Abstract
Nickel boride is generally used in the steel industry as a melting accelerator due to its feature of creating a protective and stable attribute at high temperatures. It is also used to improve the hardenability of the steel with boron addition in the production. Thus, safety studies and biocompatibility analysis of nickel boride should be performed comprehensively to understand the limitations of use in various areas. In the present study, nickel boride nanoparticles (Ni2B NPs) were synthesized by a single-step method and molecule characterizations were performed via the use of X-ray diffraction analysis (XRD), transmission electron microscopy (TEM), scanning electron microscopy (SEM), and energy dispersive X-ray (EDX) analyses. Cytotoxicity properties of Ni2B NPs were identified on human pulmonary alveolar epithelial cells (HPAEpiC) by using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), neutral red (NR), and lactate dehydrogenase (LDH) assays. Illumina human ht-12 v4.0 whole-genome microarray analysis was conducted to investigate NiB2 NPs effects on gene expression regulations of HPAEpiC cells. The database for annotation, visualization, and integrated discovery (DAVID) analysis was performed to reveal the relationship between Ni2B NP application and cellular pathway alterations. According to cytotoxicity analysis, the IC50 value for Ni2B NP application was found as 81.99 mg/L concentration. Microarray analysis of Ni2B NP application was shown for the first time that 693 gene expression changes (FC ≥ 2) occurred significantly over 40.000 gene probes and Ni2B NPs were observed to affect microtubule regulation, centrosome organization, and phosphoprotein synthesis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Nickel boride is generally used to increase the density of the material and obtain good mechanical properties at lower temperatures. Also, it is used as a sintering additive to provide high-temperature resistance for metals [1]. TiC-based materials are generally used in hard metal cutting equipment because of their wear resistance, strength, and high hardness. According to the previous study, integrating nickel boride (NiB), a low melting point boride, into the TiC-Ni composites led to densification and an increase in hardness and toughness of the materials because of the pseudo-binary eutectic reaction at lower temperatures [2]. NiB additions into stainless steel sintering procedure were found to develop the mechanical properties and increase relative density at 1280 °C at a significant rate. Moreover, tungsten-based alloys were investigated to gain higher hardness and density values after the NiB addition at various quantities [3, 4]. Previous studies mainly focused on nickel and boron-based superalloy productions because of their superior wear resistance properties. Surface structures of nickel/boron superalloys were claimed to depend on production process variables like the boriding temperature, boron source, and the exposure time that mainly give rise to NiB and Ni2B molecules [5,6,7].
In the last decade, the rapid development of nanotechnologies influenced many different areas enormously due to the enhanced physical, biological, and chemical properties of nanoparticles mainly resulted from the particle integrity, solubility, and reactivity [8]. Nanomaterials attracted great attention of many fields because of their unusual properties gained from the chemicophysical features that make them useful in wide spectrum productions such as nanosomes, dendrimers, metals, and inorganic compounds [9]. On the other hand, nanoparticles were shown to have unlimited application potentials for various industries, and increased chemical and physical properties were shown to lead detrimental health problems by enhancing reactive oxygen species (ROS) and free radicals [10]. Although nanomaterial applications were shown to have a huge impact in many production industries, there were no safety analyses for these materials yet [11]. Thus, toxicological reports should be prepared for each nanomaterial used in production industries to investigate their genotoxic, cytotoxic, and toxicogenomic properties [12].
Cells can hastily modify their transcriptomic production (gene expression pattern) in response to intracellular and extracellular environmental variations, and get adapted for their function and survival. However, usual physiological activities and biological functions can get disturbed under extreme environmental conditions. Thus, the gene expression profiling has been demonstrated to be a supportive parameter for identifying the NP toxicity and its related molecular mechanism [13,14,15]. Toxicogenomics, mainly including the hybridization machineries, has been a favored choice for modern toxicological studies. The enormous data productivity and pathway-based information characterize toxicogenomics as a prevailing approach, which has been utilized for over decades for classifying a novel mechanism of toxicity, changes in important biological pathways [16]. Recent toxicogenomic studies on boron-based nanoparticles showed that little or no cytotoxicity was investigated but, enormous modifications in gene expression patterns were observed that affect cellular phenotypes greatly. Although previous toxicogenomic investigations on boron nitride (BN), boron carbide (B4C), and tungsten boride (WB2) were shown to have no cytotoxic properties on human pulmonary alveolar epithelial cells (HPAEpiC), these molecules altered hundreds of gene expressions and changed pathway regulations remarkably [17,18,19].
In the present study, the single-step synthesis method was used to produce nickel boride (Ni2B) nanoparticles. XRD, TEM, SEM, and EDS analysis techniques were used to characterize features of the synthesized molecules. Three different toxicity tests (MTT, LDH, and NR) were conducted to observe cytotoxicity properties of Ni2B nanoparticles. Gene expression modifications were investigated via the use of microarray analysis after the application of the Ni2B nanoparticles at IC50 concentration. Finally, DAVID functional annotation software was utilized for observing differentially regulated vital pathways.
Materials and Methods
Synthesis of Nickel Boride Nanoparticles
Nickel boride was synthesized by a single-step method via dropping NiCl2 (Riedel-deHaen, 97%, 0.27 M) into NaBH4 (Alpha, 98%, 2 M) solution for 20 min until the reaction reached the saturation. The first products were urgently filtered and the compound was incubated under vacuum condition at 500 °C for 18 h. The reaction was terminated by heating treatment in an iron tube at 800 °C for 7 days [20].
Molecular Characterizations of Nickel Boride Nanoparticles
The crystal structure of Ni2B NPs was investigated via the use of X-ray diffraction (XRD) measurement by using a Rigaku/Smart Lab diffractometer at 40 kV and 30 mA with CuKα radiation (λ = 0.154059 nm) operated at room temperature. The particle size and surface morphology of Ni2B NPs were observed with a scanning electron microscope (SEM, FEI inspect S50) and transmission electron microscope (TEM, JEOL JEM-ARM200CFEG). Finally, the chemical composition of Ni2B NPs was investigated via the use of energy-dispersive X-ray spectroscopy (EDS, EDX).
Cell Culture Conditions
In the study, human pulmonary alveolar epithelial cell (HPAEpiC, Cat. #3200, ScienCell®, USA, California) cultures were used to conduct toxicological and gene expression analyses. For culture conditions, 500 mL of basal medium (alveolar epithelial cell medium, AEpiCM, ScienCell®), including 10 mL of fetal bovine serum (FBS, Cat. #0010, ScienCell®, USA, California), 5 mL of epithelial cell growth supplement, and 5 mL of penicillin/streptomycin solution, was used. HPAEpiC cultures were prepared in 48-well plates (104 cells in each well) with AEpiCM and incubated in a humidified 5% CO2 at 37 °C. Ni2B NPs were applied to the cell cultures at wide spectrum concentrations (0.625, 1.25, 2.5, 5, 10, 20, 20, 80, 160, 320, 640, and 1280 mg/L) in triple replicates and incubated for 72 h. As a positive control, hydrogen peroxide (H2O2; 25 μM Sigma-Aldrich®) was used and cell cultures without Ni2B NPs were investigated as a negative control.
MTT Assay
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) solution (Thermo Fisher®, USA) was used to analyze cell viability according to the manufacturer’s instructions. Briefly, MTT solutions were applied to the cell cultures at 5 μM concentration and the cell cultures were incubated for 3 h at 37 °C. Culture mediums were discarded and 200 μL dimethyl sulfoxide (DMSO) (Sigma-Aldrich®) was added to each well for resolving formazan crystals which were produced by viable cells. A microplate reader (Bio-Tek Instruments, USA) was used to evaluate color intensities at 570-nm wavelength.
LDH Assay
Lactate dehydrogenase cytotoxicity assay kit (Cayman Chemical Company®, USA) was used to investigate the cytotoxicity of the cell cultures. One hundred-microliter supernatant was transferred to a fresh 96-well plate and 100 μL of LDH reaction mixture was added to each well. Samples were incubated for 30 min at room temperature. Finally, a spectroscopic plate reader was used to observe color changes in the samples at 490 nm.
Neutral Red Assay
Neutral red (NR) solution (Sigma-Aldrich®, USA) was applied to the cell cultures and incubated for 2 h at 37 °C. The NR solution was discarded, and CaCl2 (0.25%) and formaldehyde (0.125%) mixture were used to wash cell cultures. A mixture of acetic acid (1%) and ethanol (50%) was added to the cell cultures and incubated at room temperature for 30 min to get rid of the NR from the HPAEpiC cells. A microplate reader was used to analyze color intensity of each sample at 540 nm [21].
Microarray Analysis
PureLink™ RNA Mini Kit (Invitrogene®, USA) was used to isolate total RNA from the cell cultures. The UV-visible spectrophotometer (NanoDrop®, USA) and bio-analyzer (Agilent Technologies, USA) were used to investigate the RNA quality. TargetAmp-Nano Labeling kit was conducted to amplify the total RNA and Illumina Expression BeadChip (EPICENTRE, Madison, USA) was used to biotinylate the cRNAs. Labeled cRNA samples were hybridized with Human HT-12 v4.0 Expression Beadchips (40,000 gene probes, Illumina Inc., USA) for 17 h at 58 °C. Amersham fluorolink streptavidin-Cy3 (GE Healthcare Bio-Sciences, UK) was used to detect array signals (Illumina array Reader).
Data Analysis
GraphPad Prism 7, Anova: Dunnett’s multiple comparison test was used for statistical calculations of all viability tests. The data were analyzed via Anova: Dunnett’s multiple comparison test at a significance level of 0.05. The overall chip performance and hybridization quality for the raw data were investigated by visual inspection of the internal quality control check. Illumina GenomeStudio v2011.1 (Gene Expression Module v1.9.0) software was used to calculate the raw data. Array data transformation into logarithm was conducted by using the quantile method. Statistically, significance expression data were converted into fold change values. Gene enrichment and functional annotation were analyzed by DAVID (http://david.abcc.ncifcrf.gov/home.jsp) online software to determine significant probes. Differentially expressed genes were statistically calculated via the use of the R 3.1.2 (www.r-project.org) program.
Results
Molecular Characterizations of Nickel Boride Nanoparticles
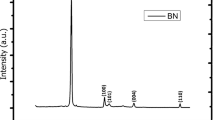
X-ray diffraction (XRD) analysis was performed to investigate the molecular structure of the synthesized molecule. According to the results, the most dominant peak was obtained at 30° (2θ) corresponding to Muller index of 110 which was correlated nickel boride (Ni2B) with the literature by using Bragg’s reflection angle law (2θ) (Fig. 1) [20]. Also, scanning electron microscope images showed that nickel boride (Ni2B) dimensions were varied from micro-sized to nanosized particle structures (Fig. 2). Besides, it could be seen from the transmission electron microscope image of nickel boride (Ni2B) particles mostly had nanolevel particle sizes (Fig. 3). Moreover, energy-dispersive X-ray spectroscopy (EDX) analysis was conducted to investigate atomic ratios (%) between boron (B) and nickel (Ni) of nickel boride (Ni2B) NPs. The analysis showed that % ratio between boron and nickel was %34.71 and %5.30, respectively. These results gave us nearly 1:2 atomic ratio for B:Ni and it was understood that the synthesized molecule mostly consisted of Ni2B molecules (Fig. 4).
Cell Viability Analysis of Human Pulmonary Alveolar Epithelial Cell Culture Against Nickel Boride Nanoparticle Applications
MTT analysis put forth that a significant difference in cell viability decrease started with 20 mg/L concentration. On the other hand, two other cytotoxicity tests (LDH and NR analysis) showed that significant inhibition in cell viability induced at 80 mg/L concentration (Fig. 5). LDH and NR cytotoxicity test results were closer to each other than MTT cell viability assay and according to this outcome, LDH and NR assay result might be more reliable than the MTT calculations. By using simple linear regression analysis in GraphPad Prism version 7.0 software, a dose inhibition equation was obtained as: Y = − 0.05180*X + 84.58 (Y = Concentration and X = Cell viability). The IC50 value was calculated as 81.99 mg/mL concentration by using the equation. As a result of this calculation, 81.99 mg/mL concentration of Ni2B NP application was further used in microarray analysis to investigate gene expression alterations in HPAEpiC cell culture.
MTT. LDH. NR assay results of nickel boride (Ni2B) NPs on the human lung alveolar epithelial cell line. Asterisk symbol represents statistically significant difference (P < 0.01) compared with negative control. (GraphPad Prism 7. Anova: Dunnett’s multiple comparison test was used to calculate the values)
In Vitro Transcriptome Analysis of Nickel Boride Nanoparticle Application
As a result of microarray analysis, it was revealed that there were alterations in the expression levels of 693 genes (fold change (FC) ≥ 2) in Ni2B NPs applied cell cultures. It was observed that there was a significant increase in gene expression in 513 of these gene probes and a decrease in gene expression in 192 gene probes. Table 1 lists the 50 genes whose expression levels vary the most in HPAEpiC cell culture treated with Ni2B NPs. IGFBP3 gene, where Ni2B NPs upregulated the level of expression, is known to be one an insulin-like growth factor [22]. EEF1A2 gene, which increased expression level with Ni2B NP application, acts as an elongation factor in translation and is thought to cause metastasis in cancer cells [23]. Another side, the expression level of the GPD1L gene decreased significantly compared with the control group, and this gene has been reported to be one of the most important factors that support or provide the viability of tumor cells in throat scale carcinomas [24]. Besides, it has been noted that the GFM1 gene, whose expression level is reduced, is involved in oxidative phosphorylation in mitochondria [25].
As a result of the DAVID analysis for Gene Ontology (the GEOTERM_BP_FAT options), it was seen that Ni2B NPs was mostly effective on oxygen level responses in cells, cell cycle regulations, and responses to organic substances (Fig. 6). Different gene pathways investigated in Gene Ontology (the GEOTERM_CC_FAT options) such as responses in chromosomal regions, centrosome organization, and microtubule regulation were also found to be affected by Ni2B NP application (Fig. 7). Moreover, DAVID annotation GO enrichment analysis for functional categories showed that Ni2B NPs generally affected reactions such as nucleus regulation and phosphoprotein synthesis in cells (Fig. 8).
Discussion
Nanoparticles were defined as small molecules having a size under 100 nm and dimension-dependent enhanced physicochemical properties. With a high surface to volume proportion, nanoparticles were known to gain higher physicochemical dynamicity and reactivity. These enhanced features attracted attention from many fields from the medical industry to cosmetics [26]. On the other hand, unique properties were shown to come with potential health risks. Today, it became possible to contact with any nanoparticle-based product in daily life such as food additives, health products, and the automotive industry [27]. To avoid detrimental consequences resulted from nanoparticle-related health problems, it was inevitable to constitute comprehensive safety reports and biocompatibility assays for nanoproducts to determine a potential area of use for these molecules.
Nickel boride (Ni2B) generally used in steel production industries for increasing the hardness of metals. It could be understood that steel products are used in every aspect of life; thus, it is possible to contact with any residues of Ni2B with even nanoforms [28]. There was no adequate information about Ni2B NP cytotoxicity in the literature, but it was mentioned that inhalation of Ni2B may cause serious health problems [29]. Also, people who work in industrial applications generally contact with NPs via inhalation. Thus, the human pulmonary alveolar epithelial cells (HPAEpiC) were used to investigate the toxicological properties of Ni2B NPs. In this study, physical, cytotoxic, and toxicogenomic properties of Ni2B NPs were investigated to understand possible toxicological consequences for human health. Ni2B NPs were synthesized via the single-step pressure method and the molecular properties of the NPs were characterized by using XRD, SEM, TEM, and EDS techniques. XRD analysis showed the dominant peak was observed at 32° (2θ) which showed the synthesized molecule most probably Ni2B particles. In addition, the fact that the other peaks were small except the dominant peak showed us that the material was quite unidirectional. This was important because it was a rather difficult growth powder crystals uniformed on any substrate, such as thin or thick films. SEM analysis showed the synthesized particles have various sizes including nanoscale molecules. Also, it could be understood from the TEM analysis most of the particles were composed of nanosized and under 20 nm. Moreover, the atomic percentage of the molecule was analyzed via the use of the EDS technique which showed the molecule consist of 2:1 ratio for Ni:B which was resulted in Ni2B formulation.
According to the cytotoxicity analyses, %50 inhibitory concentration (IC50) for HPAEpiC cell culture against Ni2B NP exposure was found 81.99 mg/mL concentration. This analysis put forth that synthesized Ni2B NPs were not highly cytotoxic molecules, but it was known that nontoxic compounds could lead to irreversible health problems by regulating gene expression patterns as claims of previous studies [30,31,32]. IC50 concentration of Ni2B NPs was used to investigate gene expression changes in HPAEpiC cells by using whole-genome microarray (Illumina human ht-12 v4.0) analysis. Gene expression analysis showed that a total of 693 genes were altered after Ni2B NP application to the cell culture. From these gene probes, 501 upregulated and 192 downregulated gene expressions were determined in HPAEpiC cell cultures.
When the upregulated gene expression profiles were examined, most prominent gene probes were found to belong IGFBP3, MT3, and EEF1A2 genes. Previous studies claimed that insulin-like growth factor binding protein 3 (IGFBP3) was found to be related to cancer progress and metastasis mediation. It was shown that IGFBP3 protein levels were highly increased in colorectal cancer cells compared with control groups and the protein could be used as a biomarker for colorectal cancer prognosis [33]. Also, in vitro transfection studies indicated that IGFBP3 overexpression in A549 cell line enhanced invasion, migration, and epithelial-mesenchymal transition [34]. Another in vitro study analyzed that IGFBP3 knockdown inhibited glioma cell proliferation in mouse intracranial xenograft models via inducing apoptosis and enhancing cell cycle G2/M arrest [35]. Moreover, overexpression analysis of metallothionein-3 (MT3) protein showed that the tumorigenesis and invasiveness were increased in prostate cancer and breast cancer cell lines [36]. Real-time reverse transcriptase-polymerase chain reaction (RT-qPCR) assays revealed that hypoxia-induced MT3 gene enhanced invasion and cell growth via inhibiting of MASPIN, NDRG2, and NDRG1 expressions in bladder carcinoma cells [37]. Also, eukaryotic elongation factor 1 alpha 2 (eEF1A2) was found to be a translation factor mainly expressed in specific organs as skeletal muscle, heart, and nervous system. It was analyzed that eEF1A2 protein expression related to tumorigenesis and poor prognosis in triple-negative breast cancers (TNBC) [38]. Clinical investigations of eEF1A2 also shown to be related to cancer progression and it was proposed to be used as a biomarker for risk-prediction in localized prostate cancer (PCa) patients [39].
Furthermore, microarray analysis showed that leading downregulated gene expressions were found at GFM1, SDF2L1, and CTPS gene probes in aspect to Ni2B NP application. G elongation factor mitochondrial 1 (GFM1) was analyzed to be a translation factor that have an important role in protein synthesis in mitochondria. Patients with downregulated GFM1 expressions were investigated to exhibit, reduced levels of oxidative phosphorylation, mitochondrial translation, progressive encephalopathy, and liver failure [25, 40]. According to the studies, stromal cell-derived factor 2 like 1 (SDF2L1) protein was highly expressed in endoplasmic reticulum (ER) stress conditions and shown to have an important role in the inhibition of misfolded ER cargoes. An in vivo study on diabetic mice analyzed that downregulation of SDF2L1 gene resulted in fatty liver and glucose intolerance [41, 42]. Cytidine 5′-triphosphate synthase (CTPS) was claimed to be a potential target for treating neointima-related disorders. A study on the mouse injury model showed that inhibition of CTPS1 might be a selective vascular repair target by preventing smooth muscle cell (SMC) proliferation and enhancing re-reendothelialization [43].
Finally, DAVID annotation GO enrichment analysis put forth that Ni2B NP application was related to various gene pathways. Pathways that regulate response to oxygen levels, cell cycle, and anti-apoptotic machinery were found to modified by Ni2B NP exposure. Also, microtubule, chromatin structures, and ER-related gene families were investigated to differentially expressed according to gene ontology analysis. Most importantly, Ni2B NP application-related functional category annotations were found to be affected phosphoprotein, DNA replication, and nuclear regulations. All in all, the investigations were taken into account together it could be concluded that although Ni2B NP application was not shown to have high cytotoxic properties on HPAEpiC cell line, improper use of the compound could affect some of the important cellular pathways that might be resulted in detrimental health problems such as cancer progressions, ER, and mitochondria-related diseases. On the other hand, further in vivo toxicology and histology experiments must be performed to better understand Ni2B NP safety conditions.
References
Acharya S, Debata M, Acharya TS, Acharya PP, Singh SK (2016) Influence of nickel boride addition on sintering behaviour and mechanical properties of TiC–Ni based cermets. J Alloys Compd 685:905–912. https://doi.org/10.1016/j.jallcom.2016.06.122
Mueller HOMESEM (2016) Binary alloy phase diagrams. In: Alloy Phase Diagrams. ASM International, pp. 89–89
Debata M, Upadhyaya A (2004) Effect of boron addition on sintering of tungsten based alloys. J Mater Sci 39:2539–2541. https://doi.org/10.1023/B:JMSC.0000020023.21159.e5
Gülsoy HÖ, Salman S (2005) Microstructures and mechanical properties of injection molded 17-4PH stainless steel powder with nickel boride additions. J Mater Sci 40:3415–3421. https://doi.org/10.1007/s10853-005-0432-2
DENG D, WANG C, LIU Q, NIU T (2015) Effect of standard heat treatment on microstructure and properties of borided Inconel 718. Trans Nonferrous Metals Soc China 25:437–443. https://doi.org/10.1016/S1003-6326(15)63621-4
Sista V, Kahvecioglu O, Kartal G, Zeng QZ, Kim JH, Eryilmaz OL, Erdemir A (2013) Evaluation of electrochemical boriding of Inconel 600. Surf Coat Technol 215:452–459. https://doi.org/10.1016/j.surfcoat.2012.08.083
Ueda N, Mizukoshi T, Demizu K, Sone T, Ikenaga A, Kawamoto M (2000) Boriding of nickel by the powder-pack method. Surf Coat Technol 126:25–30. https://doi.org/10.1016/S0257-8972(00)00517-X
Subramaniam VD, Prasad SV, Banerjee A, Gopinath M, Murugesan R, Marotta F, Sun XF, Pathak S (2018) Health hazards of nanoparticles: understanding the toxicity mechanism of nanosized ZnO in cosmetic products. Drug Chem Toxicol 42:84–93. https://doi.org/10.1080/01480545.2018.1491987
Dietz GPH, Bähr M (2004) Delivery of bioactive molecules into the cell: the Trojan horse approach. Mol Cell Neurosci 27:85–131
Brown DM, Wilson MR, MacNee W, Stone V, Donaldson K (2001) Size-dependent proinflammatory effects of ultrafine polystyrene particles: a role for surface area and oxidative stress in the enhanced activity of ultrafines. Toxicol Appl Pharmacol 175:191–199. https://doi.org/10.1006/taap.2001.9240
Lewinski N, Colvin V, Drezek R (2008) Cytotoxicity of nanoparticles. Small 4:26–49. https://doi.org/10.1002/smll.200700595
Turkez H, Arslan ME, Ozdemir O (2017) Genotoxicity testing: progress and prospects for the next decade. Expert Opin Drug Metab Toxicol 13:1–10. https://doi.org/10.1080/17425255.2017.1375097
Liu Y, Wang J (2013) Effects of DMSA-coated Fe3O4 nanoparticles on the transcription of genes related to iron and osmosis homeostasis. Toxicol Sci 131:521–536. https://doi.org/10.1093/toxsci/kfs300
Li X, He Q, Shi J (2014) Global gene expression analysis of cellular death mechanisms induced by mesoporous silica nanoparticle-based drug delivery system. ACS Nano 8:1309–1320. https://doi.org/10.1021/nn4046985
Kedziorek DA, Muja N, Walczak P, Ruiz-Cabello J, Gilad AA, Jie CC, Bulte JWM (2010) Gene expression profiling reveals early cellular responses to intracellular magnetic labeling with superparamagnetic iron oxide nanoparticles. Magn Reson Med 63:1031–1043. https://doi.org/10.1002/mrm.22290
Chen M, Zhang M, Borlak J, Tong W (2012) A decade of toxicogenomic research and its contribution to toxicological science. Toxicol Sci 130:217–228. https://doi.org/10.1093/toxsci/kfs223
Türkez H, Arslan ME, Sönmez E, Tatar A, Açikyildiz M, Geyikoğlu F (2017) Toxicogenomic responses of human alveolar epithelial cells to tungsten boride nanoparticles. Chem Biol Interact 273:257–265. https://doi.org/10.1016/j.cbi.2017.06.027
Türkez H, Arslan ME, Sönmez E, Geyikoğlu F, Açıkyıldız M, Tatar A (2019) Microarray assisted toxicological investigations of boron carbide nanoparticles on human primary alveolar epithelial cells. Chem Biol Interact 300:131–137. https://doi.org/10.1016/j.cbi.2019.01.021
Türkez H, Arslan ME, Sönmez E, Açikyildiz M, Tatar A, Geyikoğlu F (2019) Synthesis, characterization and cytotoxicity of boron nitride nanoparticles: emphasis on toxicogenomics. Cytotechnology. 71:351–361. https://doi.org/10.1007/s10616-019-00292-8
Shahbazi M, Cathey H, Danilova N, Mackinnon I (2018) Single step process for crystalline Ni-B compounds. Materials (Basel) 11:1259. https://doi.org/10.3390/ma11071259
Turkez H, Sönmez E, Di Stefano A, Mokhtar YI (2016) Health risk assessments of lithium titanate nanoparticles in rat liver cell model for its safe applications in nanopharmacology and nanomedicine. Cytotechnology 68:291–302. https://doi.org/10.1007/s10616-014-9780-6
Baxter RC (2015) Nuclear actions of insulin-like growth factor binding protein-3. Gene 569:7–13. https://doi.org/10.1016/j.gene.2015.06.028
Xu C, Hu D, Zhu Q (2013) eEF1A2 promotes cell migration, invasion and metastasis in pancreatic cancer by upregulating MMP-9 expression through Akt activation. Clin Exp Metastasis 30:933–944. https://doi.org/10.1007/s10585-013-9593-6
Feng Z, Li JN, Wang L, Pu YF, Wang Y, Guo CB (2014) The prognostic value of glycerol-3-phosphate dehydrogenase 1-like expression in head and neck squamous cell carcinoma. Histopathology 64:348–355. https://doi.org/10.1111/his.12258
Coenen MJH, Antonicka H, Ugalde C, Sasarman F, Rossi R, Heister JGAMA, Newbold RF, Trijbels FJMF, van den Heuvel LP, Shoubridge EA, Smeitink JAM (2004) Mutant mitochondrial elongation factor G1 and combined oxidative phosphorylation deficiency. N Engl J Med 351:2080–2086. https://doi.org/10.1056/NEJMoa041878
Oberdörster G, Maynard A, Donaldson K, Castranova V, Fitzpatrick J, Ausman K, Carter J, Karn B, Kreyling W, Lai D, Olin S, Monteiro-Riviere N, Warheit D, Yang H, ILSI Research Foundation/Risk Science Institute Nanomaterial Toxicity Screening Working Group (2005) Principles for characterizing the potential human health effects from exposure to nanomaterials: elements of a screening strategy. Part Fibre Toxicol 2:8
Hoet PHM, Brüske-Hohlfeld I, Salata OV (2004) Nanoparticles - known and unknown health risks. J Nanobiotechnology 2:12
Özkan Gülsoy H (2005) Influence of nickel boride additions on sintering behaviors of injection moulded 17-4 PH stainless steel powder. Scr Mater 52:187–192. https://doi.org/10.1016/j.scriptamat.2004.09.032
Nickel boride CAS 12007-01-1 | 843836. https://www.merckmillipore.com/TR/tr/product/Nickel-boride,MDA_CHEM-843836?bd=1#anchor_Safety Information. Accessed 20 Jul 2020
Rudel RA, Attfield KR, Schifano JN, Brody JG (2007) Chemicals causing mammary gland tumors in animals signal new directions for epidemiology, chemicals testing, and risk assessment for breast cancer prevention. Cancer 109:2635–2666
Sood S, Choudhary S, Wang HCR (2013) Induction of human breast cell carcinogenesis by triclocarban and intervention by curcumin. Biochem Biophys Res Commun 438:600–606. https://doi.org/10.1016/j.bbrc.2013.08.009
Bollati V, Fabris S, Pegoraro V, Ronchetti D, Mosca L, Deliliers GL, Motta V, Bertazzi PA, Baccarelli A, Neri A (2009) Imported from https://academic.oup.com/carcin/issue/36/Suppl_1. Carcinogenesis 30:1330–1335. https://doi.org/10.1093/CARCIN
Hou YL, Luo P, Ji G y, Chen H (2019) Clinical significance of serum IGFBP-3 in colorectal cancer. J Clin Lab Anal 33:e22912. https://doi.org/10.1002/jcla.22912
Yang L, Li J, Fu S, Ren P, Tang J, Wang N, Shi X, Wu J, Lin S (2019) Up-regulation of insulin-like growth factor binding protein-3 is associated with brain metastasis in lung adenocarcinoma. Mol Cell 42:321–332. https://doi.org/10.14348/molcells.2019.2441
Chen CH, Chen PY, Lin YY, Feng LY, Chen SH, Chen CY, Huang YC, Huang CY, Jung SM, Chen LY, Wei KC (2020) Suppression of tumor growth via IGFBP3 depletion as a potential treatment in glioma. J Neurosurg 132:168–179. https://doi.org/10.3171/2018.8.JNS181217
Kmiecik AM, Pula B, Suchanski J, Olbromski M, Gomulkiewicz A, Owczarek T, Kruczak A, Ambicka A, Rys J, Ugorski M, Podhorska-Okolow M, Dziegiel P (2015) Metallothionein-3 increases triple-negative breast cancer cell invasiveness via induction of metalloproteinase expression. PLoS One 10:e0124865. https://doi.org/10.1371/journal.pone.0124865
Tsui KH, Hou CP, Chang KS, et al (2019) Metallothionein 3 is a hypoxia-upregulated oncogene enhancing cell invasion and tumorigenesis in human bladder carcinoma cells. Int J Mol Sci 20:. https://doi.org/10.3390/ijms20040980
Giudici F, Petracci E, Nanni O, et al (2019) Elevated levels of eEF1A2 protein expression in triple negative breast cancer relate with poor prognosis. PLoS One 14:. https://doi.org/10.1371/journal.pone.0218030
Worst TS, Waldbillig F, Abdelhadi A, Weis CA, Gottschalt M, Steidler A, von Hardenberg J, Michel MS, Erben P (2017) The EEF1A2 gene expression as risk predictor in localized prostate cancer. BMC Urol 17:86. https://doi.org/10.1186/s12894-017-0278-3
Galmiche L, Serre V, Beinat M, Zossou R, Assouline Z, Lebre AS, Chretien F, Shenhav R, Zeharia A, Saada A, Vedrenne V, Boddaert N, de Lonlay P, Rio M, Munnich A, Rötig A (2012) Toward genotype phenotype correlations in GFM1 mutations. Mitochondrion 12:242–247. https://doi.org/10.1016/j.mito.2011.09.007
Sasako T, Ohsugi M, Kubota N, et al (2019) Hepatic Sdf2l1 controls feeding-induced ER stress and regulates metabolism. Nat Commun 10:. https://doi.org/10.1038/s41467-019-08591-6
Fujimori T, Suno R, Iemura SI, Natsume T, Wada I, Hosokawa N (2017) Endoplasmic reticulum proteins SDF2 and SDF2L1 act as components of the BiP chaperone cycle to prevent protein aggregation. Genes Cells 22:684–698. https://doi.org/10.1111/gtc.12506
Tang R, Cui XB, Wang JN, Chen SY (2013) CTP synthase 1, a smooth muscle-sensitive therapeutic target for effective vascular repair. Arterioscler Thromb Vasc Biol 33:2336–2344. https://doi.org/10.1161/ATVBAHA.113.301561
Funding
The National Boron Research Institute (BOREN) financially supported this research with Grant No: Ç0391.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Türkez, H., Arslan, M.E., Sönmez, E. et al. Safety Assessments of Nickel Boride Nanoparticles on the Human Pulmonary Alveolar Cells by Using Cell Viability and Gene Expression Analyses. Biol Trace Elem Res 199, 2602–2611 (2021). https://doi.org/10.1007/s12011-020-02374-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12011-020-02374-7