 g NaN3 was solved in deionized water by stirring for 30 min. Then 2,3 mL N2H4.H2O was added and stirred for 30 min again. After that this mixture was incubated at 300 °C temperature for 16 h in nitrogenized environment muffle furnace. When incubation completed mixture was washed with deionized water and vacuumed at 100 °C temperature for 2–3 h in vacuum pump.
Characterization of boron nitride nanoparticles
The micro-structural investigations of BN nanoparticles were made by X-ray diffraction (XRD) measurement at room temperature by using a Rigaku/Smart Lab diffractometer with CuKα radiation (λ = 0.154059 nm) operated at 40 kV and 30 mA. The measurement was taken in geometry of coupled θ–2θ varied between 20° and 90° with step of 0.02°. The surface morphology and particle size of BN nanoparticles was investigated with a scanning electron microscope (FEI inspect S50 SEM) and transmission electron microscopy (JEOL JEM-ARM200CFEG UHR-TEM). Finally, chemical characterization of BN nanoparticles was made with energy-dispersive x-ray spectroscopy (EDS, EDX).
Cell culture studies and viability tests
The human primary alveolar epithelial cells were growth in 48-well plates and incubated in a humidified 5 % CO2 at 37 °C. The cell culture was applied with BN nanoparticles for 72 h at twelve different concentrations (0.625, 1.25, 2.5, 5, 10, 20, 20, 80, 160, 320, 640 and 1280 mg/L). Each individual culture without BN nanoparticles was studied as a negative control group (Control−). Hydrogen peroxide (H2O2; 25 µM Sigma-Aldrich®) was used as the positive control in cytotoxicity testing.
MTT assay
According to the manufacturer’s instructions (Cayman Chemical Company®, Ann Arbor, MI, USA) 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) solution was applied to cell cultures. As summary, MTT was added and incubated in the cell cultures for 3 h and after incubation dimethyl sulfoxide (DMSO) (Sigma-Aldrich®) was used to solve formazan crystals, and a plate reader was used to analyze cultures at 570 nm wavelength (Turkez et al. 2012).
LDH assay
For LDH assay application, LDH cytotoxicity assay kit (Cayman Chemical Company®, Ann Arbor, MI, USA) was used according to the manufacturer’s recommendations. The cells were transferred to 96-well plates and different concentrations of BN were applied for 72 h. To get rid of BN in the wells, 96-well plate was centrifuged at 400 g for 5 min. After that 100 µL supernatant and 100 µL of reaction mixture were transferred to a fresh 96-well plate and incubated for 30 min at room temperature. Finally, the absorbance of the cultures was analyzed at 490 nm using a microplate reader (Turkez et al. 2016).
Neutral red assay
Neutral red (NR) solution was applied to the cell cultures according to the manufacturer’s recommendation (Sigma-Aldrich®, St. Louis, MO, USA). For the lysosomal dye uptake of viable cells, the human primary alveolar epithelial cells were incubated with the NR solution for 2 h at 37 °C. To wash cell culture, the mixture of formaldehyde (0.125 %) and CaCl2 (0.25 %) were used and the NR solution was removed. To extract the NR from the HPAEpiC cells, a mixture of acetic acid (1 %) and ethanol (50 %) were added to the cell cultures and incubated at room temperature for 30 min. Finally, the microplate reader (Bio-Tek Instruments, USA) was used to analyze optical density of each sample at 540 nm (Turkez et al. 2016).
Microarray analysis
To evaluate RNA purity and integrity, spectrophotometer (NanoDrop, Wilmington, USA) and bio-analyzer (Agilent Technologies, Palo Alto, USA) were used. Total RNA amplified and purified by using TargetAmp-Nano Labeling kit and Illumina Expression BeadChip (EPICENTRE, Madison, USA) was used to get biotinylated cRNA according to the manufacturer’s instructions. 500 ng of total RNA was reverse-transcribed to cDNA by using a T7 oligo (dT) primer, second-strand cDNA was synthesized, in vitro transcribed, and labeled with biotin-NTP and the ND-1000 Spectrophotometer was used to quantify cRNA. 750 ng of labeled cRNA samples were hybridized with Human HT-12 v4.0 Expression Beadchips for 17 h at 58 °C (Illumina, Inc., San Diego, USA). To detect array signal, Amersham fluorolink streptavidin-Cy3 (GE Healthcare Bio-Sciences, Little Chalfont, UK) was used according to the bead array manual. Finally, the arrays were analyzed via Illumina bead array Reader confocal scanner.
Data analysis
SPSS Software (version 18.0, SPSS, Chicago, IL, USA) was used for statistical analysis of all viability tests. The data were analyzed in Duncan’s test at a significance level of 0.05. The main method to analyze hybridization quality and overall chip performance was visual survey of raw data scanning and quality control checks for microarray analysis. To get the raw data, Illumina GenomeStudio v2011.1 (Gene Expression Module v1.9.0) software was used according to the manufacturer instructions. Normalization and array probes transformation into logarithm performed by using quantile method. Statistical significance of the expression data was determined with fold change values. Gene enrichment and functional annotation were investigated by DAVID (http://david.abcc.ncifcrf.gov/home.jsp) analysis for significant probe list. All data analysis and visualization of differentially expressed genes were statistically established with the R 3.1.2 (www.r-project.org) software.
Results
Structural, morphological and chemical characterizations of BN NPs
Structural analysis of the BN nanoparticles that it made with the x-ray diffraction (XRD) show highest peak at 26.3° corresponding to Muller indice of 200 in Fig. 1. Some of the other reflections at 2θ = 41.3°, 43.4°, 54.6°, and 75.6° corresponding to Muller indices of 100, 101, 004 and 110 detected in the XRD pattern and this result correlate with the literature. In addition to, the grain sizes of the boron nitride nanoparticles are calculated from (200) peak by using Scherrer formula (Kasar et al. 2008):
where D, is the mean grain size of nanoparticles, which may be smaller, or equal to the grain size, β is full width at half of the peak maximum (FWHM) in radians and ‘θ’ is Bragg’s angle. For (200) peak, the calculated D value is 220 nm.
The surface morphology and element composition of boron nitride nanoparticles were investigated by SEM micrographs with EDX spectra and are shown in Figs. 2 and 4, respectively. It is observed that the surface morphology of the boron nitride nanoparticles quite homogeneous. It can be seen that the BN nanoparticles are made up grains with needle-like structures. Also, transmission electron microscope (TEM) investigation support that synthesized BN nanoparticles size changes between 100 and 300 nm (Fig. 3). Energy-dispersive X-ray spectroscopy (EDX) analysis shows that BN nanoparticles have desired chemical content as a percentage (Fig. 4). Besides, these spectra clearly confirm the existence of B and N elements in the nanoparticles. EDX analyses show that the participated atomic ratios of B/N (at.%) in the starting solution are much the same ones in the nanoparticles.
Cell viability tests
When nanoparticles synthesis and characterization were completed, the human lung alveolar epithelial cell line was seeded in 24 well plates. Negative control and 12 different BN concentrations were prepared to test cytotoxicity of the molecule on the cell line. The MTT assay results were used to investigate LC20 values for BN on the cells. Probit analysis was used to calculate LC20 values, and LC20 value for BN nanoparticles was determined as 125.051 mg/L (Fig. 5). MTT, LDH and NR analysis correlated to each other, and these results proved the consistency of present investigations. Cytotoxicity level for each assay is shown in Fig. 6 and the significant concentration level for toxicity was observed at 80 mg/L in both MTT and NR assays.
Microarray analysis and DAVID annotation functional categories
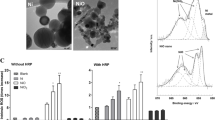
Subject to BN treatment, most up-regulated 25 genes (Fig. 7) and most down-regulated 25 genes (Fig. 8) of the alveolar epithelial cells were investigated. As a result of microarray analysis, total 2159 gene expressions changed from 40.000 analyzed genes from BN nanoparticle treatment. From these expression changes, 984 genes were determined to be up-regulated and 1175 genes were determined down-regulated at least two-fold in expression from all of the samples analyzed. According to microarray analysis, most up-regulated five genes were MIR1974, HSPA6, ATF3, HMOX1, DUSP10 and most down-regulated ones were LRRC17, CCL2, COL3A1, LRRC17, DIO2 (Figs. 7, 8). DAVID annotation was used to analyze gene enrichment and functional annotation. According to DAVID annotation KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway analysis, cell cycle, DNA replication, cancer and focal adhesion pathways were mostly affected from BN application (Fig. 9). DAVID functional annotations also show that genes regulating chromatin structure regulation, cell division, signal transduction and transcription were differentiated from BN particularly (Fig. 10). In addition to this, nucleotide binding, transcription repression and growth factor mechanisms expression level changes were investigated with GEOTERM_MF_FAT option of DAVID annotation analysis (Fig. 11).
Discussion
Past decades, there have been too many efforts on producing and characterizing nanomaterials to use in wide spectrum application area. Nanoparticles take attention of research from a very broad range of areas because of their unusual features in electronics, optical, mechanical, biomedical application and also in particular, drug delivery systems (Sousa and Sousa 2006; Tsoncheva et al. 2006). Carbon nanotubes (CNT) is always thought having unique ability to be constituted as nanotubes but, recent researches showed boron nitrides potential to form different types of nanostructures like graphite (Ahmad et al. 2015). Also, various boron nitride structures have excellent electrical and thermal properties, which can be used as ceramic material. With outstanding chemical stability, high melting point with low density and resistance to erosion makes boron nitride good candidate for variety of industrial applications (Paine and Narula 1990; Engler et al. 2007; Smith et al. 2009).
In this study, BN nanoparticles were synthesized and XRD, TEM, SEM and EDX techniques were used to characterize properties of nanoparticles. The XRD studies indicate that the preferential orientation of the boron nitride nanoparticles is (200) plane. Transmission electron microscope (TEM) analysis demonstrated size of BN nanoparticles in different interval between 50 and 300 nm (Fig. 3). Granular property of BN nanoparticles was exhibited by scanning electron microscope (SEM) investigation in Fig. 2. Moreover, Energy-dispersive X-ray spectroscopy (EDX) analysis exhibited crystal structure of BN nanoparticles, which was parallel with the literature (Fig. 4). MTT, LDH and NR cytotoxicity tests on the HPAEpiC gave similar results and analysis showed that BN nanoparticles have biocompatibility characteristic at suitable concentration for medical use. In parallel to our result, it was proven that injection of boron nitride nanotubes into rabbit circulatory system did not affect any function in blood, liver, and kidneys after 72 h treatment (Ciofani et al. 2012). Moreover, researchers synthesized boron nitride nanotubes and applied to the human neuroblastoma SH-SY5Y cells. After that, results were tested via both MTT and WST-1 viability assays, and result showed that there was no any cytotoxic effect of BNNTs on the cell line (Ciofani et al. 2012). It is seemed that previous researches and our analysis correlate each other that BN NPs is a safe material to use in any product such as electronics, food industry, cosmetic, medical and pharmaceutical use.
Over 40,000 genes were analyzed via using microarray technology after BN application to the HPAEpiC cells and significant expression change (2 ≥ fold change) was observed in 2159 genes. In these genes, 984 up-regulations and 1175 down-regulations were classified. As stated in DAVID functional annotation, cell cycle regulation, cell–cell communication, chromosomal separation, signal transductions and oncogenic pathways are immensely affected from BN exposure. When up-regulated genes analyzed individually, HSPA6 gene is one of the most up-regulated one with nearly 21-fold expression change. HSPA6 has important role in proteasome activation and cell viability, and the gene expression increases when levels of proteotoxic stress rise (Noonan et al. 2007). In addition to this, ATF3 gene expression is very high in microarray results and researchers claimed that p38-mediated transcriptional activation of ATF3 gene regulates apoptosis in human colorectal cancer cells through PARP cleavage (Lee et al. 2014). HMOX1 is one of the most up-regulated genes and deficiency and polymorphism on HMOX1 gene is known to have relationship with cancer risk, and (GT)n repeat length polymorphism in the gene can be a potential susceptibility variant for cancer in Asians (Luo et al. 2015). Researches proposed that colorectal cancer (CRC) development is influenced from deterioration of DUSP10 gene which is also overexpressed after BN application (Duan et al. 2015). These results can prove anticancer/apoptotic effect and negative impact of BN on tumor progression. When down-regulated genes are considered, LRRC17, CCL2, COL3A1 and DIO2 have the highest expression levels with over 10-fold each. LRRC17 gene has important function in bone development through preventing osteoclast differentiation and inhibits RANKL-induced osteoclastogenesis (Kim et al. 2009). Cancer development and consequent metastasis are important effects of CCL2 activation which is down-regulated by BN application in our experiment and it might be used as a potential prognostic marker of human colorectal cancer (Lim et al. 2016). COL3A1 mainly maintains the mechanical properties of arteries, forms thrombus and activates platelets but, variant of the gene expression can result in increased risk of death caused by stroke in lacunar infarct patients (Lv et al. 2014). Finally, DIO2 is one of the most down-regulated genes and it is claimed that obesity and type 2 diabetes mellitus (T2DM) in early ages can resulted from DIO2 overexpression (Nair et al. 2012). Additionally, BN may prevent type 2 diabetes mellitus (T2DM) by down-regulating specific genes. When take into consideration of these data it can be concluded that BN has (I) antitümoral potentials, (II) anti-metastatic and (III) anti-diabetic impacts, and (IV) positive effects on development.
In a conclusion, MTT, LDH and NR analysis revealed their slight toxicity potential, and all of them confirmed that BN nanoparticles (<100 mg/L) did not lead to lethal response on the HPAEpiC cells for the first time. Furthermore, microarray results indicated that anticancer/apoptotic, anti-metastatic, and development regulatory effects of BN. Again, positive impact on diabetes and stroke pathophysiology could be inference from whole genome analysis. In the light of present results we suggest the safe and effective usage potentials of boron nitride nanoparticles in nanomedicinal and nanopharmacological applications.
References
Ahmad P, Khandaker MU, Amin YM (2015) Synthesis of boron nitride nanotubes by argon supported thermal chemical vapor deposition. Phys E Low Dimens Syst Nanostruct 67:33–37. https://doi.org/10.1016/j.physe.2014.11.003
Brown DM, Wilson MR, MacNee W et al (2001) Size-dependent proinflammatory effects of ultrafine polystyrene particles: a role for surface area and oxidative stress in the enhanced activity of ultrafines. Toxicol Appl Pharmacol 175:191–199. https://doi.org/10.1006/taap.2001.9240
Bulera SJ, Eddy SM, Ferguson E et al (2001) RNA expression in the early characterization of hepatotoxicants in Wistar rats by high-density DNA microarrays. Hepatology 33:1239–1258. https://doi.org/10.1053/jhep.2001.23560
Chen X, Wu P, Rousseas M et al (2009) Boron nitride nanotubes are noncytotoxic and can be functionalized for interaction with proteins and cells. J Am Chem Soc 131:890–891. https://doi.org/10.1021/ja807334b
Ciofani G, Raffa V, Menciassi A, Cuschieri A (2008a) Cytocompatibility, interactions, and uptake of polyethyleneimine-coated boron nitride nanotubes by living cells: confirmation of their potential for biomedical applications. Biotechnol Bioeng 101:850–858. https://doi.org/10.1002/bit.21952
Ciofani G, Raffa V, Menciassi A, Dario P (2008b) Preparation of boron nitride nanotubes aqueous dispersions for biological applications. J Nanosci Nanotechnol 8:6223–6231. https://doi.org/10.1166/jnn.2008.339
Ciofani G, Danti S, Genchi GG et al (2012) Pilot in vivo toxicological investigation of boron nitride nanotubes. Int J Nanomed 7:19–24
Duan X, Gao Y, Yang H et al (2015) Polymorphisms in the DUSP10 gene are associated with sex-specific colorectal cancer risk in a Han population. Int J Clin Exp Pathol 8:2018–2025
Engler M, Lesniak C, Damasch R et al (2007) Hexagonal boron nitride (hBN): applications from metallurgy to cosmetics. CFI Ceram Forum Int 84:49–53
Hardman R (2006) A toxicologic review of quantum dots: toxicity depends on physicochemical and environmental factors. Environ Health Perspect 114:165–172
Horváth L, Magrez A, Golberg D et al (2011) In vitro investigation of the cellular toxicity of boron nitride nanotubes. ACS Nano 5:3800–3810. https://doi.org/10.1021/nn200139h
Karim MZ, Cameron DC, Murphy MJ, Hashmi MSJ (1991) Plasma deposition of cubic boron nitride films from non-toxic material at low temperatures. Surf Coat Technol 49:416–421. https://doi.org/10.1016/0257-8972(91)90093-C
Kasar RR, Deshpande NG, Gudage YG et al (2008) Studies and correlation among the structural, optical and electrical parameters of spray-deposited tin oxide (SnO2) thin films with different substrate temperatures. Phys B Condens Matter 403:3724–3729
Kim T, Kim K, Lee SH et al (2009) Identification of LRRc17 as a negative regulator of receptor activator of NF-kappaB ligand (RANKL)-induced osteoclast differentiation. J Biol Chem 284:15308–15316. https://doi.org/10.1074/jbc.M807722200
Lee JR, Lee MH, Eo HJ et al (2014) The contribution of activating transcription factor 3 to apoptosis of human colorectal cancer cells by protocatechualdehyde, a naturally occurring phenolic compound. Arch Biochem Biophys 564:203–210. https://doi.org/10.1016/j.abb.2014.10.005
Lewinski N, Colvin V, Drezek R (2008) Cytotoxicity of nanoparticles. Small 4:26–49. https://doi.org/10.1002/smll.200700595
Lim SY, Yuzhalin AE, Gordon-Weeks AN, Muschel RJ (2016) Targeting the CCL2–CCR2 signaling axis in cancer metastasis. Oncotarget 7: 28697–28710. https://doi.org/10.18632/oncotarget.7376
Luo H, Shao Y, Yao N et al (2015) Association of heme oxygenase-1 polymorphisms with cancer risk: a systematic review and meta-analysis. J BUON 20:1142–1153.
Lv W, Lin Y, Song W et al (2014) Variants of COL3A1 are associated with the risk of stroke recurrence and prognosis in the Chinese population: a prospective study. J Mol Neurosci 53:196–203. https://doi.org/10.1007/s12031-014-0283-x
Nair S, Muller YL, Ortega E et al (2012) Association analyses of variants in the DIO2 gene with early-onset type 2 diabetes mellitus in Pima Indians. Thyroid 22:80–87. https://doi.org/10.1089/thy.2010.0455
Noonan EJ, Place RF, Giardina C, Hightower LE (2007) Hsp70B’ regulation and function. Cell Stress Chaperones 12:393–402
Oberdörster G, Stone V, Donaldson K (2009) Toxicology of nanoparticles: a historical perspective. Nanotoxicology 1: 2-25. https://doi.org/10.1080/17435390701314761.
Paine RT, Narula CK (1990) Synthetic routes to boron nitride. Chem Rev 90:73–91. https://doi.org/10.1021/cr00099a004
Reilly TP, Bourdi M, Brady JN et al (2001) Expression profiling of acetaminophen liver toxicity in mice using microarray technology. Biochem Biophys Res Commun 282:321–328. https://doi.org/10.1006/bbrc.2001.4576
Singh S, Nalwa HS (2007) Nanotechnology and health safety–toxicity and risk assessments of nanostructured materials on human health. J Nanosci Nanotechnol 7:3048–3070. https://doi.org/10.1166/jnn.2007.922
Smith MW, Jordan KC, Park C et al (2009) Very long single- and few-walled boron nitride nanotubes via the pressurized vapor/condenser method. Nanotechnology 20:505604. https://doi.org/10.1088/0957-4484/20/50/505604
Sousa A, Sousa EMB (2006) Influence of synthesis temperature on the structural characteristics of mesoporous silica. J Non Cryst Solids 352:3451–3456. https://doi.org/10.1016/j.jnoncrysol.2006.03.080
Tsoncheva T, Rosenholm J, Teixeira CV et al (2006) Preparation, characterization and catalytic behavior in methanol decomposition of nanosized iron oxide particles within large pore ordered mesoporous silicas. Microporous Mesoporous Mater 89:209–218. https://doi.org/10.1016/j.micromeso.2005.10.028
Turkez H, Geyikoglu F, Mokhtar YI, Togar B (2012) Eicosapentaenoic acid protects against 2,3,7,8-tetrachlorodibenzo-p-dioxin-induced hepatic toxicity in cultured rat hepatocytes. Cytotechnology. https://doi.org/10.1007/s10616-011-9386-1
Turkez H, Sönmez E, Di Stefano A, Mokhtar YI (2016) Health risk assessments of lithium titanate nanoparticles in rat liver cell model for its safe applications in nanopharmacology and nanomedicine. Cytotechnology 68:291–302. https://doi.org/10.1007/s10616-014-9780-6
Waring JF, Gum R, Morfitt D et al (2002) Identifying toxic mechanisms using DNA microarrays: evidence that an experimental inhibitor of cell adhesion molecule expression signals through the aryl hydrocarbon nuclear receptor. Toxicology 181–182:537–550. https://doi.org/10.1016/S0300-483X(02)00477-8
Acknowledgements
This research was supported by National Boron Research Institute (BOREN) (Grant Number: Ç0391).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Türkez, H., Arslan, M.E., Sönmez, E. et al. Synthesis, characterization and cytotoxicity of boron nitride nanoparticles: emphasis on toxicogenomics. Cytotechnology 71, 351–361 (2019). https://doi.org/10.1007/s10616-019-00292-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10616-019-00292-8