Abstract
Our study aimed to explore associations between microRNA-21 (miR-21) and PTEN/PI3K/AKT signaling pathway and, further, to elucidate the regulation of miR-21 on biological behaviors in human esophageal cancer cells. The expressions of miR-21, PTEN, PI3K, and AKT were detected in 89 esophageal cancer samples and 58 adjacent normal tissues respectively. The human esophageal cancer cells (TE11) were grouped as following: blank (TE11 cells without transfection), negative (TE11 cells with miR-21 negative inhibitor), and Inhibition-miR21 (TE11 cells with miR-21 inhibitor). Western blot was used for detection of PTEN, P13K, and AKT protein expressions, MTT method for cell proliferation, Transwell assay for cell migration and invasion, and flow cytometry for cell cycle and apoptosis. MiR-21, PI3K, and AKT have higher expressions, but PTEN has lower expression in esophageal cancer tissues compared with adjacent normal tissues. The esophageal cancer tissues with lymph node metastasis and poor differentiation showed significantly low positive rate of PTEN protein, but high positive rates of PI3K and AKT proteins. Compared with blank and negative groups, PTEN expression of TE11 cells in Inhibition-miR21 group was significantly up-regulated, but PI3K and AKT were down-regulated. Further, PTEN was a target gene of miR-21. Besides, compared with blank and negative groups, the proliferation, migration, and invasion of TE11 cells were less active in Inhibition-miR21 group. TE11 cells were significantly increased in the G0/G1 phase of cell cycles, but decreased in the S and G2/M phase in Inhibition-miR21 group. The TE11 cells exhibited significantly increased apoptosis rates. MiR-21 targets key proteins in PTEN/PI3K/AKT signal pathway, promoting proliferation, migration, invasion, and cell cycle, and inhibiting apoptosis of human esophageal cancer cells. It may serve as a novel therapeutic target in esophageal cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Esophageal cancer is a common malignant tumor of the digestive tract, and an estimated 456,000 new cases and 400,000 deaths were attributed to this disease worldwide in 2012 [1]. China is one of high-incidence areas of esophageal cancer, and it has been reported approximately 208,473 people die of esophageal cancer in 2010 [2]. Currently, the pathogeny of esophageal cancer has not been figured out yet, but it is related to various factors including chemical stimulation, inflammation and trauma, genetic factors, and lifestyles [3–6]. The traditional treatment for esophageal cancer includes radiation therapy, chemotherapy, surgical treatment, and comprehensive treatment, while these therapies cannot always achieve satisfactory therapeutic effects [7–9]. Therefore, an in-depth understanding of pathogenesis as well as biological changes of esophageal cancer is of profound significance for esophageal cancer treatment [10].
With the extensive research of molecular biology of cancer, researchers have found that PTEN/PI3K/AKT signal pathways are closely associated with growth, proliferation, infiltration, expansion, and metastasis of malignant cells [11]. Currently, the abnormity of this signal pathway has been found in diseases such as cerebral cavernous malformations, triple-negative breast cancer, thyroid cancer, and prostate cancer [12–15]. MicroRNAs (miRNAs) are small, non-coding, 21–23 nucleotide long RNAs that negatively regulate gene expression through translational inhibition as well as destabilization of mRNA [16]. Genome-wide study has reported that miRNAs are commonly located at cancer-related genomic regions or in fragile sites, and in minimal regions of loss of heterozygosity or of amplifications, or in frequent breakpoint [17]. It has shown to be that miR-21 participates in the process of cell division, differentiation, and apoptosis in gastric cancer, lung cancer, and breast cancer [17–19]. The latest researches show that miR-21 which works on PTEN/PI3K/AKT signal pathways influences the biological behaviors of malignant cells [20, 21]. The purpose of this study was to explore associations between miR-21 and PTEN/PI3K/AKT signaling pathway and, further, to elucidate the regulation of miR-21 on biological behaviors in human esophageal cancer cells.
Materials and methods
Ethical statement
The study was approved by the ethics committee of Union Hospital Affiliated to Tongji Medical College of Huazhong University of Science and Technology and the informed consents were signed by all participants. All the experimental procedures in this study were performed on the basis of Declaration of Helsinki [22].
Study Subjects
This study included 89 paraffin specimens (case group) radically excised from 89 patients who were pathologically diagnosed with esophageal cancer in Union Hospital Affiliated to Tongji Medical College of Huazhong University of Science and Technology between October 2012 and October 2014. All patients, 58 males and 31 females, were not treated with radiotherapy and chemotherapy before surgery, and the average age was 62.7 ± 10.2 years old. Among these patients, there were 31 with lymph node metastasis and 58 without, and there were 32 cases of high differentiation, 34 cases of moderate differentiation, and 23 cases of poor differentiation. In addition, 58 adjacent normal tissues (more than 5 cm from lesions) which were pathologically confirmed to be normal squamous epithelium with a small amount of lamina propria were collected as control group.
Immunohistochemistry streptavidin-peroxidase method
The immunohistochemical technique was used according to streptavidin-peroxidase (SP) kit instruction. Paraffin sections were dewaxed and hydrated; 3 % of H2O2–methanol was used to inhibit enzymatic activity of endogenous peroxidase, and 0.5 % bovine serum albumin was used for sealing non-specific antibody. First antibodies (PTEN: ab31392, Abcam, Cambridge, UK; PI3k: ab86714, Abcam, Cambridge, UK; p-AKT: ab38449, Abcam, Cambridge, UK) were added to react in 4 °C humidity chamber overnight. Then we added biotin labeling second antibody [horseradish peroxidase (HRP)-labeled goat anti-rabbit, A0208, Beyotime, Shanghai, China] for incubation and added third antibody (HRP-labeled streptavidin working fluid, Zhongshan Jinqiao Co., Ltd., Beijing, China). In the three steps, we used phosphate buffered saline (PBS) for irrigation three times and used diaminobenzidine (DAB) chromogen and hematoxylin for counterstain and went through regular dehydration/clearing and mounting.
We got 5 high power fields (×400) of each section at random and took the average figure of each section. The calculation method took positive cell percentage ratio and tinting strength into consideration. The sections were classified in terms of positive cell percentage ratio. The number of positive cells is 0, score 0 point. The positive cells are less than 10 %, score 1 point. The number of positive cells is between 11 and 50 %, score 2 points. The positive cells are 50–75 %, score 3 points. The number of positive cells is more than >70 %, score 4 points. The sections were classified in terms of cell tinting strength. There is no tint in the cells, score 0 point. There is light tint (wax yellow) in the cells, score 1 point. There is medium tint (pale brown), score 2 points. There is strong tint (brown), score 3 points. The sections were classified in terms of the multiplication of tinting strength and positive cell percentage ratio. The result is between 0 and 2 points, mark (−). The result is between 3–5 points, mark (+). The result is between 10–12, mark (+++) [23].
Real-time polymerase chain reaction
The total RNA was extracted according to Trizol (ThermoFisher Scientific, Shanghai, China) instruction. Ultraviolet spectrophotometer was used to test the concentration of RNA: A260/280 = 1.8–2.1 which was adjusted to 500 ng/μL. Real-time polymerase chain reaction (PCR) was used to analyze the expression level of miR-21 of tissue samples and human esophageal cancer TE11 cells after transfection using miRNA cDNA Synthesis kit and miR-21 qualification real time-PCR kit (Takara Biotechnology (DALIAN) CO., LTD., China). Real-time PCR was performed on an ABI7500 real-time PCR system (Applied Biosystems, Foster City, CA, USA). The reaction conditions were as follows: 95 °C at 10 s, 1 cycle; 95 °C at 5 s, 60 °C at 34 s, 45 cycles. U6RNA was taken as internal reference, and amplification primer sequence was shown in Table 1. 2-△△CT was used to calculate the relative quantitative expression level of miR-21.
Cell culture
The human esophageal cancer cells (TE11) were provided by Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences. RPMI-1640 culture medium (Gibco company) supplemented with 10 % FBS, 100 μg/mL of streptomycin, and 100 U/mL of penicillin was used to culture the cells in 5 % CO2 cell incubator at 37 °C and changed the fluid every 48 h and then transfected the cells when the cell fusion was up to 70–80 %.
Cell transfection
We divided the TE11 cells into 3 groups including the Inhibition-miR21, negative and blank groups. Instead of using miR-21 inhibitor, we used miR-21 inhibitor negative control (Ambion Inc., Austin, TX, USA) in negative and used PBS in Blank.
Cell transfection steps are as follows: 50 μL of serum-free RPMI-1640 culture medium was used to dilute 20 pM of miR-21 inhibitor and incubated the cells at room temperature for 5 min, and 1 μL Lipofectamine 2000 (ThermoFisher Scientific, Shanghai, China) was put in 50 μL of serum-free RPMI-1640 culture medium without antibiotics for dilution and incubated the cells for 5 min. Then miR-21 inhibitor and Lipofectamine 2000 diluted were mixed into tranfection fluid. One hundred microliters of transfection fluid was added in every well of TE11 cells and placed the culture plates in 5 % CO2 incubator at 37 °C for 6 h. The serum-free culture medium was replaced without antibiotics by common culture medium, and the cells were tested again 48–72 h later.
Luciferase assay
The binding site of miR-21 and PTEN 3′UTR was predicted with the help of miRNA target prediction software of bioinformatics. The promoter sequence of PTEN 3′UTR containing the binding site of miR-21 was synthetized, with construction of wild-type (WT) plasmid (PTEN 3′UTR-WT). Based on this plasmid, the binding site of miR-21 was mutated and the mutant-type (MUT) plasmid (PTEN 3′UTR–MUT). According to the instructions of plasmid extraction kit (Promega Corporation, Beijing, China), 0.75 % agarose gel electrophoresis was prepared after the content of these plasmids was measured using Nenodrop. In our experiment, non-sequence group (NC) and miR-21 transfection group were set. The cells were maintained in 24-well plate at 37 °C 5 % CO2 for 24 h to obtain 50–60 % cell fusion. Also, according to the instructions of Lipofectamine 2000 (Thermo Fisher Scientific, USA), Lipofectamine 2000 transfection reagent and oligonucleotide were mixed and incubated for 15 min. Then the mixture was placed in 24-well plate. After 6 h, the solution was changed and the mixture continued to culture for 24 h, and then plasmid transfection was repeated, with culture of 24 h again. After 48 h, microwell plate light device was used to detect fluorescence content.
Western blot
The cells which grew well in the three groups were split by using pre-cooling cell lysis solution and extracted total protein. Bicinchoninic acid (BCA) protein assay was used to test protein concentration. Total protein of cells were blended with 2 × SDS gel electrophoresis sample loading buffer and then boiled for 5 min. With 15 % running gel and 5 % concentrated latex prepared, tris-glycine running buffer was put in electrophoresis tank and added 5 μg of protein and moved electrophoresis and electricity to nitrocellulose filter (NC) membrane, and 1 × Ponceau S was used to tint the cells for 2–3 min and used 5 % skimmed milk powder to seal NC membrane and put it at 37 °C for 2 h. Tris-buffered saline containing 0.2–0.4 % Tween-20 (TBST) was used to wash it every 5 min and added coating antibody which were rabbit anti PTEN-specific antibody (ab31392, 1:1000, Abcam, Cambridge, UK), rabbit anti PI3K-specific antibody (ab86714, 1:1000, Abcam, Cambridge, UK), and rabbit anti AKT-specific antibody (ab38449, 1:1000, Abcam, Cambridge, UK) and incubated the cells at 4 °C overnight. After washing (3 × 10 min TBST) the membrane, HRP-labeled anti-rabbit IgG detection antibody (A0208, 1:2000, Beyotime Company, Shanghai, China) was added and incubated the cells for 1 h at room temperature and washed (3 × 10 min TBST) the membrane. Equivalent amount of detection reagents 1 and 2 of an ECL Western Blotting Analysis System was added and mixed and put onto the membrane for 1 min. The extra fluid was removed from the edge, and plastic wrap was used to cover it and the images were exposed, developed, and fixed. Western blot result image was fed into the computer, and BEIHANG CM-2000 Biomedicine Image Analysis System was used to test integrated grey level of stripes. β-Actin (ab194952, 1:1000, Abcam, Cambridge, UK) was taken as internal reference for correction, and semi-quantitative ratio of optical density (OD) of stripes to OD of β-actin protein bands was used for indication.
The methyl-tetrazolium method
The cells in the three groups were seeded on 96-well plate at a density of 5 × 103 cells/well, with each well filled with 200 μL. After 24 h of culture, 20 μL methyl-tetrazolium (MTT) fluid (5 mg/mL) was added for incubation at 7 °C for 4 h and then supernatant liquid was removed. Dimethyl sulphoxide (DMSO, 150 μL) continued to be added and then vibrated at low speed for 10 min. Enzyme-linked immuno sorbent assay (ELISA) Reader was used to test OD570 (once every 1 day for 4 days).
Transwell assay
A volume of 60 μL of Matrigel matrix (5 mg/mL) (Becton Dickinson Biosciences, Bedford, Mass., USA) was added in Transwell upper cab and was shaken horizontally until it covered the bottom of the cab according to Transwell instruction. Pancreatin/EDTA (0.05 %) was used to digest the cells of three groups, and serum-free RPMI 1640 culture medium was used to wash and resuspend cells and the density was adjusted to 1 × 106/mL. Two hundred microliters of cell suspension was added in the upper cab and 600 μL of RPMI culture medium which contained 10 % fetal bovine serum was added and put in CO2 cell incubator for 24 h in accordance with routine culture. Cotton swabs were used to wipe out the residual Matrigel matrix, and the cells in the upper cab and 0.1 % crystal violet were used to tint the cells in the lower cab for 30 min and 10 % acetic acid was used to wash the crystal violet. Coated wells were used to test OD570 and every group repeated 3 times. In the migration experiment, there was no need to add Matrigel matrix in the upper cab and other conditions were the same as the invasion experiment.
Flow cytometry
Cell apoptosis test
PBS was used to wash the cells twice, and 400 μL of binding buffer was used to resuspend cells and 5 μL of AnnexinV-FITC was added and blended for reaction at room temperature for 10 min. Ten microliters 20 μg/mL of PI was added for incubation away from light at 4 °C for 30 min, and flow cytometry (GUAVA Technologies, Inc., Billerica, MA, USA) was used to test for analysis. Ten thousand fluorescence signals of each sample were collected.
Cell cycle test
Pancreatin/EDTA (0.05 %) was used to digest the cells of three groups, and PBS was used to wash the cells twice and 75 % ethanol was used to fix them (−20 °C). Four hours later, 400 μL of binding buffer was used to resuspend cells and 10 μL 20 μg/mL PI (with RnaseA) dye liquor was added and placed in refrigerator at 4 °C for dyeing away from light and flow cytometry was used to test for analysis. A total of 10,000 fluorescence signals of each sample were collected.
Statistical analysis
All data were analyzed using SPSS.19.0 (SPSS Inc., Chicago, IL, USA). Categorical data were presented with percentage or rate, and their comparison was examined using chi-square test. The quantitative data were expressed as mean ± standard deviation, among which comparison between groups were conducted using t test, and among groups using one-way analysis of variance (ANOVA), with pairwise comparison tested by Ducan method (ɑ = 0.05). P < 0.05 was considered as statistically significant.
Results
The expression of PTEN, PI3K, and AKT proteins in esophageal cancer tissue and adjacent normal tissues
PTEN, PI3K, and AKT proteins are three key proteins of PTEN/PI3K signal pathways. The results of immunohistochemical staining were shown in Fig. 1. The positive expression of PTEN protein was mainly in endochylema and cell membrane, and the positive rate of PTEN protein expression in esophageal cancer tissue was significantly lower than those in adjacent normal tissue (P < 0.05). The positive expression of PI3K and AKT protein were mainly in endochylema and karyon, and the positive rate of PI3K and AKT protein in esophageal cancer tissue was significantly higher than those in adjacent normal tissue (P < 0.05). The results were shown in Table 2.
Immunohistochemical staining of PTEN, PI3K, and AKT proteins in esophageal cancer tissues and adjacent normal tissues. a PTEN protein in cancer issues presented negative expression (SP × 400). b PTEN protein in adjacent normal tissues presented positive expression (SP × 400). c PI3K protein in cancer issues presented positive expression (SP × 400). d PI3K protein in adjacent normal tissues presented positive expression. e AKT proteins in cancer tissues presented positive expression (SP × 400). f AKT protein in adjacent normal tissues presented positive expression (SP × 400)
Table 3 showed the associations of the expressions of PTEN, PI3K, and AKT proteins in esophageal cancer with clinical features. There was no significant difference of the positive expressions of PTEN, PI3K, and AKT proteins among cancer issues in terms of age and gender (P > 0.05). The positive rate of PTEN protein in cancer tissue with lymph node metastasis was lower than that without lymph node metastasis (P < 0.05), while the positive rates of PI3K and AKT proteins in cancer tissue with lymph node metastasis were higher than that without lymph node metastasis (P < 0.05). The higher differentiation of cancer tissue indicated the lower positive rate of PTEN protein, but the higher positive rates of PI3K and AKT proteins (all P < 0.05).
miR-21 expression in esophageal cancer and adjacent normal tissues
The total RNA of human esophageal cancer tissues and adjacent normal tissues were extracted, and U6RNA was taken as internal reference. Two-step RT-PCR was used to test the expression level of miR-21, which was shown in Fig. 2, and the relative expression rate of miR-21 in human esophageal cancer tissues was significantly higher than that in adjacent normal tissues (P < 0.05). In addition, we also found that miR-21 expression bore no relationship with age, gender, lymph node metastasis, and differentiation degree (all P > 0.05) (Table 4).
The relative expression rate of miR-21 in esophageal cancer tissues and adjacent normal tissues. The relative expression rate of miR-21 in esophageal cancer tissues (experimental group) was higher than that in adjacent normal tissues (control group). a in the figure represents that there is significant difference when compared with the control group (P < 0.05)
miR-21 expression in TE11 cells after transfection
As shown in Fig. 3, the expression of miR-21 in the Inhibition-miR21, negative, and blank groups were 0.615 ± 0.037, 1.039 ± 0.050, and 1.050 ± 0.063, respectively. The expression level of miR-21 in the Inhibition-miR21 was significantly lower than that in the blank and negative groups (P < 0.05), suggesting the successful transfection.
The expression of miR-21 in TE11 cells when the expression of miR-21 was inhibited. The expression level of miR-21 in the Inhibition-miR-21 group was significantly lower than the blank and negative group. a in the figure represents that there is significant difference when compared with the blank and negative control groups (P < 0.05)
Down-regulation of miR-21 up-regulates PTEN but down-regulates PI3K and AKT proteins
The result of Western blotting test was shown in Fig. 4. After down-regulation of miR-21, the expression level of PTEN protein in TE11 cells was significantly up-regulated and the expression level of PI3K and AKT proteins were significantly down-regulated (all P < 0.05).
The expression levels of PTEN, PI3K, and AKT proteins when the expression of miR-21 was inhibited. The expression level of PTEN protein in TE11 cells was significantly up-regulated, and the expression level of PI3K and AKT proteins was significantly down-regulated. a in the figure represents that there is significant difference when compared with the blank and negative control groups (P < 0.05)
Target association between miR-21 and PTEN
MiRNA target gene prediction online demonstrated that PTEN is one of target genes of miR-21 (Fig. 5a). Luciferase reporter gene activity assay indicated that after PTEN 3′UTR-WT co-transfected with mimic-miR-21, luciferase was more active (P < 0.01, while co-transfection of PTEN 3′UTR-MUT and mimic-miR-21 failed to change luciferase activity (Fig. 5b). The results were consistent with prediction of bioinformatics and further demonstrated miR-21 bound with basic group in PTEN mRNA 3′UTR, suggesting that miR-21 could negatively target PTEN.
Down-regulation of miR-21 inhibits cell proliferation
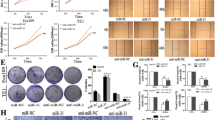
Compared with the blank and negative groups (Fig. 6), after 1 day, the OD value of the cells in Inhibition-miR21 was significantly decreased (P < 0.05) and there was no difference between the blank and negative groups (P > 0.05). The down-regulation of miR-21 could efficiently inhibit the proliferation of human esophageal cancer TE11 cells.
Down-regulation of miR-21 suppresses cell migration and invasion
The number of migrated/invasive cells represented the migration and invasion capacity of cancer cells. The result in Fig. 7 showed that the migration and invasion capacity of cells in the Inhibition-miR21 were significantly lower than the blank and negative groups (both P < 0.05), and there was no significant difference between the blank and negative groups (P > 0.05).
The migration and invasion of TE11 cells in the Inhibition-miR-21, blank, and negative groups. The migration and invasion capacity of cells in the Inhibition-miR21 was significantly lower than the blank and negative groups. a in the figure represents that there is significant difference when compared with the blank and negative control groups (P < 0.05)
Down-regulation of miR-21 reduces cell cycle
The results were shown in Fig. 8 that there was no significant difference between the cells in the blank and negative groups in G0/G1 phase, S phase, and G2/M phase (P > 0.05). Compared with the cells in the blank and negative groups, the cells of Inhibition-miR21 in G0/G1 phase were significantly increased and those in S phase and G2/M phase were significantly decreased (both P < 0.05), which indicated that, mitotically, competence of cells was weakened.
The analysis of flow cytometry chart and cell cycle of cells in the Inhibition-miR-21, blank, and negative groups. Compared with the cells in the blank and negative groups, cells of Inhibition-miR21 in G0/G1 phase were significantly increased and those in S phase and G2/M phase were significantly decreased. a in the figure represents that there is significant difference when compared with the blank and negative control groups (P < 0.05)
Down-regulation of miR-21 increases cell apoptosis
The results were shown in Fig. 9. There was no significant difference in cell apoptosis rate between the blank and negative groups (P > 0.05). Compared with the blank and negative groups, TE11 cell apoptosis rate in the Inhibition-miR21 group was significantly increased (P < 0.05).
The analysis of flow cytometry chart and apoptosis rate of cells in the Inhibition-miR-21, blank, and negative groups. Compared with the cells in the blank and negative groups, TE11 cell apoptosis rate in the Inhibition-miR21 group was significantly increased. a in the figure represents that there is significant difference when compared with the blank and negative control groups (P < 0.05)
Discussion
In this study, we found, compared with normal tissues, the positive rate of PTEN protein expression in esophageal cancer tissue was significantly decreased while the positive rate of PI3K and AKT protein expression was significantly increased. Then we found that the higher differentiation of cancer tissue indicated the lower positive rate of PTEN protein, but the higher positive rates of PI3K and AKT proteins. The latest research showed that in the PTEN/PI3K/AKT signal pathway, PI3K can catalyze 3, 4, 5-phosphatidylinositol trisphosphate phosphorylate when it was activated and then activated protein kinase AKT to promote the proliferation and growth of cells [24]. The protein expression level of PI3K and AKT in malignant cells was too high to lose control of cell proliferation; PTEN could antagonize the activity of PI3K in the process which made 3, 4, 5-phosphatidylinositol trisphosphate dephosphorylate and then inhibited the activity of AKT. When lacking PTEN expression, it may lead to continuous activation of signal pathway and losing control of cell growth [25].
Importantly, we found that the positive rate of miR-21 expression in esophageal cancer cells was significantly higher than normal issues, indicating that miR-21 might participate in adjusting the proliferation and growth of cancer cells. We took esophageal cancer TE11 cells as study subject and found that when the expression of miR-21 was inhibited, the expression level of PTEN was significantly increased and the expression level of PI3K and AKT proteins was significantly decreased. Also, in our study, miRNA target gene prediction online demonstrated that PTEN is one of target genes of miR-21 and miR-21 that can inhibit the expression of PTEN and then enhance PI3K/AKT protein expressions and finally promote proliferation and growth of tumor cells. Yang et al. found that the sequence of miR-21 had higher homology with the 3′UTR of PTEN mRNA by comparing sequences and proved that miR-21 could be attached to 3′UTR of PTEN mRNA by experiment and decreased the expression level of PTEN and then increased the expression level of PI3K and AKT proteins [26]. Also, consistent with our study, Luo et al. reported that PTEN is the target gene of miR-21, and miR-21 simultaneously enhances cell proliferation, apoptosis, and tumor invasiveness by targeting PTEN in lung cancer [27].
As mentioned above, when the expression level of PTEN is decreased, the expression level and activity of PI3K and AKT proteins will be increased and the growth and division will be promoted [24]. When the expression of miR-21 is inhibited by experiment method, the expression level of PTEN increase and the cell mitosis will be inhibited and the cell cycle will be extended so more cells will be stay in G0/G1 phase and the cell apoptosis will be promoted. The expression level of PTEN is also associated with the biological behaviors of cancer cells including infiltration and metastasis. Transforming growth factor (TGF-β) can promote infiltration and metastasis of cancer cells and PTEN can inhibit TGF-β and control cancer cells. When the expression level of PTEN is decreased, the inhibitory effect of TGF-β will be weakened which leads to biological behaviors of cancer cells including infiltration and metastasis [28]. Therefore, miR-21 promotes the proliferation, invasion, and migration of tumor cells by regulating the expression level of PTEN and a series of chain reaction.
Collectively, MiR-21 targets key proteins in PTEN/PI3K/AKT signal pathway, promoting proliferation, migration, invasion, and cell cycle, and inhibiting apoptosis of human esophageal cancer cells. It may serve as a novel therapeutic target in esophageal cancer. The association of miR-21 and PTEN, PTEN/PI3K/Akt signal pathways may become new molecular markers and potential therapeutic targets which will provide certain theoretical basis for related research.
References
Zhang SK, Guo LW, Chen Q, Zhang M, Liu SZ, Quan PL, et al. The association between human papillomavirus 16 and esophageal cancer in Chinese population: a meta-analysis. BMC Cancer. 2015;15:1096.
Chen W, Zheng R, Zhang S, Zhao P, Zeng H, Zou X, et al. Annual report on status of cancer in China, 2010. Chin J Cancer Res. 2014;26(1):48–58.
Fedotovskikh GV, Potapov VA. [Clinico-morphological evaluation of pathogenetic mechanisms of bouginage and intubation on cicatricial esophageal stenosis after chemical burns]. Grud Serdechnososudistaia Khir 1990 (10):65–8.
Scarpa M, Cavallin F, Saadeh LM, Pinto E, Alfieri R, Cagol M, et al. Hybrid minimally invasive esophagectomy for cancer: impact on postoperative inflammatory and nutritional status. Dis Esophagus. 2015. doi:10.1111/dote.12418.
Song B, Cui H, Li Y, Cheng C, Yang B, Wang F, et al. Mutually exclusive mutations in notch1 and pik3ca associated with clinical prognosis and chemotherapy responses of esophageal squamous cell carcinoma in China. Oncotarget. 2015;7:3599–613.
Horna Strand A, Franzen T. Influence of life style factors on Barrett’s oesophagus. Gastroenterol Res Pract. 2014;2014:408470.
Kobayashi R, Yamashita H, Okuma K, Shiraishi K, Ohtomo K, Nakagawa K. Salvage radiation therapy and chemoradiation therapy for postoperative locoregional recurrence of esophageal cancer. Dis Esophagus. 2014;27(1):72–8.
Sugimura K, Miyata H, Tanaka K, Takahashi T, Kurokawa Y, Yamasaki M, et al. High infiltration of tumor-associated macrophages is associated with a poor response to chemotherapy and poor prognosis of patients undergoing neoadjuvant chemotherapy for esophageal cancer. J Surg Oncol. 2015;111(6):752–9.
Dvoretskii S, Levchenko EV, Karachun AM, Komarov IV, Pelipas’ Iu V, Avanesian AA, et al. [Experience of the use of endovideotechnology in surgical treatment of esophageal cancer]. Vestn Khir Im I I Grek. 2014;173(6):54–9.
Liu X, Liu Z, Hou W, Wang K, Ding W, Chen D, et al. [Changes in mitochondria fusion protein-2 hepatic expression in conditions of liver cirrhosis and acute on chronic liver failure]. Zhonghua Gan Zang Bing Za Zhi. 2014;22(9):671–5.
Yang Z, Fang S, Di Y, Ying W, Tan Y, Gu W. Modulation of nf-kappab/mir-21/pten pathway sensitizes non-small cell lung cancer to cisplatin. PLoS One. 2015;10(3):e0121547.
Kar S, Samii A, Bertalanffy H. Pten/pi3k/akt/vegf signaling and the cross talk to krit1, ccm2, and pdcd10 proteins in cerebral cavernous malformations. Neurosurg Rev. 2015;38(2):229–36. discussion 36–7.
Wang J, Zhang C, Chen K, Tang H, Tang J, Song C, et al. Erbeta1 inversely correlates with pten/pi3k/akt pathway and predicts a favorable prognosis in triple-negative breast cancer. Breast Cancer Res Treat. 2015;152(2):255–69.
de la Chapelle A, Jazdzewski K. Micrornas in thyroid cancer. J Clin Endocrinol Metab. 2011;96(11):3326–36.
Bertrand FE, McCubrey JA, Angus CW, Nutter JM, Sigounas G. Notch and pten in prostate cancer. Adv Biol Regul. 2014;56:51–65.
Kloosterman WP, Plasterk RH. The diverse functions of micrornas in animal development and disease. Dev Cell. 2006;11(4):441–50.
Zhang Z, Li Z, Gao C, Chen P, Chen J, Liu W, et al. Mir-21 plays a pivotal role in gastric cancer pathogenesis and progression. Lab Investig. 2008;88(12):1358–66.
Zhang JG, Wang JJ, Zhao F, Liu Q, Jiang K, Yang GH. Microrna-21 (mir-21) represses tumor suppressor pten and promotes growth and invasion in non-small cell lung cancer (nsclc). Clin Chim Acta. 2010;411(11–12):846–52.
Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. Mir-21-mediated tumor growth. Oncogene. 2007;26(19):2799–803.
Yan-nan B, Zhao-yan Y, Li-xi L, Jiang Y, Qing-jie X, Yong Z. Microrna-21 accelerates hepatocyte proliferation in vitro via pi3k/akt signaling by targeting pten. Biochem Biophys Res Commun. 2014;443(3):802–7.
Hong L, Han Y, Zhang Y, Zhang H, Zhao Q, Wu K, et al. Microrna-21: a therapeutic target for reversing drug resistance in cancer. Expert Opin Ther Targets. 2013;17(9):1073–80.
Mastroleo I. Post-trial obligations in the declaration of helsinki 2013: classification, reconstruction and interpretation. Dev World Bioeth 2015.
Rahman MA, Dhar DK, Yamaguchi E, Maruyama S, Sato T, Hayashi H, et al. Coexpression of inducible nitric oxide synthase and cox-2 in hepatocellular carcinoma and surrounding liver: possible involvement of cox-2 in the angiogenesis of hepatitis c virus-positive cases. Clin Cancer Res. 2001;7(5):1325–32.
Ying J, Xu Q, Liu B, Zhang G, Chen L, Pan H. The expression of the pi3k/akt/mtor pathway in gastric cancer and its role in gastric cancer prognosis. Oncol Targets Ther. 2015;8:2427–33.
Li P, Mao WM, Zheng ZG, Dong ZM, Ling ZQ. Down-regulation of pten expression modulated by dysregulated mir-21 contributes to the progression of esophageal cancer. Dig Dis Sci. 2013;58(12):3483–93.
Yang H, Kong W, He L, Zhao JJ, O'Donnell JD, Wang J, et al. Microrna expression profiling in human ovarian cancer: Mir-214 induces cell survival and cisplatin resistance by targeting pten. Cancer Res. 2008;68(2):425–33.
Xu LF, Wu ZP, Chen Y, Zhu QS, Hamidi S, Navab R. Microrna-21 (mir-21) regulates cellular proliferation, invasion, migration, and apoptosis by targeting pten, reck and bcl-2 in lung squamous carcinoma, Gejiu city, China. PLoS One. 2014;9(8):e103698.
Hong Q, Yu S, Geng X, Duan L, Zheng W, Fan M, et al. High concentrations of uric acid inhibit endothelial cell migration via mir-663 which regulates phosphatase and tensin homolog by targeting transforming growth factor-beta1. Microcirculation. 2015;22(4):306–14.
Acknowledgments
We would like to acknowledge the helpful comments from our reviewers on this paper.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
None.
Rights and permissions
About this article
Cite this article
Wu, YR., Qi, HJ., Deng, DF. et al. MicroRNA-21 promotes cell proliferation, migration, and resistance to apoptosis through PTEN/PI3K/AKT signaling pathway in esophageal cancer. Tumor Biol. 37, 12061–12070 (2016). https://doi.org/10.1007/s13277-016-5074-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-016-5074-2