Abstract
The tumor suppressor PTEN is involved in the regulation of cell proliferation, lineage determination, motility, adhesion and apoptosis. Loss of PTEN in the bone mesenchymal stem cells (BMSCs) was shown to change their function in the repair tissue. So far, the CRISPR/Cas9 system has been proven extremely simple and flexible. Using this system to manipulate PTEN gene editing could produce the PTEN-Knocking-out (PTEN-KO) strain. We knocked out PTEN in MSCs and validated the expression by PCR and Western blot. To clarify the changes in proliferation, CCK-8 assay was applied. In support, living cell proportion was assessed by Trypan blue staining. For osteogenic and adipogenic induction, cells were cultured in different media for 2 weeks. Oil red staining and alizarin red staining were performed for assessment of osteogenic or adipogenic differentiation. The expression of Id4, Runx2, ALP and PPARγ was examined by qPCR and immunocytochemistry staining. The PTEN-KO strain was identified by sequencing. The PTEN-KO cells had an increased cell viability and higher survival compared with the wild type. However, decreased expression of Runx2 and PPARγ was found in the PTEN loss strain after induction, and consistently decreased osteogenic or adipogenic differentiation was observed by alizarin and oil red staining. Together, PTEN-KO strain showed an increased proliferation capability but decreased multi-directional differentiation potential. When BMSCs serve as seed cells for tissue engineering, the PTEN gene may be used as an indicator.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Mesenchymal stem cells (MSCs) reside mainly within the stromal compartment of bone marrow, and have multiple differentiation potentials under suitable conditions. MSCs can migrate to the injury sites and perform site-specific differentiation, immune regulation, support hematopoiesis. Therefore, they are ideal seeding cells in gene engineering and extensively applied in experimental and clinical researches for regeneration of bone and cartilage, heart, skin, central nervous and so on, owning an excellent application prospect in the field of tissue repair. However, many theoretical and technical problems exit in the present application of MSCs. Further studies are needed to determine how to effectively control their directional differentiation, proliferation, migration and integration in vivo.
The clustered, regularly interspaced, short palindromic repeats (CRISPR) system is a prokaryotic adaptive immune system which widely exists in bacteria and archaea. So far, the CRISPR/Cas9 system has been proven extremely simple and flexible. Perhaps most importantly, this system has achieved highly efficacious alteration of the genome in a number of cell types and organisms (Hwang et al. 2013; Wang et al. 2013; Wei et al. 2013; Niu et al. 2014). Using CRISPR/Cas9 technology, the limitations of transgenic and RNAi-mediated editing could be overcome in all probability.
The tumor suppressor PTEN (phosphatase and tensin homolog deleted from chromosome 10) is involved in the regulation of cell proliferation, lineage determination, motility, adhesion and apoptosis. Also, PTEN serves in the maintenance of intracellular homeostasis through coordinating with the PI3 K (phosphatidylinositol-3-kinase) signaling. Loss or mutation of PTEN has been implicated in various hereditary disorders, as well as several sporadic human cancers. Importantly, PTEN was known to confer MSCs increased migration ability (Guo et al. 2016). In this study,we aimed to clear what role might PTEN play in the in vivo proliferation and differentiation through a gene-editing approach. We applied CRISPR/Cas9 to knockout the PTEN gene in MSCs and observed the changes in proliferation and differentiation in vivo. The findings provide suggestions for PTEN regulating according to different purposes of MSC seeding.
Materials and methods
Cell culture
Rabbit bone marrow mesenchymal stem cells (BMSC) were obtained from Cyagen Corporation (Guangzhou, China) and cultured in F12 medium with 10% fetal bovine serum (FBS) (Gibco, Grand Island, USA). All media contained 2 mM glutamine, 100 units of penicillin/ml and 100 mg of streptomycin/ml. Cells were incubated at 37 °C under 5% CO2 atmosphere in a humidified chamber.
Plasmid construction
We inserted a 20-bp sequence specific to the target locus into the empty vector and created a plasmid expressing the gRNA (Fig. 1a). For this purpose, an empty gRNA expression vector was obtained from Addgene (Fig. 1b). For single gRNA cloning we purchased a set of self-complementary oligos from Invitrogen and then ligated them into pSpCas9(BB)-2A-GFP (px458, Plasmid #48138). The gRNA design was based on CRISPR design (http://crispr.mit.edu/). Six complementary oligos (5′-caccGAAACTATTCCAATGTTCAG-3′, 5′-aaacCTGAACATTGGAATAGTTTC-3′, 5′-caccgAATCCAGATGATTCTTTAAC-3′, 5′-aaacGTTAAAGAATCATCTGGATTc-3′, 5′-caccgCAAGTTCCGCCACTGAACAT-3′, 5′-aaacATGTTCAGTGGCGGAACTTGc-3′) targeting DNA within the exon of PTEN were selected, which were predicted with high-specificity to protospacer adjacent motif (PAM) target sites in the rabbit exome. Two complementary oligos (5′-CACCGCACTCTGCGGGACAGCCGC-3′ and 5′-AAACGCGGCTGTCCCGCAGAGTGC-3′) containing the PTEN guide sequence and BbsI ligation adapters were synthesized by Invitrogen. The sequence of the construct was verified by automated DNA sequence analysis performed at Invitrogen. The wild-type and the knockout sequences were also aligned by the Clustal Omega program. All PCR products were verified by DNA sequencing, namely pSpCas9(BB)-2A-GFP-T1 (px458-T1), pSpCas9(BB)-2A-GFP (px458-T2) and pSpCas9(BB)-2A-GFP-T3 (px458-T3).
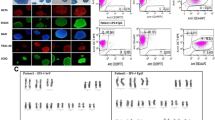
PTEN-KO validation of the MSCs. a The designed target site exon region. b The map of empty vector (from Addgene, Massachusetts, USA). c T7E1 digestion products were analyzed by agarose gel electrophoresis. The pSpCas9(BB)-2A-GFP-T2 (px458-T2) plasmid showed the highest Knock-out performance. d DNA was extracted from plasmid-transfected BMSCs and amplified through PCR. The PTEN bands did not exist after transfection e Western blot confirmed PTEN was knocked out
DNA mismatch-specific endonuclease assay
BMSC cells in six-well dishes were grown to 70–80% confluence. Cells (2000/well) were transfected using 5 μl Lipofectamine with 1 μg single-guide RNA plasmid constructs (px458-T1, px458-T2 and px458-T3). About 48 h post transfection, cells were harvested, and genomic DNAs were extracted (using QuickExtract DNA extraction solution, Epicentre, Sweden). A region of exon of the PTEN gene was amplified with genomic DNA-specific primers as follows: forward primer: TAGATTTTTCTCTTTAGATA; reverse primer: TTGCTGAACTCCTCTGG.
Homoduplex PCR products were denatured and rehybridized using stepdown annealing conditions to generate homo and heteroduplexes. The mixture of duplexes was treated with T7E1 nuclease for 15 min at 37 °C (New England Biolabs, Beijing, China). The reaction was stopped using 1.5 μl of 0.25 M EDTA, and the products were analyzed on a 2% agarose gel (Fig. 1b) (Ran et al. 2013).
Transfection and cell clone
BMSCs were cultured in six-well dishes to 70–80% confluence. Cells were cotransfected with 1 μg of pSpCas9 (BB)-2A-GFP-T2 plasmid plus 1 μg of pEGFP-N1 (Clontech, CA, USA) and 5 μl of Lipofectamine 2000/well. pEGFP-N1-derived enhanced green fluorescent protein (EGFP) was used as a fluorescent marker to sort transfected cells. Forty-eight hours posttransfection, cells were pelleted in PBS+ 2% FBS and sorted in 96-well plates using fluorescence-activated cell sorting (FACS) with a FACS-Aria II cell sorter (BD BioSciences, MD, USA). GFP-expressing cells were expanded to obtain individual clones.
Clone validation
For clone validation, DNAs were extracted for PCR amplification. The PCR products of the PTEN gene target site exon region (primer sequence) were annealed and digested by T7E1 (New England Biolabs, Beijing, China) for 15 min at 37 °C. The digested products were assayed after 2% agarose gel electrophoresis (Ran et al. 2013). As shown in Fig. 1d, the positive clones of PTEN knockout were verified by sequencing the PCR products. Further, knockout was confirmed by Western blot analysis (Fig. 1e).
CCK-8 assay
The CCK-8 assay was used to detect the cellular proliferation. The cells (PTEN-knockout and wild type) were counted separately. Cells were seeded into the 96-well plate (4800 cells per well). At 0, 24, 48, 72 and 96 h, the cellular viability was determined through adding the CCK-8 solution in 10% volume. The plate was incubated for 1 h in dark and then the absorbance at 450 nm was measured by a microplate reader. Three replications were analyzed in parallel.
Colony formation assay
Cells in the logarithmic phase were inoculated in 10-cm dishes at a density of 200 cells/dish. Culture medium was changed every 3 days. Cells were allowed to grow for 20 days to form colonies. When the colonies were clearly visible, cells were washed with PBS twice and stained with Giemsa dye (Merck, Etobicoke, CA). After washing with water for several times, the dishes were captured by a camera and the number of colonies was counted. The colony formation ratio was calculated as follow: colony number/200.
Trypan blue staining
BMSCs were digested with 0.25% trypsin and single cell suspension (1 × 106 cells/ml) was prepared. Nine volumes (180 μl) of cell suspension and one volume (20 μ) of 0.4% trypan blue were mixed. Three minutes later, living (not stained) and dead cells (dyed blue) were counted. Live cell rate (%) = the number of living cells/total cell number.
Identification of BMSC
The BMSCs were identified by the multi-directional differentiation ability. Cells were inoculated in the 0.01% type I collagen (Sigma-Aldrich, MO, USA) coated 24-well plates at a density of 5 × 103 cells/cm2. For osteogenic and adipogenic induction, cells were cultured in different media (Cyagen, China. Rabbit Mesenchymal Stem Cell Osteogenic Differentiation Basal Medium RBXMX-90021 for osteogenic induction; Adipogenic Differentiation Basal Medium BXMX-9003 for adipogenic induction) for 2–3 weeks, and the medium was refreshed every 3 days. Finally, Oil Red staining and alizarin red staining were performed for assessment of osteogenic or adipogenic differentiation. Oil-Red was prepared with fresh isopropanol (purchased from Sigma-Aldrich). The attached cells (in 6-well plate) were washed with PBS and fixed with 60% isopropanol solution for 5 min, and then incubated with Oil-Red solution (about 2 ml/well) for 5–10 min. Staining was observed under a microscope. For alizarin red staining, cells were washed with PBS and fixed with 70% ethanol, followed by rehydration for 5 min. For each well, 1 ml of alizarin red (1%, pH 4.2) was added. Ten min later, cells were washed with water for 3 times and placed at 25 °C overnight. Images were captured next day under a microscope.
Immunocytochemistry
Cells were fixed by 4% formalin for 2 h, washed by PBS for 3 times, and treated with 0.3% hydrogen peroxide at room temperature for 30 min. After washing with PBS again, cells were incubated with the primary antibodies (anti-Id4, Runx2, ALP and PPARγ, 1: 500) overnight. The next day, cells were washed and the HRP-labeled secondary antibodies (1: 100, with 1% BSA) was added followed by incubation at room temperature for 2 h, and then again washed for 3 times. DAB staining was performed. After washing again, hematoxylin staining was performed for 3 min. For negative control, PBS was applied instead of the primary antibodies.
Western blot
The protein lysate was added into each well and proteins were extracted. The amount of protein was quantified by the BCA Kit. For electrophoresis, 50 μg protein was loaded and separated. The protein was transferred to the PVDF membrane under a constant current (250 mA) at 4 °C. The membranes were blocked in 5% TBST-skim milk for 1 h at 37 °C, and then incubated overnight at 4 °C with primary antibodies. The blots were then incubated with the secondary antibodies. After washing for 3 times, immunoblots were visualized using Millipore ECL Western Blotting Detection System (Millipore). The primary and secondary antibodies were as follow: rabbit anti-PTEN (Abcam, 1: 1000, San Francisco, USA), rabbit anti-RUNX2 (Abcam, 1: 1000, San Francisco, USA), rabbit anti-PPARγ (Abcam, 1: 1000, San Francisco, USA), rabbit anti-GAPDH (Abcam, 1: 1000, San Francisco, USA), goat anti-rabbit IgG H&L-HRP (Abcam, 1: 1000, San Francisco, USA).
Real-time qPCR
Trizol (Merck, Etobicoke, CA) was added to a pellet of cells. All tubes were centrifuged (12,000 r/min) for 10 min at 4 °C. The supernatant was transferred to a new EP tube and placed on ice for 5 min. The supernatant was added with one-fifth volume of chloroform (compared to Trizol), rapidly vortex shocked for 15 s, and then centrifuged (23,000 g) for 15 min at 4 °C. The upper layer was transfered to a new EP tube and an equal volume of isopropyl alcohol was added. Again, the mixture was centrifuged (23,000 g) for 15 min at 4 °C, and the supernatant was discarded. 75% ethanol (in DEPC water) was added and mixed. After centrifugation at 4 °C (23,000 g) for 5 min, the supernatant was discarded. After drying naturally at room temperature, the RNA was dissolved by 20 µl DEPC water and stored at − 80 °C.
The total RNA was reverse transcribed into cDNA. Quantitative real-time PCR (qPCR) reactions were performed using the SYBR Green detection systems (Bio Rad, CA, USA), according to the following protocol: (95 °C for 3 min) + 40 cycles of (95 °C for 30 s + 52 °C for 30 s + 70 °C for 30 s). Each gene (Id4, Runx2, ALP and PPARγ) was subjected to three parallel samples. GAPDH was used as a house-keeping control for qualification.
Statistical analysis
All cell experiments were performed at least three times and the values were expressed as mean ± SEM. Statistical analysis was performed with One-way ANOVA or Two-way ANOVA. Differences were considered significant when *P < 0.05 and highly significant when **P < 0.01.
Results
PTEN-KO clone validation
As Fig. 1c shown, the T7E1 digestion products were analyzed by agarose gel electrophoresis. The pSpCas9 (BB)-2A-GFP-T2 plasmid showed the highest Knock-out performance (Fig. 1c, T2 group had the highest relative density of lower two digested bands/three bands). For validation, DNA was extracted from plasmid-transfected BMSCs and amplified through PCR. Also, Western blot analysis confirmed that PTEN was truly knocked out, because the PTEN band did not exist after transfection (Fig. 1e). Sequencing validation of the PTEN exon was as follow.
Wild type:
TCCACCAGGGAGTAACTATTCCCAGTCAGAGGCGCTATGTGTATTATTATAGCTACCTG. The allel with a 4-bp loss:
TCCACCAGGGAGT—TATTCCCAGTCAGAGGCGCTATGTGTATTATTATAGCTACCTG. The allel with a 13-bp loss:
TCCACCAGGGAGTAA———GAGGCGCTATGTGTATTATTATAGCTACCTG.
PTEN-KO BMSCs have an increased cell proliferation
The cell viability was assessed by both CCK-8 assay and Trypan blue staining. PTEN-KO cells showed an increased proliferation in four days’ growing compared with the WT strain (Fig. 2a, on day 1–4, t-test P < 0.05 vs WT). This conclusion was also supported by Trypan blue staining, which demonstrated that living cells accounted for 96.3% in PTEN-KO cells while for 91.8% in the WT. Moreover, the colony formation (Fig. 2b) of the PTEN-KO cells (87.36%) was much higher than for WT cells (58.21%). These results suggested that PTEN loss may confer an up-regulation in cell proliferation.
PTEN-KO BMSCs have an increased proliferation and decreased multipotent capability. a CCK-8 assay indicated an increased viable cell number compared to the WT (day 0 = 100% for each group). b Colony formation assay. The identical seeding number of PTEN-KO cells formed much more clonies than WT. c Alizarin red staining. d Oil red staining. e PTEN-KO BMSCs showed a decreased Runx2 mRNA level (normalized by the WT level). f PTEN-KO BMSCs showed a decreased PPARγ mRNA level (normalized by the WT level). g PTEN-KO BMSCs showed a decreased Runx2 protein level assessed by Western blot. h PTEN-KO BMSCs showed a decreased PPARγ protein level assessed by Western blot. i PTEN-KO BMSCs showed a decreased Runx2 protein level assessed by immunochemistry. j PTEN-KO BMSCs showed a decreased PPARγ protein level assessed by immunochemistry. *P < 0.05 versus WT, **P < 0.01 versus WT
PTEN-KO decreases the multipotent capability
For two strains, alizarin red and oil red staining was performed for assessment of multipotent capability after PTEN knocking out. On one hand, osteogenic differentiation level (assessed by alizarin red staining) of the PTEN-KO cells was obviously decreased compared with the wild type (Fig. 2c); on the other hand, the adipogenic differentiation level (assessed by oil red staining) was also decreased (Fig. 2d). For further confirmation, the osteogenic marker Runx2 and adipogenic marker PPARγ were examined. Real-time qPCR results demonstrated that the Runx2 and PPARγ mRNA levels were significantly down-regulated after gene editing (Fig. 2e, f; Runx2: PTEN-KO vs WT P < 0.01; PPARγ: PTEN-KO vs WT P < 0.05). Consistently, the protein expression of Runx2 and PPARγ were decreased in the PTEN-KO BMSCs compared with wild type, as shown by Western blot and immunochemistry analysis (Fig. 2g–j). These findings suggested that PTEN loss increases the proliferation ability when it suppresses the multi-directional differentiation potential of BMSCs.
Discussion
In this study, we exploited the CRISPR/Cas technology for genetic manipulations in BMSCs. Our results demonstrated that rabbit BMSCs are amenable to precise genetic manipulations, and using this technology for PTEN knockout cells were obtained, with an increased proliferation capability and decreased osteogenic and adipogenic differentiation ability.
The tumor suppressor PTEN preferentially dephosphorylates phosphoinositide substrates. It negatively regulates intracellular levels of phosphatidylinositol-3,4,5-trisphosphate in cells and functions as a tumor suppressor by negatively regulating AKT/PKB signaling pathway. Previously, its role in BMSCs was reported that PTEN-PI3 K/Akt/mTOR pathway, which could be negatively regulated by Notch signaling, promoted the adipogenic differentiation (Song et al. 2015). Our result, that PTEN-KO BMSCs had a suppressed adipogenic differentiation, was consistent with this report. Besides, PTEN/Akt signaling was known to play an essential role in the senescent and apoptotic BMSCs from systemic lupus erythematosus patients by up-regulating p27 expression (Tan et al. 2015). Another work exposed MSCs to hyperglycemic conditions and found a decreased miR-32-5p expression and disturbed cell cycle progression through a PTEN-mediated inhibitory effect on the PI3 K/Akt signaling pathway (Zhu et al. 2013). The above two conclusions are also supported by our data showing the increased proliferation of the PTEN-KO strain.
PTEN is not a classic osteogenic promoter, but some independent studies commonly suggested it is positively relating to osteogenic differentiation. A mechanism of Runx2 as the osteogenic marker was reported that it up-regulates PTEN and thus inhibits pAKT/pGSK3β/β-catenin signaling pathways (Zhou et al. 2016a, b). In vascular smooth muscle cells, an increase in PTEN induces early osteogenic differentiation (Kajimoto et al. 2015). However, opposite conclusions exit as well. For example, BMP9 down-regulates PTEN, and the PTEN inhibitor VO-Ohpic increases osteogenic markers induced by BMP9 in embryonic fibroblasts (Huang et al. 2014). Similarly, adiponectin downregulated PTEN expression, and SF1670 (a PTEN inhibitor) increased the expression of osteogenic markers such as osteocalcin, ALP, and RUN2 in adiponectin-treated hBMSCs (Liu et al. 2017). Together, PTEN’s role in osteogenic differentiation may be decided by the cell types. In our rabbit BMSC model, the decreased Runx2 expression and reduced alizarin red staining in PTEN-KO cells were more an indicator of impaired multi-directional differentiation potential under specific induction medium.
In the adipogenic differentiation field, besides the study showing PTEN/PI3 K/Akt/mTOR pathway was negatively regulated by Notch signaling (Song et al. 2015), some inconsistent evidences are also needed to mention. A recent work found that miR-26a targets (inhibits) PTEN and hence promotes adipogenesis (Trohatou et al. 2017). This is opposite to our findings about the phenotype of PTEN-KO MSCs. PTEN was also reported as a target of microRNA-26b, which was positively correlated with mRNA levels of adipocyte-specific molecular markers (Song et al. 2014). These challenging conclusions merit further confirmation, and at least they were not performed on BMSCs, which indicates distinct roles of PTEN located in different cells. Our further study would probe the expression of miR-26a and miR-26b in PTEN-KO MSCs.
The effect of PTEN on proliferation was widely studied, especially in tumor researches. Overwhelming views suggested an antiproliferative role of PTEN via PTEN/PI3 K/Akt/mTOR (PTEN suppresses Akt/mTOR) (Song et al. 2016; Wu et al. 2016; Zhou et al. 2016a, b; Tian et al. 2017). Furthermore, it has also been reported that MiR-429 regulates PTEN and hence controls the Wnt and AKT signaling pathways in the process of cancer cell proliferation and tumor formation (Tang et al. 2015). Our result also demonstrated an increased proliferation of BMSCs after PTEN loss.
In conclusion, we manipulated the PTEN gene through the CRISPR/Cas9 system and acquired the PTEN-KO BMSCs from rabbit. The PTEN-KO strain showed an increased proliferation capability and decreased multi-directional differentiation potential compared with the WT. These results indicate that when BMSCs, as the seed cells for tissue engineering, showed a low expression of PTEN, it suggests an impaired differentiation and tissue repair function.
Abbreviations
- CRISPR:
-
Clustered, regularly interspaced, short palindromic repeats
- FBS:
-
Fetal bovine serum
- MSCs:
-
Mesenchymal stem cells
- PTEN:
-
Phosphatase and tensin homolog deleted from chromosome 10
- SEM:
-
Standard error of mean
- TALENs:
-
Transcription activator-like effector nucleases
- ZFNs:
-
Zinc-finger nucleases
References
Guo XR, Hu QY, Yuan YH, Tang XJ, Yang ZS, Zou DD, Bian LJ, Dai LJ, Li DS (2016) PTEN-mRNA engineered mesenchymal stem cell-mediated cytotoxic effects on U251 glioma cells. Oncol Lett 11:2733–2740
Huang J, Yuan SX, Wang DX, Wu QX, Wang X, Pi CJ, Zou X, Chen L, Ying LJ, Wu K, Yang JQ, Sun WJ, Deng ZL, He BC (2014) The role of COX-2 in mediating the effect of PTEN on BMP9 induced osteogenic differentiation in mouse embryonic fibroblasts. Biomaterials 35:9649–9659
Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JR, Joung JK (2013) Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol 31:227–229
Kajimoto H, Kai H, Aoki H, Uchiwa H, Aoki Y, Yasuoka S, Anegawa T, Mishina Y, Suzuki A, Fukumoto Y, Imaizumi T (2015) BMP type I receptor inhibition attenuates endothelial dysfunction in mice with chronic kidney disease. Kidney Int 87:128–136
Liu X, Chen T, Wu Y, Tang Z (2017) Role and mechanism of PTEN in adiponectin-induced osteogenesis in human bone marrow mesenchymal stem cells. Biochem Biophys Res Commun 483:712–717
Niu J, Zhang B, Chen H (2014) Applications of TALENs and CRISPR/Cas9 in human cells and their potentials for gene therapy. Mol Biotechnol 56:681–688
Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F (2013) Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8:2281–2308
Song G, Xu G, Ji C, Shi C, Shen Y, Chen L, Zhu L, Yang L, Zhao Y, Guo X (2014) The role of microRNA-26b in human adipocyte differentiation and proliferation. Gene 533:481–487
Song BQ, Chi Y, Li X, Du WJ, Han ZB, Tian JJ, Li JJ, Chen F, Wu HH, Han LX, Lu SH, Zheng YZ, Han ZC (2015) Inhibition of Notch Signaling Promotes the Adipogenic Differentiation of Mesenchymal Stem Cells Through Autophagy Activation and PTEN-PI3 K/AKT/mTOR Pathway. Cell Physiol Biochem 36:1991–2002
Song H, Zhang Y, Liu N, Wan C, Zhang D, Zhao S, Kong Y, Yuan L (2016) miR-92b regulates glioma cells proliferation, migration, invasion, and apoptosis via PTEN/Akt signaling pathway. J Physiol Biochem 72:201–211
Tan W, Gu Z, Shen B, Jiang J, Meng Y, Da Z, Liu H, Tao T, Cheng C (2015) PTEN/Akt-p27(kip1) Signaling Promote the BM-MSCs Senescence and Apoptosis in SLE Patients. J Cell Biochem 116:1583–1594
Tang J, Li L, Huang W, Sui C, Yang Y, Lin X, Hou G, Chen X, Fu J, Yuan S, Li S, Wen W, Tang S, Cao D, Wu M, Chen L, Wang H (2015) MiR-429 increases the metastatic capability of HCC via regulating classic Wnt pathway rather than epithelial-mesenchymal transition. Cancer Lett 364:33–43
Tian DD, Zhang RX, Wu N, Yuan W, Luo SH, Chen HQ, Liu Y, Wang Y, He BC, Deng ZL (2017) Tetrandrine inhibits the proliferation of human osteosarcoma cells by upregulating the PTEN pathway. Oncol Rep 37:2795–2802
Trohatou O, Zagoura D, Orfanos NK, Pappa KI, Marinos E, Anagnou NP, Roubelakis MG (2017) miR-26a Mediates Adipogenesis of Amniotic Fluid Mesenchymal Stem/Stromal Cells via PTEN, Cyclin E1, and CDK6. Stem Cells Dev 26:482–494
Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R (2013) One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153:910–918
Wei C, Liu J, Yu Z, Zhang B, Gao G, Jiao R (2013) TALEN or Cas9–rapid, efficient and specific choices for genome modifications. J Genet Genom 40:281–289
Wu YR, Qi HJ, Deng DF, Luo YY, Yang SL (2016) MicroRNA-21 promotes cell proliferation, migration, and resistance to apoptosis through PTEN/PI3 K/AKT signaling pathway in esophageal cancer. Tumour Biol 37:12061–12070
Zhou J, Hu Y, Chen Y, Yang L, Song J, Tang Y, Deng F, Zheng L (2016a) Dicer-dependent pathway contribute to the osteogenesis mediated by regulation of Runx2. Am J Transl Res 8:5354–5369
Zhou XM, Sun R, Luo DH, Sun J, Zhang MY, Wang MH, Yang Y, Wang HY, Mai SJ (2016b) Upregulated TRIM29 promotes proliferation and metastasis of nasopharyngeal carcinoma via PTEN/AKT/mTOR signal pathway. Oncotarget 7:13634–13650
Zhu G, Chai J, Ma L, Duan H, Zhang H (2013) Downregulated microRNA-32 expression induced by high glucose inhibits cell cycle progression via PTEN upregulation and Akt inactivation in bone marrow-derived mesenchymal stem cells. Biochem Biophys Res Commun 433:526–531
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Shen, Y., Zhang, J., Yu, T. et al. Generation of PTEN knockout bone marrow mesenchymal stem cell lines by CRISPR/Cas9-mediated genome editing. Cytotechnology 70, 783–791 (2018). https://doi.org/10.1007/s10616-017-0183-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10616-017-0183-3