Abstract
The Ras-Association Domain Family 1 (RASSF1) gene, which is located on the small arm of chromosome 3, contains two CpG islands and generates seven transcripts (RASSF1A-RASSF1G) by differential promoter usage and alternative splicing. As the main transcript, RASSF1A, B and C may play different roles in tumorigenesis. The present study was to detect the role of RASSF1A, B and C in esophageal squamous cell carcinoma (ESCC) and clarify the critical CpG sites of RASSF1A, in order to clarify more information on the role of RASSF1 with regard to the pathogenesis of ESCC. Frequent silencing of RASSF1A but not RASSF1B and RASSF1C were found in esophageal cancer cell lines and the silencing of RASSF1A may be reversed by 5-Aza-dC treatment. The aberrant promoter and exon 1 especially exon 1 methylation of RASSF1A induces silencing of its expression in TE13 cell line. Decreased mRNA and protein expression of RASSF1A was observed in ESCC tumor tissues and was associated with RASSF1A promoter and exon 1 methylation status. Unlike RASSF1A, methylation and expression variation of RASSF1B was not found in ESCC tissues. However, RASSF1C is highly expressed in ESCC tissues. RASSF1A methylation and protein expression were independently associated with ESCC patients’ survival. These data indicated that the inactivation of RASSF1A through promoter and exon 1 methylation may play an important role in ESCC carcinogenesis and reactivation of RASSF1A gene may has therapeutic potential and may be used as a prognostic marker for ESCC patients.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Esophageal cancer is the eighth most common malignancy and the sixth most common cause of cancer-related death worldwide [1], its prevalence and death rate are continuously increasing and thus has become a major health concern [2]. Esophageal squamous cell carcinoma (ESCC) is the predominant type of esophageal cancer and has a striking geographic distribution in China, especially in some counties bordering Henan, Hebei, and Shanxi Provinces, where nutritional deficiencies, intake of pickled vegetables, nitrosamine-rich or mycotoxin-contaminated foods and low socioeconomic status are likely to contribute to ESCC [3]. There is also a strong tendency toward familial aggregation of ESCC in these high-risk areas [4], suggesting that genetic susceptibility, in combination with exposure to environmental risk factors, may contribute to the high rates of ESCC in these areas. However, the precise molecular mechanisms of development and/or progression of ESCC remain unknown, although multiple genetic and epigenetic alterations have been detected in ESCC [5, 6]. Therefore, additional elucidation of the molecular mechanisms involved in ESCC is urgently needed for more effective treatment.
Ras-Association Domain Family 1 (RASSF1) was first identified in a yeast two-hybrid screen through its interaction with the human DNA excision repair protein XPA [7]. The RASSF1 gene is located on chromosome 3p21.3 and consists of eight exons (1α, 1β, 2αβ, 2γ, 3, 4, 5 and 6) spanning ∼ 11 kb [8]. Further studies have identified seven different RASSF1 isoforms (RASSF1A to RASSF1G) that are generated by differential usage of two promoters (distance 3.5 kb) and through alternative splicing [9]. The 1.9 kb RASSF1A transcript initiates from a promoter located in the first CpG island and transcription initiates with exon 1α followed by exon 2αβ. RASSF1B has the same exon 2αβ as RASSF1A but utilizes a different 5′ exon (exon 1β). The 1.7 kb RASSF1C transcript initiates from a promoter located in the second CpG island and transcription initiates with a single N-terminal exon (exon 2γ), the protein sequence of which has no significant similarity to any known protein. The remaining 4 isoforms (RASSF1D-G) are all splice variants of RASSF1A [10]. So far however, except for RASSF1A, the biological relevance of other isoforms has not been fully demonstrated.
Epigenetic alterations, including DNA methylation and histone modifications, play important roles in regulating gene transcription [11, 12]. Our along with others studies have previously shown that tumor suppressor genes can be transcriptionally silenced by epigenetic modifications in ESCC and epigenetically modified genes can be used for both ESCC diagnosis and prognosis [13–15]. Loss of RASSF1A expression is one of the most common events in human cancers, with aberrant promoter methylation reported in different tumor types, including bladder, brain (neuroblastoma, glioblastoma, medulloblastoma), breast, cervical, colon, gastric, head and neck, hepatocellular, Hodgkin’s lymphoma, kidney, melanoma, nasopharyngeal, osteosarcoma, ovarian, pancreatic, prostate, etc. [16, 17]. Recent studies have demonstrated that methylation frequency of individual CpG sites within a CpG island may differ and CpG sites showing close association of methylation and inhibition of transcription are called critical CpG sites. Although the hypermethylation of RASSF1A in ESCC has been reported [18–20], the critical CpG sites and its role on ESCC progression and prognosis have not been clarified. In addition, the effects of other epigenetic alterations such as histone acetylation on the expression of RASSF1A in ESCC are still remaining unknown. Decreased expression of RASSF1B was found in bladder carcinoma [21], gastric adenocarcinoma [22], and osteosarcoma [23], and no other reports about its role in tumors. RASSF1C did not show hypermethylation of its promoter region and was expressed in almost all lung, breast, pediatric, and pancreatic endocrine tumors and tumor cell lines tested [7, 24–26]. However, RASSF1C expression was almost undetectable in the KRC/Y renal cell carcinoma cell line [27]. Thus RASSF1C may have a tissue-specific effect. Although not reported, expression of RASSF1D, RASSF1E, RASSF1F, and RASSF1G is also likely to be linked with expression of RASSF1A as they share the same promoter region. To our best knowledge, the role of RASSF1B and RASSF1C in ESCC has not been investigated. In the present study, we attempted to detect the role and methylation status of the main isoforms of RASSF1, including RASSF1A, RASSF1B, and RASSF1C, in esophageal squamous cell carcinoma to clarify more information on the role of different isoforms of RASSF1 with regard to the pathogenesis and prognosis of ESCC.
Materials and methods
Cell culture and treatment
The human esophageal cancer cell lines TE-1, TE-13, Yes-2, Eca109, and T.Tn were cultured in RPMI-1640 medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10 % heat-inactivated FBS (Invitrogen, Carlsbad, CA, USA), 100 U/mL penicillin, and 100 μg/mL streptomycin. Cells were seeded at a low density and incubated for 24 h prior to treatment with DNA methyltransferase inhibitor 5-aza-2′-deoxycytidine (5-Aza-dC) and histone deacetylase (HDAC) inhibitor trichostatin A (TSA). All five esophageal cancer Cells (2 × 105/mL) were treated with 5 μmol/L 5-Aza-dC (Sigma, St Louis, MO, USA) for 72 h and medium containing 5-Aza-dC was changed every 24 h or with 0.3 μmol/L TSA for 24 h or with the combination of 5 μmol/L 5-Aza-dC for 48 h followed by 0.3 μmol/L TSA for an additional 24 h. The dose and timing of 5-Aza-dC and/or TSA was based on similar preliminary studies as well as published studies showing optimal reactivation of gene expression [28, 29]. Control cells received no drug treatment. DNA and RNA were isolated from these cells.
Patients and specimens
All study subjects were ethnically homogeneous Han nationality and residents of Hebei Province and its surrounding regions. Tumor and corresponding adjacent normal tissues were obtained from 141 ESCC cases, which were all inpatients for surgical treatment in the Fourth Affiliated Hospital, Hebei Medical University between the years of 2005 and 2008. All subjects were interviewed by professional interviewers for their gender, age, histopathological diagnosis, and upper gastrointestinal cancers (UGIC) family history. The patients included 100 males and 41 females, mean age 58.9 years (ranged from 37 to 78 years). Individuals with at least one first-degree relative or at least two second-degree relatives having esophageal/cardia/gastric cancer were defined as having family history of upper gastrointestinal cancers (UGIC). Tumor and corresponding normal tissues were divided into two parallel parts, one part were frozen and stored at −80 °C until DNA and RNA was extracted, the other part were formalin-fixed and paraffin-embedded. Histological tumor typing of the cases was carried out on the basis of resected specimens in the department of pathology of the same hospital. Information on clinicopathologic characteristics was available from hospital recordings and pathological diagnosis. Recurrence and survival data were ascertained through the Tumor Registry and Hospital chart review (Supplementary Table 1). The study was approved by the Ethics Committee of Hebei Cancer Institute and informed consent was obtained from all recruited subjects.
Stable gene transfections
Transfection was done using FuGENE HD transfection reagent (Roche, Swiss) as recommended by the manufacturer’s instructions. For overexpression of RASSF1A, exponentially grown TE13 cells cultured in six-well plates were transfected with RASSF1A expression plasmid (pcDNA3.1-RASSF1A) or the relevant empty vector (pcDNA3.1-EV) as control. The expression plasmid contained the full-length cDNA of RASSF1A. After transfection, cells were incubated in antibiotic-free medium for 5 h, followed by replacement with fresh normal growth medium for a further incubation of 43 h by adding 800 μg/mL of G418 (Life Technologies, Carlsbad, CA, USA) to the supplemented culture medium. Resistant cell clones were isolated and expanded for further characterization.
RASSF1A, RASSF1B, and RASSF1C mRNA expression via regular reverse transcription-polymerase chain reaction (RT-PCR) and quantitative real-time RT-PCR assays
Total RNA was extracted from 5-Aza-dC or TSA treated and untreated cell lines, the stable transfected TE13 clones, frozen tumor and corresponding normal tissues by standard methods using trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions and quantified by UV absorbance at 260–280 nm. Two μg RNA was used to synthesize cDNA using the RT-for-PCR kit (Clontech, Palo Alto, CA, USA) with oligo (dT) priming as recommended in the protocol provided. cDNA from each sample was used as regular RT-PCR and quantitative real-time RT-PCR template and primers for RASSF1A, RASSF1B, and RASSF1C were designed according to different alternative splicing. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene was used as internal control. All of the primers and reaction conditions were listed in Supplementary Table 2. For regular RT-PCR, the PCR products were tested in 2 % agarose gel in electrophoresis and visualized with ethidium bromide staining, and the reaction was repeated once with each of the samples for quality control. For quantitative real-time RT-PCR, SYBR Green PCR Master mix (Life Technology, Foster City, CA, USA) was used as amplification reaction mixture in a 10 μL reaction volume, which contained 5 μL of SYBR Green PCR mix, 0.2 μM each of forward and reverse primers, 1 μL of diluted cDNA template and appropriate amounts of sterile double-distilled water. The PCR reaction was conducted at 95 °C for 5 min and followed by 40 cycles of 95 °C for 30 s and 53–56 °C for 45 s in the stepone plus thermal cycler (Applied Biosystems, Foster City, CA, USA). The melting curve analysis was performed to confirm PCR product specificity and the qRT-PCR results were analyzed and expressed as relative mRNA expression of CT (threshold cycle) value, which was then converted to fold changes. The expression levels of target genes were normalized with GAPDH using the 2−ΔΔCT method [30]. All experiments were repeated in triplicate.
Cell proliferation assay
Cell proliferation was measured with MTT assay. Briefly, TE13 cells and the stable transfected clones were plated in 96-well plates at a density of 2 × 103 per well in 200 μL of RPMI 1640 with 10 % FBS under standard tissue culture conditions (six wells/group). Assessment of TE13 cell proliferation was respectively measured using MTT after the cells were treated with 5-Aza-dC for 72 h, or treated with TSA for 24 h, or treated with the combination of 5-Aza-dC for 48 h and TSA for additional 24 h. Briefly, the cells were incubated with 100 μL of MTT solution (0.5 mg/mL; Sigma, St Louis, MO, USA) for 4 h at 37 °C. After centrifugation, 100 μL of 0.04 mol/L HCl-isopropanol were added. The absorbance was measured at 490 nm using ELISA microplate reader. This experiment was repeated three times.
RASSF1A luciferase constructs
To explore the transcriptional regulation of RASSF1A, three promoter reporter plasmids (RASSF1A-R1 spanned the −801 to +150 bp; RASSF1A-R2 spanned the −501 to +150 bp; RASSF1A-R3 spanned the −250 to +150 bp) were constructed. The amplified fragments were inserted into the pGL3-basic vector (Promega, Madison, WI, USA) between the KpnI and HindIII sites. These recombination plasmids were then sequenced for confirmation.
Luciferase assay
1 × 105 TE13 cells per well were seeded in 24-well dish 24 h before transfection. In all, 200 ng of RASSF1A deletion construct (RASSF1A-R1 to R3), pGL3-control vector (positive control) or pGL3-basic vector (negative control) constructs was cotransfected with 10 ng of pRL-TK vector using lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). Luciferase activity was measured with the dual-luciferase reporter assay system (Promega, Madison, WI, USA). Promoter activities were expressed as the ratio of firefly luciferase to renilla luciferase activity and each data point was done in triplicate.
In vitro DNA methylation
Construct pGL3-R2 was in vitro methylated as described previously [31]. 200 ng of mock or SSI-treated vector were transfected in TE13 cells. After 48 h, luciferase and renilla activity were assayed as described above.
Methylation analysis of RASSF1A, RASSF1B, and RASSF1C
Total DNA was isolated from 5-Aza-dC treated and untreated cells using DNAzol (Invitrogen, Carlsbad, CA, USA) according to manufacturer’s recommendation. Genomic DNA from tumor and corresponding normal sections was isolated from flash frozen tissues using a simplified proteinase K digestion method. To examine the DNA methylation patterns, genomic DNA was treated with sodium bisulfite as described previously [32]. The methylation status of RASSF1A, RASSF1B, and RASSF1C was then determined by bisulfite genomic sequencing (BGS) and bisulfite conversion-specific and methylation-specific polymerase chain reaction method (BS-MSP) as described previously [33]. For BGS, primers were designed to recognize sodium bisulfite converted DNA and encompassing CpG Island within the human RASSF1A gene promoter and exon 1 (from −199 to +160, and +139 to +434 bp, respectively). For BS-MSP, three regions (from −159 to −36 bp, from +103 to +302 bp, from +337 to +494 bp, respectively) of RASSF1A and one region of main CpG island of RASSF1B (from +299 to +552) and RASSF1C (from −470 to −217) were analyzed. Briefly, BS-MSP consists of two-step PCR amplifications. In the first step of BS-MSP, 100 ng of bisulfite-treated DNA was amplified and a primer set that does not contain any CpG but contains many cytosines of non-CpG sites at the 3′ position was used. Only the sequence that is fully converted by bisulfite is amplified. The second step of BS-MSP used the conventional MSP primer sets that contain many cytosines of CpG sites at the 3′ position specific for methylated and unmethylated sequences. The primers and reaction conditions were listed in Supplementary Table 2. Genomic DNA, methylated in vitro by CpG methyltransferase (Sss I) following the manufacturer’s directions (New England BioLabs, Beverly, MA, USA), was used as a positive control and water blank was used as a negative control. BS-MSP products were analyzed on 2 % agarose gel with ethidium bromide staining, and were determined to have methylation if a visible band was observed in the methylation reaction. Reactions were performed in duplicate with each of the samples, in order to ensure the reproducibility and consistency of the results.
RASSF1A protein expression via immunohistochemical staining
RASSF1A protein expression was determined by immunostaining using the avidin–biotin complex immunoperoxidase method, which was performed on parallel histopathological sections from paraffin-embedded tumor section and corresponding normal tissues. After blocking of endogenous peroxidase and non-specific reactions, the primary antibody against RASSF1A (1:100 dilution, goat anti-human polyclonal antibody, SC-18724, Santa Cruz Biotechnology, San Diego, CA, USA) was applied to sections, which were then incubated with biotinylated secondary antibody and ABC reagent. 0.5 % 3, 3′-Diaminobenzidine (Sigma, St Louis, MO, USA) was used as the chromagen. For a negative control, the primary antibody was replaced with mouse IgG. Slides with positive staining of RASSF1A were used as positive control.
Immunohistochemical staining was evaluated according to a scoring method reported previously [34]. Scoring accounted for both representation of the areas and intensities of the stains. Briefly, the score is the sum of the percentage of positive cells (0, less than 25 % positive cells; 1, 26–50 % positive cells; 2, 51–75 % positive cells, and 3, more than 75 % positive cells) and the staining intensity (0, negative; 1, weak; 2, moderate; 3, strong). Sums between 0 and 2 were scored as negative; sums of 3 and 6 were scored as positive. All slides were examined and scored by three independent observers, who were blinded to the clinical data. All of the slides were reviewed concurrently by three experienced pathologists.
Statistical analysis
Statistical analysis was performed using SPSS13.0 software package (SPSS Company, Chicago, Illinois, USA). The results of RT-PCR, and real-time RT-PCR were expressed as the mean ± S.D. The means were compared using the Student’s t test. Chi square test was used to compare the frequency of gene methylation and protein expression between ESCC tissues and corresponding normal tissues. Relationships between variables were tested by Spearman correlation analysis. Kaplan–Meier survival curves were constructed and the Log-rank or the Breslow tests were used as needed for the univariate comparison of RASSF1A expression and methylation categories. Cox’s multivariate test applied in a stepwise forward method was used to adjust for potentially confounding variables (e.g., stage and UGIC family history) and to evaluate the role of RASSF1A as independent predictors of patient prognosis. Two-sided tests were used to determine significance; and P values less than 0.05 were regarded as statistically significant for all statistic tests.
Results
Frequent silencing of RASSF1A but not RASSF1C in esophageal cancer cell lines
The mRNA expression of RASSF1A, RASSF1B, and RASSF1C were examined in 5 esophageal cancer cell lines to determine whether there are similar expression of these genes in esophageal cancer. As shown in Fig. 1, mRNA expression of RASSF1A was remarkably silenced or reduced in TE13, Yes-2 and T.Tn cell lines, while the mRNA expression levels of RASSF1C remained high in virtually all cell lines.
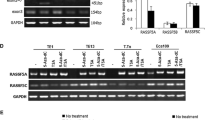
RASSF1A, RASSF1B, and RASSF1C mRNA expression in five human esophageal cancer cell lines and the effect of RASSF1A on TE13 cell line. A mRNA expression of RASSF1A, RASSF1B, and RASSF1C in five esophageal cancer cell lines treated with 5-Aza-dC, TSA, 5-Aza-dC/TSA. B Relative expression of RASSF1A, RASSF1B, and RASSF1C mRNA in five esophageal cancer cell lines treated with 5-Aza-dC, TSA, 5-Aza-dC/TSA. a Relative expression of RASSF1A mRNA in five esophageal cancer cell lines. b Relative expression of RASSF1B mRNA in five esophageal cancer cell lines. c Relative expression of RASSF1C mRNA in five esophageal cancer cell lines. C MTT assay showed that 5-Aza-dC, 5-Aza-dC/TSA treatment and stable transfection of RASSF1A resulted in a significant inhibition of TE13 cell proliferation
Up-regulation of RASSF1A by 5-Aza-dC treatment in esophageal cancer cell lines
In order to know whether epigenetic mechanisms such as methylation and acetylation were associated with the expression of RASSF1A, RASSF1B, and RASSF1C in esophageal cancer, we further detected the mRNA expression of RASSF1A, RASSF1B, and RASSF1C in 5-Aza-dC, TSA or a combination of 5-Aza-dC and TSA treated esophageal cancer cell lines. TE13 cell line was also used to reconstitute the expression of RASSF1A by stably transfected with RASSF1A to provide more evidence for the epigenetic role of RASSF1A in esophageal cancer cell. As shown in Fig. 1, treatment with 5-Aza-dC significantly increased the level of RASSF1A mRNA in esophageal cancer cells, especially in TE13, Yes-2 and T.Tn cell lines. However, TSA treatment did not result in significant up-regulation of RASSF1A mRNA in esophageal cancer cell lines. The combination of 5-Aza-dC/TSA treatment resulted in significant up-regulation of RASSF1A mRNA in TE13, Yes-2 and T.Tn cell lines. 5-Aza-dC, TSA or a combination of 5-Aza-dC and TSA treatment did not result in any significant mRNA expression variation of RASSF1B and RASSF1C in 5 esophageal cancer cell lines.
Inhibition of proliferation in TE13 Cell line after treatment with 5-Aza-dC
MTT assay was used to assess the inhibitory effects of 5-Aza-dC/TSA/5-Aza-dC + TSA and stable transfection of RASSF1A on the proliferation of TE13. As shown in Fig. 1c, 5-Aza-dC treatment and stable transfection of RASSF1A resulted in a significant inhibition of TE13 cell proliferation. This observation suggested that RASSF1A may act as a tumor suppressor gene in esophageal cancer through inhibition of cell proliferation.
The aberrant promoter and exon 1 methylation of RASSF1A induces silencing of RASSF1A expression
The MethPrimer program [35] and the CpG island searcher [36] were used to determine whether the sequence of RASSF1A, RASSF1B and RASSF1C contain CpG islands. As shown in Fig. 2a, 1 CpG island is found to be located in RASSF1A promoter and exon 1 (from −205 to +570 bp). As shown in Fig. 2b, RASSF1B and RASSF1C are shown to have the same CpG islands which locate in promoter and exon 1 region. BGS assay showed hypermethylation of promoter and exon 1 of RASSF1A in TE13 cell line (Fig. 2c). The results were further verified by BS-MSP assay (Fig. 2d). Fully methylation of 3 regions of RASSF1A was observed in TE13 cells, while incomplete methylation of RASSF1A was observed in Yes-2 and T.Tn cell lines. After treatment with 5-Aza-dC, demethylation of RASSF1A was observed in these cells, together with the results of silenced mRNA expression of RASSF1A in TE13 cell line (Fig. 1), indicating that promoter and exon 1 methylation of RASSF1A may suppress the expression of RASSF1A in TE13 esophageal cells. As shown in Fig. 2d, BS-MSP assay did not show any methylation of RASSF1B and RASSF1C in 5 esophageal cancer cell lines.
Methylation analysis of RASSF1A, RASSF1B, and RASSF1C in esophageal cancer cell lines. a A 775 bp CpG island of RASSF1A is shown and the BS-MSP regions analyzed is indicated. b Four CpG islands of RASSF1B and RASSF1C are shown and the BS-MSP region analyzed is indicated. c High-resolution mapping of the methylation status of every CpG site in the RASSF1A promoter and exon 1 by BGS in five esophageal cancer cell lines. Each CpG site is shown at the top row as an individual number. Percentage methylation was determined as percentage of methylated cytosines from 8 to 10 sequenced colonies. The color of circles for each CpG site represents the percentage of methylation. d Methylation status of the three regions of RASSF1A detected by BS-MSP in various tumor cell lines with or without 5-Aza-dC treatment. M, methylated; U, unmethylated. e Luciferase activity of promoter constructs. pGL3-control vector was used as a positive (POS), and empty pGL3-basic vector as a negative control (EV). RASSF1A-R2 showed the highest relative luciferase activity. f The promoter region −501 to +150 bp was in vitro methylated, cloned into pGL3 vector and luciferase activity was determined and compared with that of unmethylated RASSF1A-R2. In vitro methylation of RASSF1A-R2 led to a significant decrease in luciferase activity
In vitro methylation of RASSF1A leads to a significant decrease in luciferase activity
Three constructs (RASSF1A-R1–R3) were designed for functional characterization of the RASSF1A promoter. RASSF1A-R2 had the highest relative luciferase activity providing a potential explanation for the importance of promoter methylation of RASSF1A on the control of RASSF1A transcription (Fig. 2e). As a direct evidence for the role of methylation in this region, in vitro methylation of RASSF1A-R2 led to a 90 % decrease in luciferase activity (Fig. 2f).
mRNA expression of RASSF1A, RASSF1B, and RASSF1C in clinical specimens
mRNA expression of RASSF1A, RASSF1B, and RASSF1C was detected in 141 ESCC tumor tissues and corresponding normal tissues. As shown in Fig. 3a, RASSF1A mRNA expression in ESCC tumor tissues was significantly decreased compared to corresponding normal tissues (P < 0.01). When stratified for clinicopathologic characteristics, RASSF1A mRNA expression was associated with TNM stage, pathological differentiation, and LN metastasis (P < 0.05). RASSF1B mRNA expression in ESCC tumor tissues was not significantly different from that in corresponding normal tissues (P > 0.05). However, RASSF1C mRNA expression in ESCC tumor tissues was significantly increased compared to corresponding normal tissues (P < 0.05) and RASSF1C mRNA expression was not found to be associated with any clinicopathologic characteristics (P > 0.05). No correlation was found between mRNA levels of RASSF1A and RASSF1C in ESCC tumor tissues.
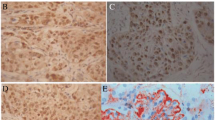
mRNA expression of RASSF1A, RASSF1B, and RASSF1C in ESCC tissues and methylation status, and immunohistochemical staining of RASSF1A in ESCC tissues. A Relative expression of RASSF1A, RASSF1B, and RASSF1C mRNA in tumor tissues and corresponding normal tissues. *P<0.05. B RT-PCR analysis of RASSF1A, RASSF1B, and RASSF1C in ESCC tumor tissues. C The methylation status of the RASSF1A regulatory CpG site determined by the BS-MSP analysis in ESCC tumor tissues. u indicates the presence of unmethylated genes; m indicates the presence of methylated genes. D Immunohistochemical staining of of RASSF1A in ESCC tumor tissues (SP × 400). a positive staining of RASSF1A in ESCC tissue; b negative staining of RASSF1A in ESCC tissue
Decreased protein expression of RASSF1A in clinical specimens
The pattern of immunohistochemical staining of RASSF1A was cytoplasmic (Fig. 3d). Positive protein expression of RASSF1A in tumor tissues (55.3 %, 78/141) was significantly lower than that in corresponding normal tissues (95.0 %, 134/141) (P < 0.01). According to our scoring method, no tumor tissues showed higher protein expression than corresponding normal tissues, only 35 tumor tissues showed similar positive protein expression when compared to corresponding normal tissues. The sums of the percentage of positive cells and the staining intensity in the other 43 tumor tissues which showed positive protein expression of RASSF1A were lower than that in corresponding normal tissues. When stratified for clinicopathologic characteristics, RASSF1A protein expression was associated with TNM stage, pathological differentiation, LN metastasis, and distant metastasis or recurrence (P < 0.05) (Table 1).
Aberrant promoter and exon 1 methylation of RASSF1A in clinical specimens
The methylation analysis was successfully performed by BS-MSP method in all specimens. The methylation frequency of RASSF1A region 1, region 2 and region 3 in tumor tissues (37.6 %, 53/141; 51.8 %, 73/141; and 55.3 %, 78/141; respectively) was significantly higher than that in corresponding normal tissues (4.2 %, 6/141) (P < 0.05). When stratified for clinicopathologic characteristics, methylation status of the three regions was associated with TNM stage, pathological differentiation, distant metastasis or recurrence (P < 0.05). Methylation status of region 2 and region 3 was also found to be associated with LN metastasis and UGIC family history (P < 0.05) (Table 1).
Association between RASSF1A methylation status and expression
RASSF1A mRNA expression in ESCC tumor tissues with no detectable RASSF1A protein was significantly reduced compared to ESCC tumor tissues with detectable RASSF1A protein (P < 0.05). RASSF1A mRNA expression in ESCC tumor tissues where promoter or exon 1 of RASSF1A was methylated was significantly reduced compared to that in ESCC tumor tissues without methylation of the gene (P < 0.05). The correlation of RASSF1A methylation and protein expression was shown in Table 2; a close correlation was noted between RASSF1A promoter or exon 1 methylation and the loss of protein expression of the gene in ESCC (P < 0.01).
Survival analysis of RASSF1A in ESCC
As shown in Fig. 4, RASSF1A protein expression was positively correlated with ESCC patients’ survival. In the RASSF1A-expression ESCC tumors, the 5-year overall survival rates (OS) were 48 % (median survival time not reached) as opposed to the RASSF1A-negative ESCC tumors displaying 5-year survival rates of 23 % (median survival time, 31 months; P < 0.05; Log-rank test). RASSF1A region 3 methylation was inversely correlated with ESCC patients’ survival. In the RASSF1A-methylation ESCC tumors, the 5-year OS were 21 % (median survival time, 30 months; P < 0.05; Log-rank test) as opposed to the RASSF1A-unmethylation ESCC tumors displaying 5-year survival rates of 51 % (median survival time not reached). ESCC patients with both negative protein expression and methylation of region 3 of RASSF1A showed poor patient survival. ESCC patients in stage III and IV, with positive UGIC family history, negative expression and hypermethylation of RASSF1A were most likely to develop metastatic disease and also showed the worse survival.
Kaplan–Meier univariate survival analysis of RASSF1A expression and methylation in ESCC. a Kaplan–Meier curves for cumulative survival stratified by RASSF1A expression status: showing a direct correlation between negative RASSF1A expression and poor patient survival of ESCC cases. b Kaplan–Meier curves for cumulative survival stratified by RASSF1A methylation status: showing a direct correlation between RASSF1A methylation and poor patient survival of ESCC cases. c Kaplan–Meier curves for cumulative survival stratified by RASSF1A expression and methylation status: ESCC patients with both negative protein expression and methylation of RASSF1A showing poor patient survival. d Kaplan–Meier curves for cumulative survival stratified by RASSF1A expression and TNM stages: stage III and IV ESCC patients with negative protein expression of RASSF1A showing poor patient survival. e Kaplan-Meier curves for cumulative survival stratified by RASSF1A expression and TNM stages: stage III and IV ESCC patients with RASSF1A methylation showing poor patient survival
Cox multivariate analysis was done using RASSF1A methylation, expression, tumor stage, as well as other confounding variables such as UGIC family history, age, and patient gender. RASSF1A methylation status, protein expression, and TNM stage were independently associated with ESCC patients’ survival (Table 3).
Discussion
Several studies have suggested that chromosome 3p21 contains one or more candidate tumor suppressor genes involved in various tumor types [8]. The RASSF1 gene is a novel candidate tumor suppressor gene that was isolated from this region [7]. The seven different RASSF1 isoforms (RASSF1A to RASSF1G) have four common C-terminal exons (exons 3–6) which encode a RalGDS/AF6 or Ras association (RA) domain [37] and a Sav/RASSF/Hpo (SARAH) domain [38]. Exon 3 also contains a putative ataxia telagectasia mutant (ATM) kinase phosphorylation consensus sequence motif. The RASSF1A transcript also contains a diacylglycerol/phorbol ester-binding (DAG) domain, which contains a central zinc finger (zinc-binding domain). RASSF1A is one of the most frequently inactivated proteins ever identified in human cancer. Loss of RASSF1A expression is largely attributed to promoter hypermethylation, as somatic mutations of RASSF1A are uncommon. In more than 200 samples lung, breast, kidney and nasopharyngeal carcinomas and cell lines analyzed, only one frame-shift mutation (at codon 277 in the RA domain) and one missense mutation (at codon 201 in the RA domain) have been identified [7, 24, 39]. However, numerous polymorphisms have been identified in these tumours and cell lines, many of which are located in the functional domains of RASSF1A (five in the C1 domain, four in the ATM phosphorylation site consensus sequence, and five in the RA domain) [9], and many of them have proven to encode a functionally impaired mutant RASSF1A.
RASSF1A methylation has the potential to be an ideal cancer biomarker as it occurs at moderate to high frequency in a very wide range of tumor types, yet is comparatively rarely found in normal tissues [16, 17]. In the present study, we found silenced or decreased mRNA expression of RASSF1A in TE13, Yes-2 and T.Tn cell lines, the up-regulation of RASSF1A mRNA and inhibition of proliferation in TE13 cells after treatment with 5-Aza-dC or stable transfected of RASSF1A, suggesting that RASSF1A may act as a tumor suppressor gene in esophageal cancer and aberrant methylation of RASSF1A may suppress RASSF1A expression in esophageal cancer cell. The observation was further verified by BGS and BS-MSP analysis of promoter and exon 1 methylation status of RASSF1A in TE13 cell, and the hypermethylation of promoter and exon1 in RASSF1A can influence transcriptional activity. However, TSA treatment did not result in significant up-regulation of RASSF1A mRNA in esophageal cancer cell lines, indicating histone acetylation may not play crucial roles in the inactivation of RASSF1A. BGS assay has found the density of methylated CpG sites of RASSF1A is higher in exon 1 than in promoter, suggesting the critical CpG sites may located in exon 1 of RASSF1A which is further verified by BS-MSP assay in clinical specimens. Hypermethylation of 3 regions has been found in ESCC tumor tissues with region 3 showing the highest methylation frequency. In addition, similar to previous studies in esophageal cancer [18–20] and other carcinomas such as bladder, lung, breast cancer, gastric adenocarcinoma and osteosarcoma [21–24], there was a significant concordance between RASSF1A methylation status and its expression in the present study. These findings indicate that methylation-mediated inactivation of RASSF1A may attribute to methylation of dense CpG sites in the exon 1 region.
Methylation patterns have been found to be useful to assess clinical outcomes or response to chemotherapeutic agents. High levels of DNA methylation are associated with a poor prognosis such as in lung cancer [40]. In the present study, we showed that hypermethylation and expression of RASSF1A was significantly and directly correlated with ESCC patients’ survival. In multivariate analysis, the combination of tumor stage, UGIC family history, and RASSF1A methylation and expression provided independent predictive information on ESCC patients’ metastasis and poor survival. Patients with hypermethylation and down expression of RASSF1A, in stage III and IV, with positive UGIC family history had the worst 5-year overall survival time. When methylation and expression of RASSF1A were combined to analysis, ESCC patients with both negative protein expression and methylation of RASSF1A showed poor patient survival, thereby indicating that RASSF1A silencing through hypermethylation may confer a growth advantage in ESCC. Thus, hypermethylation and loss of RASSF1A expression may be considered to be useful markers of ESCC tumor progression and poor prognosis.
RASSF1B expression has been detected in cells from the hematopoetic systems [7]. Lee et al. [21] identified that RASSF1B was undetectable in 60 % (3 of 5) of bladder cell lines and in 31 % (17 of 55) of primary bladder carcinomas. Byun et al. [22] found RASSF1B transcripts were not expressed in 33 % (5 of 15) of gastric carcinoma cell lines and Lim et al. [23] found that RASSF1B expression was absent in 30 % (3/10) of primary osteosarcomas. However, Malpeli et al. [41] demonstrated that RASSF1B was always expressed in both normal and pancreatic endocrine tumor with no significant difference. In the present study, we found absent expression of RASSF1B in TE13 cell line and this inactivation can not be restored by 5-Aza-dC of TSA treatment, indicating that methylation or histone acetylation may not play crucial roles in the expression of RASSF1B and was further verified by BS-MSP assay. No methylation of RASSF1B was found in ESCC tumor tissues, however, decreased mRNA expression of RASSF1B in tumor tissues was found although it did not reach significant difference when compared with corresponding normal tissues. Further studies need to be done to clarify the exact effect of RASSF1B in ESCC.
For RASSF1C, the other major RASSF1 isoform which sharing the same promoter with RASSF1B, there are less consistent studies. Although no hypermethylation of the promoter was detected [7, 24], its transcript was missing in some cancer cell lines, and this could be attributed to deletions of 3p21.3, which are frequently observed in cancer [42]. In one report, it was stated that the inactivation of RASSF1C during in vivo tumor growth identified it as a tumor suppressor gene [27]. In contrast in other studies, it was reported that RASSF1C stimulates human lung cancer cell proliferation and breast cancer cell migration [43, 44]. Malpeli et al. [41] found RASSF1C expression was 11.4 times higher in pancreatic endocrine tumor tissues than in normal tissues. Byun et al. [22] reported that RASSF1C was detectable in 15 gastric carcinoma cell lines and Lee et al. [21] reported that RASSF1C transcript was detected in 10 bladder and prostate cancer cell lines and primary bladder carcinomas. In the present study, we found high expression levels and no methylation of RASSF1C in virtually all 5 esophageal cell lines. RASSF1C mRNA expression in ESCC tumor tissues was significantly increased compared to corresponding normal tissues, indicating that RASSF1C, unlike RASSF1A, is not a tumor suppressor, but instead may play a role in promoting progression of ESCC.
In conclusion, the present study suggested that promoter and exon 1 especially exon 1 of RASSF1A were frequently methylated in ESCC and the methylation was related to reduced mRNA and protein expression. Hypermethylation may be one of the mechanisms for inactivation of RASSF1A in ESCC. Unlike RASSF1A, methylation and expression variation of RASSF1B was not found in ESCC tissues and corresponding normal tissues. RASSF1C is highly expressed in ESCC tissues suggesting that RASSF1C might play a pathogenetic role in tumor development. Additionally, hypermethylation and down expression of RASSF1A, in stage III and IV, with positive UGIC family history is highly predictive of metastasis and poor prognosis in ESCC. RASSF1A may be a functional tumor suppressor and may serve as a prognostic methylation biomarker for ESCC.
References
Enzinger PC, Mayer RJ (2003) Esophageal cancer. N Engl J Med 349:2241–2252
Shahbaz Sarwar CM, Luketich JD, Landreneau RJ, Abbas G (2010) Esophageal cancer: an update. Int J Surg 8:417–422
Sun G, Wang S, Hu X, Su J, Huang T, Yu J, Tang L, Gao W, Wang JS (2007) Fumonisin B1 contamination of home-grown corn in high-risk areas for esophageal and liver cancer in China. Food Addit Contam 24:181–185
Guohong Z, Min S, Duenmei W, Songnian H, Min L, Jinsong L, Hongbin L, Feng Z, Dongping T, Heling Y, Zhicai L, Shiyong L, Quansheng G, Xiaoyun L, Yuxia G (2010) Genetic heterogeneity of oesophageal cancer in high-incidence areas of southern and northern China. PLoS ONE 5:e9668
Guo W, Zhou RM, Wan LL, Wang N, Li Y, Zhang XJ, Dong XJ (2008) Polymorphisms of the DNA repair gene xeroderma pigmentosum group A and C and risk of esophageal squamous cell carcinoma in a population of high incidence region of North China. J Cancer Res Clin Oncol 134:263–270
Guo W, Wang G, Dong Y, Guo Y, Kuang G, Dong Z (2013) Decreased expression of WWOX in the development of esophageal squamous cell carcinoma. Mol Carcinog 52:265–274
Dammann R, Li C, Yoon JH, Chin PL, Bates S, Pfeifer GP (2000) Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3. Nat Genet 25:315–319
Lerman MI, Minna JD (2000) The 630 kb lung cancer homozygous deletion region on human chromosome 3p21.3: identification and evaluation of the resident candidate tumor suppressor genes. The international lung cancer chromosome 3p21.3 tumor suppressor gene consortium. Cancer Res 60:6116–6133
Dammann R, Schagdarsurengin U, Strunnikova M, Rastetter M, Seidel C, Liu L, Tommasi S, Pfeifer GP (2003) Epigenetic inactivation of the Ras-association domain family 1 (RASSF1A) gene and its function in human carcinogenesis. Histol Histopathol 18:665–677
van der Weyden L, Adams DJ (2007) The Ras-association domain family (RASSF) members and their role in human tumourigenesis. Biochim Biophys Acta 1776:58–85
Bird A (2001) Molecular biology methylation talk between histones and DNA. Science 294:2113–2115
Jones PA, Baylin SB (2007) The epigenomics of cancer. Cell 128:683–692
Guo W, Dong Z, Lin X, Zhang M, Kuang G, Zhu T (2012) Decreased expression and aberrant methylation of Raf kinase inhibitory protein gene in esophageal squamous cell carcinoma. Cancer Invest 30:703–711
Guo W, Wang G, Dong Y, Guo Y, Kuang G, Dong Z (2013) Decreased expression of WWOX in the development of esophageal squamous cell carcinoma. Mol Carcinog 52:265–274
Zhu YH, Fu L, Chen L, Qin YR, Liu H, Xie F, Zeng T, Dong SS, Li J, Li Y, Dai Y, Xie D, Guan XY (2013) Downregulation of the novel tumor suppressor DIRAS1 predicts poor prognosis in esophageal squamous cell carcinoma. Cancer Res 73:2298–2309
Dammann R, Schagdarsurengin U, Seidel C, Strunnikova M, Rastetter M, Baier K, Pfeifer GP (2005) The tumor suppressor RASSF1A in human carcinogenesis: an update. Histol Histopathol 20:645–663
Hesson LB, Cooper WN, Latif F (2007) The role of RASSF1A methylation in cancer. Dis Markers 23:73–87
Kuroki T, Trapasso F, Yendamuri S, Matsuyama A, Alder H, Mori M, Croce CM (2003) Promoter hypermethylation of RASSF1A in esophageal squamous cell carcinoma. Clin Cancer Res 9:1441–1445
Lo PH, Xie D, Chan KC, Xu FP, Kuzmin I, Lerman MI, Law S, Chua D, Sham J, Lung ML (2007) Reduced expression of RASSF1A in esophageal and nasopharyngeal carcinomas significantly correlates with tumor stage. Cancer Lett 257:199–205
Mao WM, Li P, Zheng QQ, Wang CC, Ge MH, Hu FJ, Fang XH, Dong ZM, Ling ZQ (2011) Hypermethylation-modulated downregulation of RASSF1A expression is associated with the progression of esophageal cancer. Arch Med Res 42:182–188
Lee MG, Kim HY, Byun DS, Lee SJ, Lee CH, Kim JI, Chang SG, Chi SG (2001) Frequent epigenetic inactivation of RASSF1A in human bladder carcinoma. Cancer Res 61:6688–6692
Byun DS, Lee MG, Chae KS, Ryu BG, Chi SG (2001) Frequent epigenetic inactivation of RASSF1A by aberrant promoter hypermethylation in human gastric adenocarcinoma. Cancer Res 61:7034–7038
Lim S, Yang MH, Park JH, Nojima T, Hashimoto H, Unni KK, Park YK (2003) Inactivation of the RASSF1A in osteosarcoma. Oncol Rep 10:897–901
Burbee DG, Forgacs E, Zöchbauer-Müller S, Shivakumar L, Fong K, Gao B, Randle D, Kondo M, Virmani A, Bader S, Sekido Y, Latif F, Milchgrub S, Toyooka S, Gazdar AF, Lerman MI, Zabarovsky E, White M, Minna JD (2001) Epigenetic inactivation of RASSF1A in lung and breast cancers and malignant phenotype suppression. J Natl Cancer Inst 93:691–699
Harada K, Toyooka S, Maitra A, Maruyama R, Toyooka KO, Timmons CF, Tomlinson GE, Mastrangelo D, Hay RJ, Minna JD, Gazdar AF (2002) Aberrant promoter methylation and silencing of the RASSF1A gene in pediatric tumors and cell lines. Oncogene 21:4345–4349
Malpeli G, Amato E, Dandrea M, Fumagalli C, Debattisti V, Boninsegna L, Pelosi G, Falconi M, Scarpa A (2011) Methylation-associated down-regulation of RASSF1A and up-regulation of RASSF1Cin pancreatic endocrine tumors. BMC Cancer 11:351
Li J, Wang F, Protopopov A, Malyukova A, Kashuba V, Minna JD, Lerman MI, Klein G, Zabarovsky E (2004) Inactivation of RASSF1C during in vivo tumor growth identifies it as a tumor suppressor gene. Oncogene 23:5941–5949
Kondo Y, Shen L, Issa JP (2003) Critical role of histone methylation in tumor suppressor gene silencing in colorectal cancer. Mol Cell Biol 23:206–215
Meng CF, Zhu XJ, Peng G, Dai DQ (2007) Re-expression of methylation-induced tumor suppressor gene silencing is associated with the state of histone modification in gastric cancer cell lines. World J Gastroenterol 13:6166–6171
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408
Yu L, Liu C, Vandeusen J, Becknell B, Dai Z, Wu YZ, Raval A, Liu TH, Ding W, Mao C, Liu S, Smith LT, Lee S, Rassenti L, Marcucci G, Byrd J, Caligiuri MA, Plass C (2005) Global assessment of promoter methylation in a mouse model of cancer identifies ID4 as a putative tumor-suppressor gene in human leukemia. Nat Genet 37:265–274
Guo W, Dong Z, Chen Z, Yang Z, Wen D, Kuang G, Guo Y, Shan B (2009) Aberrant CpG Island hypermethylation of RASSF1A in gastric cardia adenocarcinoma. Cancer Invest 27:459–465
Sasaki M, Anast J, Bassett W, Kawakami T, Sakuragi N, Dahiya R (2003) Bisulfite conversion-specific and methylation-specific PCR: a sensitive technique for accurate evaluation of CpG methylation. Biochem Biophys Res Commun 309:305–309
Umemoto M, Yokoyama Y, Sato S, Tsuchida S, Al-Mulla F, Saito Y (2001) Carbonyl reductase as a significant predictor of survival and lymph node metastasis in epithelial ovarian cancer. Br J Cancer 85:1032–1036
Li LC, Dahiya R (2002) MethPrimer: designing primers for methylation PCRs. Bioinformatics 18:1427–1431
Takai D, Jones PA (2003) The CpG island searcher: a new WWW resource. In Silico Biol 3:235–240
Yamamoto T, Taya S, Kaibuchi K (1999) Ras-induced transformation and signaling pathway. J Biochem 126:799–803
Scheel H, Hofmann K (2003) A novel interaction motif, SARAH, connects three classes of tumor suppressor. Curr Biol 13:R899–R900
Dreijerink K, Braga E, Kuzmin I, Geil L, Duh FM, Angeloni D, Zbar B, Lerman MI, Stanbridge EJ, Minna JD, Protopopov A, Li J, Kashuba V, Klein G, Zabarovsky ER (2001) The candidate tumor suppressor gene, RASSF1A, from human chromosome 3p21.3 is involved in kidney tumorigenesis. Proc Natl Acad Sci USA 98:7504–7509
Brock MV, Hooker CM, Ota-Machida E, Han Y, Guo M, Ames S, Glöckner S, Piantadosi S, Gabrielson E, Pridham G, Pelosky K, Belinsky SA, Yang SC, Baylin SB, Herman JG (2008) DNA methylation markers and early recurrence in stage I lung cancer. N Engl J Med 358:1118–1128
Malpeli G, Amato E, Dandrea M, Fumagalli C, Debattisti V, Boninsegna L, Pelosi G, Falconi M, Scarpa A (2011) Methylation-associated down-regulation of RASSF1A and up-regulation of RASSF1C in pancreatic endocrine tumors. BMC Cancer 11:351
Vos MD, Ellis CA, Bell A, Birrer MJ, Clark GJ (2000) Ras uses the novel tumor suppressor RASSF1 as an effector to mediate apoptosis. J Biol Chem 275:35669–35672
Amaar YG, Minera MG, Hatran LK, Strong DD, Mohan S, Reeves ME (2006) Ras association domain family 1C protein stimulates human lung cancer cell proliferation. Am J Physiol Lung Cell Mol Physiol 291:L1185–L1190
Reeves ME, Baldwin SW, Baldwin ML, Chen ST, Moretz JM, Aragon RJ, Li X, Strong DD, Mohan S, Amaar YG (2010) Ras-association domain family 1C protein promotes breast cancer cell migration and at tenuatesapoptosis. BMC Cancer 10:562
Acknowledgments
We thank the patients and control individuals for taking part in this study. Supported by Grants from the National Natural Science Foundation (No. 81101854).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Guo, W., Cui, L., Wang, C. et al. Decreased expression of RASSF1A and up-regulation of RASSF1C is associated with esophageal squamous cell carcinoma. Clin Exp Metastasis 31, 521–533 (2014). https://doi.org/10.1007/s10585-014-9646-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10585-014-9646-5