Abstract
Imaging is broadly used in biomedical research, but signal variation complicates automated analysis. Using the Pulmonary Metastasis Assay (PuMA) to study metastatic colonization by the metastasis suppressor KISS1, we cultured GFP-expressing melanoma cells in living mouse lung ex vivo for 3 weeks. Epifluorescence images of cells were used to measure growth, creating large datasets which were time consuming and challenging to quantify manually due to scattering of light from outside the focal plane. To address these challenges, we developed an automated workflow to standardize the measurement of disseminated cancer cell growth by applying statistical quality control to remove unanalyzable images followed and a filtering algorithm to quantify only in-focus cells. Using this tool, we demonstrate that expression of the metastasis suppressor KISS1 does not suppress growth of melanoma cells in the PuMA, in contrast to the robust suppression of lung metastasis observed in vivo. This result may suggest that a factor required for metastasis suppression is present in vivo but absent in the PuMA, or that KISS1 suppresses lung metastasis at a step in the metastatic cascade not tested by the PuMA. Together, these data provide a new tool for quantification of metastasis assays and further insight into the mechanism of KISS1 mediated metastasis suppression in the lung.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Imaging plays a critical role in assessing components of living systems and understanding their biology as a whole. Qualitative imaging data are striking and give insight into biologic function, but quantitative analysis of imaging data remains challenging. Large variations in signal intensity, shape, or distribution can complicate objective and quantitative image analysis [1]. Such complex variations are often present within living systems and increase with the range of imaging depth [2]. While methods such as confocal microscopy allow for the independent imaging of each focal plane for subsequent data compilation, data collected using standard epifluorescence microscopy, in vivo imaging and time-lapse microscopy often capture images containing regions that are both in- and out-of-focus. Thus, out-of-focus objects within an image or light scattered as it is transmitted through tissues can skew quantification efforts [3, 4]. In metastasis research, image data from intact lungs, epifluorescence photomicrographs, in vivo luciferase, and Pulmonary Metastasis Assays (PuMA) [5] data all present such challenges. In these cases, objectively distinguishing between in-focus and out-of-focus fluorescent particles within the same image remains challenging.
PuMA is a powerful tool to study mechanisms of pulmonary metastasis ex vivo [6]. However, we found manual quantification of fluorescent image data to be time consuming and potentially subjective. In order to overcome these challenges, we designed an automated workflow to identify images suitable for automated analysis and quantify in-focus regions of interest (ROI) by adapting a method originally developed to study sediment deposition [7]. The goal of this workflow is not to replace one method of microscopy with another (i.e. confocal with epifluorescence), as we recognize each imaging platform has unique strengths and limitations. Instead, we sought to improve the objectivity and speed with which complex datasets containing images and features of varying quantity and quality can be stratified and analyzed.
We developed this tool to measure the growth of cancer cells disseminated to the lung in order to use the PuMA as a platform to further dissect how the metastasis suppressor KISS1 suppresses melanoma lung metastasis [8]. We hypothesized that melanoma cells expressing KISS1 would be growth suppressed (i.e., dormant) in the PuMA, just as they appear in vivo [9]. Surprisingly, we did not see any difference in the rate of growth in the PuMA as measured by area of GFP positive cells in each lung slice. Nonetheless, these experiments were useful in developing a tool for automated quantification of PuMA and other imaging datasets containing images and data varying in quality and intensity.
Methods
Pulmonary metastasis assay
All animal studies were conducted in accordance with the Guide for the Care and Use of Laboratory Animals (National Institutes of Health). Protocol (#2014–2208) was approved by University of Kansas Medical Center Institutional Animal Care and Use Committee. The PuMA was performed as described [5] with modifications. Female nude mice aged 6–8 weeks were intravenously injected with 50,000 enhanced green fluorescent protein- (GFP) expressing C8161.9 cells (human amelanotic melanoma, clone 9) [10, 11] suspended in 200 µl of 0–4 °C Hanks Buffered Salt Solution (HBSS, Life Technologies, #14175-103). Cells circulated for 20 min and lodged in lung capillaries. Mice were euthanized using CO2 before lungs were insufflated with a 1:1 mixture of media and agarose using an 18-gauge (GA) needle and 10 ml syringe. After tying off the trachea with suture, lungs were extracted and placed in sterile phosphate buffered saline (PBS) on ice. Lungs were cut into ~ 0.5–1 mm sections using sterile forceps and microdissection scissors. Sections were placed on media-saturated Gelfoam® (Pfizer-Pharmacia & Upjohn Co., #09-0315-08) in a 6-well cell culture plate for incubation and imaging. Media was replaced every 2 days.
Experimental metastasis assay
In order to seed the lungs with disseminated melanoma cells, injections were performed as previously described [12]. Briefly, 50,000 C8161.9 cells were suspended in 0–4 °C HBSS and injected into the tail vein in a volume of 100 µl using a 27 GA needle and 1 ml syringe. Cells were allowed to grow in vivo for 5 weeks or until the animal was moribund. After CO2 induced euthanasia, lungs were imaged with the aid of a fluorescence dissecting microscope and tumor and lung tissues were collected for analysis. Macroscopic metastases were quantified by using the multi-point tool in ImageJ to count each metastasis visible on the surface of the lung.
Immunofluorescence
Samples of lung growing in PuMA culture were collected weekly for 3 weeks and flash frozen in OCT (Fisher # 23-730-571). Frozen sections (7 µm) were cut using a cryostat and fixed 4% paraformaldehyde in PBS. Fish gelatin (4% in PBS) was used for blocking and dilution of the primary (GFP, Abcam #6556, 1:100) and secondary (Alexa 594, #R37117, ThermoFisher, 1:500) antibodies. Nuclei were counterstained with DAPI in mounting medium (#H-1200, Vectashield).
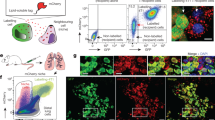
Image acquisition
Lung sections containing disseminated GFP-expressing melanoma cells were imaged at days 0, 7, 14 and 21. Each Gelfoam® sponge containing lung sections and cancer cells were placed lung side down on a culture dish (MatTek, #P35-G-1.5-20-C) for imaging. A Nikon Eclipse TS100 Inverted Microscope, QImaging QIClick monochrome CCD camera and Metamorph software were used to take ~ 30 non-redundant images per experimental group at 20× magnification for each time point (Fig. 1). GFP-expressing cancer cells were present throughout the lung section. The focal plane which optimized the number of in-focus cancer cells was chosen for each image. Immunofluorescence (IF) images were collected using a Nikon Eclipse 80i microscope, QImaging QIClick 8 bit monochrome camera and an XCite120PC light source. Five images per time point were collected and quantified using ImageJ. After imaging, Gelfoam® with lung sections was returned to the 6 well dish and media was replaced.
PuMA image acquisition and automated image analysis workflow. PuMA image acquisition and automated image analysis workflow. a Mouse lung sections (0.5–1 mm thick) containing disseminated GFP-expressing melanoma cells were placed on media-saturated Gelfoam® sponges cultured ex vivo for up to 3 weeks. Using an inverted microscope, an average of 30 non-redundant photomicrographs were recorded per experimental group every 7 days. Scale bar for cell culture dish is 1 cm. Scale bar for fluorescent image is 50 µm. b Cross-section views of lung sections containing GFP-expressing melanoma cells within lung section atop Gelfoam® with respect to the focal plane of the image. c Schematic of the automated workflow used for image analysis. Output from a logistic regression model selects analyzable images for subsequent automated analysis. To standardize measurement of melanoma growth, only in-focus fluorescent cell clusters/regions of interest (ROI) were quantified. In-focus ROI were selected by applying a Gaussian gradient and selecting ROI with a clarity value above the in-focus threshold for quantification. Scale bars are 50 µm
Image analysis
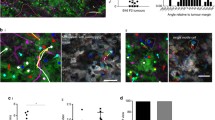
In collecting and analyzing PuMA data, we noted that while some images were easily analyzed by automated processing, images with high background skewed analysis because the software was not able to recognize all GFP-expressing cells or groups of cells. Hereafter, these cell clusters will be referred to as regions of interest (ROI) (Fig. 2a). A step-by-step protocol for the method described (Supplementary 1) and all operating scripts required for the protocol are freely available online (https://drive.google.com/drive/folders/1zmjp_VW_Pnw7BdyeAut36DS1J6AoJ-QG?usp=sharing). To develop a training set for machine learning, we used a large set of images and manually stratified analyzable images by how accurately the automated threshold identified ROI. ImageJ [13] was used for image processing and cell measurements. R was used for statistical analysis, and MATLAB was used to identify out-of-focus particles.
Validation of Image Analysis to Measure Tumor Cell Growth. Validation of image analysis measurements and image stratification. a Image of disseminated GFP-expressing melanoma cells in living lung tissue. Raw image (monochrome), image negative, image after thresholding, and cancer cell(s) identified by ImageJ are shown. In-focus cell clusters/ROI are retained while out-of-focus particles are excluded. Width of cell cluster measured by ImageJ is scaled correctly. Scale bar is 50 µm. b Comparison of representative analyzable and unanalyzable images stratified by the logistic regression model. Raw images, negatives and ROI identified after applying the Gaussian gradient to identify in-focus ROI are shown. Analyzable images show correctly identified ROI, while unanalyzable images do not. Scale bars are 50 µm. c Microscopy images of GFP+ control cells on Day 0 and Day 21 as seen by IF (upper panels) staining for GFP or live cell inverted epifluorescence microscopy (lower panels). Scale bars are 20 µm (IF) and 50 µm (image analysis). d Quantification of tumor cell growth in the PuMA at Day 0 and Day 21 as measured by IF (upper graph) and image analysis (lower graph). Comparison of image analysis quantification with immunofluorescence (IF) show similar growth trends
To process the images, first the MaxEntropy threshold [14] from ImageJ was applied to images from the PuMA. Then, we manually classified images into two groups: those which the MaxEntropy threshold accurately identified as fluorescent cells (“analyzable”) and those which were not (“unanalyzable”). In both the analyzable and unanalyzable image sets, parameters from ImageJ served as explanatory variables (numROI, avgArea, avgPerim, avgWidth, avgHeight, avgMajor, avgMinor, avgAngle, avgCirc, avgFeret, avgIntDen, avgMinFeret, avgAr, avgRound, avgSolidity). Parameter descriptions are provided in Table 1 and Supplemental Fig. 1a.
Next, we used these explanatory variables to fit a multivariate logistic regression against the outcome variable of whether an image was analyzable or unanalyzable. Features which contributed significantly to distinguishing analyzable and unanalyzable images were used to stratify future image datasets (Table 1). Examples of analyzable and unanalyzable images identified by the model are shown in Fig. 2b.
The outcome of our logistic regression was used to set a threshold for classifying which images were analyzable. To determine this threshold and assess prediction performance, we conducted a cross-validation using our test dataset by comparing original estimates of image analyzability from the multivariate analysis to the cross-validated model (Supplemental Fig. 1b). The probability threshold was set by constructing a 2 × 2 table comparing images determined to be analyzable by manual or automated stratification and determining which image probability value (0.57) gave the fewest misclassified images. Images with a predicted probability below this threshold were excluded from further analysis, while images with a probability above it were retained for further analysis. Comparison of automated and manual image stratification is summarized in Table 2. Overall, automated stratification performed well as a test for whether or not an image was analyzable with a positive and negative predictive values of 0.957 and 0.678 respectively (Table 3).
Finally, we selected only in-focus ROI for analysis using a method originally developed to remove out-of-focus ROI from images in sediment deposition research [7]. First, a clarity value is calculated for each ROI by applying a Gaussian gradient smoothing function. The threshold for in-focus ROI was determined by comparing a series of gradient-produced images with the original image. A clarity value threshold was set which distinguished between in-focus and out-of-focus ROI (Figs. 1b, c, 2a, b). ROI with a sharper gradient (larger value) than the clarity value were retained and the area was quantified, while ROI with a more gradual (smaller) gradient than the clarity value were considered out-of-focus and excluded. To verify our image analysis measured cell growth accurately, we compared image analysis measurements to direct measurements of cell growth using IF of GFP positive cells at Day 0 and 21 (Fig. 2c, d). We observed similar growth trends using these techniques, validating the measurements made by image analysis. Thus, we standardized the ROI which were selected before calculating the surface area of the ROI. Surface area of the in-focus fluorescent ROI as compared to the total area imaged was then used as a proxy for cell growth ex vivo and we were able to automate the measurement of the tumor explants in living lung.
Statistical analysis
Logistic regression, one-way ANOVA, Student’s t-tests and graphing were performed using R (Vienna, Austria) [15] and MATLAB [16].
Results
Our goal was to develop an automated workflow whereby epifluorescence images could be objectively quantified (Fig. 1a). Three tasks had to be automated in order to achieve this objective: (1) classification of an image as “analyzable” or “unanalyzable”; (2) measurement of ROI/cancer cell features in the image; and, (3) removal of identified out-of-focus ROI that might bias the data.
First, images were classified as “analyzable” or “unanalyzable”. We found that some images were amiable to automated cell measurement (task 2) while some were not (Fig. 1c). All images unable to be measured must be removed from the analysis to avoid biased ensemble results (Fig. 2b). From our test dataset, we used several features measured by ImageJ (Table 1, Supplemental Fig. 1a) to stratify analyzable from unanalyzable images (Fig. 2b). Features varied in their ability to identify analyzable images and we found that ROI Width, Feret’s Diameter, Height and Aspect Ratio were most significant in distinguishing analyzable from unanalyzable images (Pr (>|z|) of 4.300E−06, 1.650E−05, 1.760E−04, and 0.001 respectively). Also statistically significant were the ROI perimeter, solidity, and roundness (Pr (>|z|) of 0.005, 0.018, and 0.024 respectively). These features are a result of the statistical model built around our test data set and we would expect these features may vary for different data sets.
Recognizing that several of the above referenced features may be potentially correlated (e.g. perimeter and area), we performed a cross-validation of our test set to check for over-fitting of our model and found that the model performed equally well on unique subsets of our data (Supplemental Fig. 1a). Next, we determined which probability threshold would result in the fewest misclassified images and found this value to be 0.57. This threshold was then used to distinguish analyzable and unanalyzable images in future experiments. Common causes of unanalyzable images included decreased image contrast due to absence of fluorescent cancer cells, pixel oversaturation due to robust cancer cell growth, cancer cells outside the focal plane, and increased tissue density surrounding bronchioles producing increased background. We also compared the automated analysis to manual stratification (Table 2) and found that automated stratification was able to identify similar numbers of analyzable images as when the same dataset was analyzed manually.
Next, we wanted to ensure that there was not bias in our model between any of the biologically distinct groups analyzed. Over the course of a 3-week PuMA experiment, lung sections from three biologically distinct lines of GFP-expressing C8161 melanoma cells were imaged at 0, 7, 14 and 21 days. Approximately 30 images (range, 14–58) were taken per experimental group at each time with a median and average of 25.5 and 29.8 images respectively (standard deviation, 10.9). To demonstrate that analyzable and unanalyzable images were stratified in an unbiased manner from distinct biological groups, we recorded the number of excluded unanalyzable images at each day of analysis and compared the means using one-way ANOVA and Tukey’s post hoc test (Supplemental Fig. 1c). No statically-significant differences were identified between any of the groups. These data suggest that the image stratification is functioning equitably between biologic groups.
The second task was the measurement of the ROI features. The key steps involved in this process were: (i) removing broad-scale trends in the grayscale color of the image; (ii) creating a binary image of black and white pixels through thresholding; and (iii) applying particle measurement routines to obtain quantitative data on the cell population (e.g., area, perimeter, long and short axis length and the orientation of long axis) for each ROI. Similarity between the ROI observed in the raw image and the ROI particles identified by the set thresholds (Fig. 2a, b) suggests that the threshold is able to correctly identify ROI of interest.
The third and final step was to exclude from further analysis any ROI which were out-of-focus and might bias the data. In this way, we could objectively select high quality images and quantify the area of in-focus ROI to standardize analysis between samples.
This workflow was developed while we attempted to elucidate the mechanism of KISS1 suppression. While the ability of KISS1 to strongly suppress metastasis [8] has been demonstrated in multiple tumor types [9] and there have been hints related to upstream and downstream regulatory pathways [17], the biochemical underpinnings of KISS1 metastasis suppression remain largely unknown. However, previous studies showed that KISS1 allows all steps prior to colonization of secondary sites [18]. As a result, we sought to use the PuMA as a model for understanding KISS1 mediated suppression in the lung so that we could test what regulates KISS1’s metastasis-suppressing activity.
To our surprise, there was no statistical difference in the growth of KISS1-expressing cells compared to controls (Fig. 3a). In some cases, growth in lung appeared even greater (Fig. 3b). Since secretion of KISS1 is required to suppress metastasis [18], we confirmed that KISS1 was still secreted in the PuMA at Day 21 by immunoblot (Fig. 3c). Together, these data show that the outgrowth of KISS1-expressing cells is not due to loss of KISS1 expression. In parallel studies, the same KISS1-expressing C8161 cells were still robustly suppressed for metastasis despite seeding lung in vivo (Fig. 3d, e).
KISS1 expression does not suppresses growth in the PuMA, but suppresses robustly in vivo. Comparison of KISS1 Growth Suppression in vivo and ex vivo (PuMA). a Representative images of growth of GFP control (top) or KISS1 expressing (bottom) C8161.9 melanoma cells at days 0, 7, 14 and 21 in the PuMA. Scale bar for fluorescent image is 50 µm. b Quantification of PuMA three independent experiments by image analysis. No significant differences. c Immunoblot for KISS1 from conditioned media from either GFP control or KISS1-expressing melanoma cells grown in the PuMA demonstrates KISS1 remains expressed in the PuMA to D21. d Representative images of gross lungs containing vector control (top) or KISS1 expressing (bottom) C8161.9 melanoma cells 5 weeks after tail vein injection demonstrating suppression of lung outgrowth by KISS1 in vivo. n = 5 mice/group. e Quantification of macroscopic lung metastases
Discussion
While the PuMA assay did not faithfully replicate what we had observed in vivo for KISS1-expressing cells, the data set obtained afforded an opportunity to refine the imaging workflow so that future experiments would be more readily analyzed. Objective and quantitative analysis of images containing data from a range of focal planes is difficult due to variations in signal intensity and distribution. We recognize that all microscopy methods have unique strengths and limitations [19,20,21]. Methods such as confocal microscopy may not suffer as much from large variations in signal intensity. Indeed, this workflow is not intended to substitute one microscopy for another. Rather, our goal was to improve the capacity for epifluorescence microscopy data, nearly ubiquitous in many labs, to be more objectively quantified. Automating image selection may also help reduce (remove) bias by applying a standardized method to image selection for analysis, rather than leaving the decision to a single observer. While we acknowledge that our initial stratification was empiric and could be a source of potential bias, we also demonstrate that the complex signal differences between analyzable and unanalyzable image parameters were quantifiable and could be used objectively select of images useful for automated analysis in future experiments. This platform mirrors other machine learning approaches such as non-negative matrix factorization, Random Forest classification, or Potts models [22, 23].
Using this workflow, we measured growth of disseminated cancer cells as a function of in-focus GFP-positive surface area in lung tissue. This method has immediate utility in the PuMA, but might also be applied to in vivo data such as the quantification of luciferase signal in whole animals with metastases [20]. While our model identified a number of features within an image to be significant in stratifying images, the features identified for other types of data are likely to vary greatly. This flexibility lends itself to the objective analysis of a potentially broad range of biological imaging data. For example, comparing relative roundness versus spindle morphologies could assess epithelial-to-mesenchymal transition, which contributes to some cells’ metastatic or invasive potential [24, 25]. Additionally, using multiple features of the ROI creates a stronger tool for distinguishing in-focus from out-of-focus ROI than would a single modality, such as a gradient based on pixel saturation alone [26].
Despite the disappointment that PuMA did not mimic in vivo results in the C8161 KISS1 melanoma model, the results may still have provided clues regarding mechanism of action. KISS1-expressing cells still proliferated. Since we previously showed that C8161 cells do not express the KISS1 receptor [18] eliminating autocrine feedback of KP54 as a potential mechanism, the hypotheses related to paracrine mechanisms or alternative feedback [27] are supported. Three possibilities exist for the lack of consistency between the in vitro and in vivo data: (1) a factor required to suppress growth of KISS1-expressing cells is missing in PuMA; (2) a molecule that promotes growth of KISS1-expressing cells is ‘uncovered’ in PuMA; or, (3) KISS1 suppresses at a step other than proliferation at the secondary site and that step is not measured by PuMA. These data, taken together with results from all of the other assays measuring steps of metastasis in vitro, emphasize that the ‘gold standard’ for studying metastasis is in vivo.
References
Heller D, Hoppe A, Restrepo S, Gatti L, Tournier AL, Tapon N, Basler K, Mao Y (2016) EpiTools: an open-source image analysis toolkit for quantifying epithelial growth dynamics. Dev Cell 36(1):103–116. https://doi.org/10.1016/j.devcel.2015.12.012
Li C, Guo J, Cong R, Pang Y, Wang B (2016) Underwater image enhancement by dehazing with minimum information loss and histogram distribution prior. IEEE Trans Image Process. https://doi.org/10.1109/TIP.2016.2612882
Henriquez NV, van Overveld PG, Que I, Buijs JT, Bachelier R, Kaijzel EL, Lowik CW, Clezardin P, van der Pluijm G (2007) Advances in optical imaging and novel model systems for cancer metastasis research. Clin Exp Metastasis 24(8):699–705. https://doi.org/10.1007/s10585-007-9115-5
Phillips KG, Baker-Groberg SM, McCarty OJ (2014) Quantitative optical microscopy: measurement of cellular biophysical features with a standard optical microscope. J Vis Exp. https://doi.org/10.3791/50988
Mendoza A, Hong S-HH, Osborne T, Khan MA, Campbell K, Briggs J, Eleswarapu A, Buquo L, Ren L, Hewitt SM, Dakir EHlH, Dakir E-Hl-H, Garfield S, Walker R, Merlino G, Green JE, Hunter KW, Wakefield LM, Khanna C (2010) Modeling metastasis biology and therapy in real time in the mouse lung. J Clin Investig 120(8):2979–2988. https://doi.org/10.1172/JCI40252
Lizardo MM, Sorensen PH (2018) Practical considerations in studying metastatic lung colonization in osteosarcoma using the pulmonary metastasis assay. J Vis Exp. https://doi.org/10.3791/56332
Keyvani A, Kyle S (2013) A fully-automated image processing technique to improve measurement of suspended particles and flocs by removing out-of-focus objects. Comput Geosci 52:189198. https://doi.org/10.1016/j.cageo.2012.08.018
Lee JH, Miele ME, Hicks DJ, Phillips KK, Trent JM, Weissman BE, Welch DR (1996) KiSS-1, a novel human malignant melanoma metastasis-suppressor gene. J Natl Cancer Inst 88(23):1731–1737
Beck BH, Welch DR (2010) The KISS1 metastasis suppressor: a good night kiss for disseminated cancer cells. Eur J Cancer (Oxford England 1990) 46(7):1283–1289. https://doi.org/10.1016/j.ejca.2010.02.023
Welch DR, Chen P, Miele ME, McGary CT, Bower JM, Stanbridge EJ, Weissman BE (1993) Microcell-mediated transfer of chromosome 6 into metastatic human C8161 melanoma cells suppresses metastasis but does not inhibit tumorigenicity. Oncogene 9(1):255–262
Nash KT, Phadke PA, Navenot J-MM, Hurst DR, Accavitti-Loper MA, Sztul E, Vaidya KS, Frost AR, Kappes JC, Peiper SC, Welch DR (2007) Requirement of KISS1 secretion for multiple organ metastasis suppression and maintenance of tumor dormancy. J Natl Cancer Inst 99(4):309–321. https://doi.org/10.1093/jnci/djk053
Goldberg SF, Harms JF, Quon K, Welch DR (1999) Metastasis-suppressed C8161 melanoma cells arrest in lung but fail to proliferate. Clin Exp Metastasis 17(7):601–607
Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9(7):671–675
Siritantikorn S, Jintaworn S, Noisakran S, Suputtamongkol Y, Paris DH, Blacksell SD (2012) Application of ImageJ program to the enumeration of Orientia tsutsugamushi organisms cultured in vitro. Trans R Soc Trop Med Hyg 106(10):632–635. https://doi.org/10.1016/j.trstmh.2012.05.004
R Development Core Team (2015) R: a language and environment for statistical computing. R Development Core Team, Vienna
MATLAB (2015). vol 8.5.0. The MathWorks Inc., Natick
Goldberg SF, Miele ME, Hatta N, Takata M, Paquette-Straub C, Freedman LP, Welch DR (2003) Melanoma metastasis suppression by chromosome 6: evidence for a pathway regulated by CRSP3 and TXNIP. Cancer Res 63(2):432–440
Nash KT, Phadke PA, Navenot JM, Hurst DR, Accavitti-Loper MA, Sztul E, Vaidya KS, Frost AR, Kappes JC, Peiper SC, Welch DR (2007) Requirement of KISS1 secretion for multiple organ metastasis suppression and maintenance of tumor dormancy. J Natl Cancer Inst 99(4):309–321. https://doi.org/10.1093/jnci/djk053
Kimura H, Hayashi K, Yamauchi K, Yamamoto N, Tsuchiya H, Tomita K, Kishimoto H, Bouvet M, Hoffman RM (2010) Real-time imaging of single cancer-cell dynamics of lung metastasis. J Cell Biochem 109(1):58–64. https://doi.org/10.1002/jcb.22379
Leong HS, Lizardo MM, Ablack A, McPherson VA, Wandless TJ, Chambers AF, Lewis JD (2012) Imaging the impact of chemically inducible proteins on cellular dynamics in vivo. PLoS ONE 7(1):e30177. https://doi.org/10.1371/journal.pone.0030177
Hoffman RM (2001) Visualization of GFP-expressing tumors and metastasis in vivo. Biotechniques 30(5):1016–1022
Hedberg-Buenz A, Christopher MA, Lewis CJ, Fernandes KA, Dutca LM, Wang K, Scheetz TE, Abramoff MD, Libby RT, Garvin MK, Anderson MG (2016) Quantitative measurement of retinal ganglion cell populations via histology-based random forest classification. Exp Eye Res 146:370–385. https://doi.org/10.1016/j.exer.2015.09.011
Jones TR, Carpenter AE, Lamprecht MR, Moffat J, Silver SJ, Grenier JK, Castoreno AB, Eggert US, Root DE, Golland P, Sabatini DM (2009) Scoring diverse cellular morphologies in image-based screens with iterative feedback and machine learning. Proc Natl Acad Sci USA 106(6):1826–1831. https://doi.org/10.1073/pnas.0808843106
Lambert AW, Pattabiraman DR, Weinberg RA (2017) Emerging Biological Principles of Metastasis. Cell 168(4):670–691. https://doi.org/10.1016/j.cell.2016.11.037
Chaffer CL, San Juan BP, Lim E, Weinberg RA (2016) EMT, cell plasticity and metastasis. Cancer Metastasis Rev 35(4):645–654. https://doi.org/10.1007/s10555-016-9648-7
Jerome Friedman TH, Robert Tibshirani (2000) Additive logistic regression: a statistical view of boosting. Ann Stat 28(2):337–407
Angeliki M, Nikolaos P, Peter L, Constantine P, Michael K (2008) The kisspeptin (KiSS-1)/GPR54 system in cancer biology. Cancer Treat Rev 34(8):682–692. https://doi.org/10.1016/j.ctrv.2008.05.007
Acknowledgements
The authors acknowledge Drs. Chand Khanna, DVM, PhD, Michael Lizardo, PhD and Arnulfo Mendoza, DVM for their training in the PuMA. The authors also acknowledge support from the Biostatistics and Informatics Shared Resource of the KU Cancer Center.
Funding
The authors received support from the following organizations: Melanoma Research Foundation (EDY), National Cancer Institute 1F30CA216998 (EDY), CA134981 (DRW) and CA168524 (DRW), the National Foundation for Cancer Research (DRW) and the Hall Family Professorship in Molecular Medicine (DRW).
Author information
Authors and Affiliations
Contributions
Conceived and designed experiments: EDY, KS, DRW. Analyzed data: EDY, AFT, JLU, SM, JTM. Wrote first draft of the manuscript: EDY. Contributed to manuscript writing: EDY, KS, AFT, JLU, DRW. Agree with manuscript results and conclusions: EDY, KS, AFT, JLU, SM, JTM, DRW. Developed the structure and arguments for the paper: EDY, KS, JLU, DRW. Made critical revisions and approved final version of the manuscript: EDY, KS, AFT, JLU, JTM, DRW.
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Young, E.D., Strom, K., Tsue, A.F. et al. Automated quantitative image analysis for ex vivo metastasis assays reveals differing lung composition requirements for metastasis suppression by KISS1. Clin Exp Metastasis 35, 77–86 (2018). https://doi.org/10.1007/s10585-018-9882-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10585-018-9882-1