Abstract
Human TH2 cell differentiation results in the selective demethylation of several specific CpG dinucleotides in the IL-4 and IL-13 genes, which are expressed in activated TH2, but not TH1, cells. This demethylation is accompanied by the appearance of six DNase I hypersensitive sites within 1.4 kb at the 5'-end of the IL-4 gene. Micrococcal nuclease (MNase) digestion revealed that in both TH1 and TH2 cells nine nucleosomes with a repeat length of 201 bp are identically positioned around the 5'-end of the IL-4 gene. However, only in TH2 cells are six out of the eight intervening linkers exposed to DNase I. This suggests that a major perturbation of the higher-order chromatin structure occurs above the level of the nucleosome in vivo. It is observed in cells that are poised for expression but which are not actively expressing the gene (i.e. resting TH2 cells). Notably, all the demethylated CpGs in TH2 cells are found in DNA that is accessible to DNase I. This may suggest that the opening of the chromatin structure allows binding of specific trans-acting factors that prevent de novo methylation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
DNA in the eukaryotic nucleus is packaged as chromatin at several levels. Most of it remains highly condensed, but cell differentiation and/or gene activation leads to a more open chromatin structure of the genes that are poised for expression in particular tissues (reviewed in Elgin and Workman (2000) and Wolffe (1998)). The first level of packing, the nucleosome, allows transcription after remodelling and/or histone modifications and substitutions. The second level, the ‘30 nm chromatin fibre’, is transcriptionally dormant chromatin that forms a regular helix of about seven nucleosomes per turn with a repeat length in the region of 200 bp. It represents the bulk of the silent chromatin of avian erythrocytes, and all the high-molecular-weight chromatin that diffuses out of MNase-digested nuclei is in a fibre conformation. Hence, the fibre itself is a thermodynamically stable structure (van Holde 1988). Variegation effect studies have shown that gene silencing is accompanied by higher compaction, with more regularly spaced nucleosomes (Wallrath and Elgin 1995). This suggests that most of the nucleosome positions are defined by structural constraints due to nucleosome interactions, rather than by the DNA sequence. However, it is not known whether the fibre is unfolded in the active genes or whether it unfolds immediately before transcription.
Conversely, the chromatin structure of active genes is less compact. Sedimentation coefficients of chromatin fragments enriched for active genes are lower than those of the correspondingly sized dormant genes (Kimura et al. 1983; Gilbert et al. 2004). They are digested faster by DNase I and MNase (Weintraub and Groudine 1976; Hebbes et al. 1994; Ravindra et al. 1999; Wang and Simpson 2001). There are local perturbations of their structure caused by missing, remodelled, or unfolded nucleosomes, which are manifested as DNase I hypersensitive sites (DHSs) (Gross and Garrard 1988; Workman and Kingston 1998; Boeger et al. 2003). The histones are modified posttranscriptionally (Avni et al. 2002) and DNA is demethylated (Elgin and Workman 2000; Meehan and Stancheva 2001) and is transcribed intergenically (Ashe et al. 1997; Rogan et al. 1999; Gribnau et al. 2000; Rogan et al. 2004).
The role of the higher-order chromatin structure(s) of transcriptionally active genes—the interactions of consecutive nucleosomes along the DNA and the path of the linker DNA within them–has attracted less attention and is not yet fully understood. Nevertheless, a general picture was established in which nucleosomes in the active genes resemble a ‘beads-on-a-string’ or ’11 nm fibre’ (filament) conformation, which folds into a 30 nm fibre when the genes are silenced (Wolffe 1998). Several cartoons have been published illustrating this (Felsenfeld and Groudine 2003). However, the ‘11 nm fibre (filament)’ or ‘beads-on-a-string’, which is more open than the 30 nm fibre, has not been observed under physiological conditions, but only at extremely low salt concentrations (van Holde 1988) or after depletion of H1 histone or partial trypsinization. No intermediate structures between the nucleosome and the 30 nm fibre have been observed and recently general opinion has begun to change amid suggestions that active genes may be in a 30 nm fibre (Bulger and Groudine, 1999).
General DNase I and MNase sensitivity is assumed to distinguish between transcriptionally active and inactive chromatin. For some loci, like the β-globin locus and the yeast silent mating type locus HMLα, these methods reveal clear-cut correlations with the transcriptional status and/or the level of histone acetylation (Weintraub and Groudine 1976; Hebbes et al. 1994; Ravindra et al. 1999; Wang and Simpson 2001). However, reports showing no direct correlation between general DNase I sensitivity and transcriptional status have also been published for other genes; for review see Higgs (1998). This is because the general nuclease sensitivity does not relate directly to any structural feature of chromatin.
MNase is a small molecule that penetrates the condensed chromatin and always digests linker DNA first, producing a nucleosome ladder. If nucleosomes are identically positioned in both alleles in the majority of cells, their positions can be visualized using indirect end-labelling. Conversely, the bulky DNase I molecule cannot access DNA linkers and digests only the parts of DNA that are exposed on the outside of the fibre (Staynov 1983; Staynov and Proykova 1998, 2007; Staynov 2000; Woodcock and Dimitrov 2001). If the fibre is open after partial trypsinization or depletion of H1 histone, the internucleosomal linkers are exposed to DNase I and it produces an identical nucleosome ladder to that from MNase (Staynov 2000). In reconstituted oligonucleosomes without H1, DNase I also mimics MNase digestion (Cirillo et al. 2002). However, when DNase I is used in vivo it produces only a few DHSs and does not produce a nucleosome ladder even from highly active genes such as β-globins, which show considerable general sensitivity to this enzyme and presumably must be in some kind of ‘more open’ structure (Reitman and Felsenfeld 1990). One possible explanation is that the nucleosomes are positioned not uniformly but randomly in different cells, or that they have alternative positions in the two alleles. However, even when positioned nucleosomes are observed in some active genes, DNase I still introduces only a few cuts around the start of transcription (Bert et al. 2007). An alternative explanation is that the active genes are also in a fibre structure, which is marked for transcription by several alternative mechanisms. It opens in front of the transcriptional machinery, accompanied by temporal displacement of nucleosomes, and it closes immediately after transcription. This might contribute to a higher general DNase I sensitivity without showing footprints of the nucleosomes there (Higgs 1998; Bulger and Groudine 1999). If this is the case, the highly transcribed genes will exhibit more pronounced ‘general DNase I sensitivity’ than genes that fire irregularly. Indeed, recent works suggest that the active or potentially active genes can also be in a fibre, or a modified fibre (Sapojnikova et al. 2008). For a recent review of facultative heterochromatin see Trojer and Reinberg (2007).
Here we studied the unusual higher-order chromatin structure of the gene encoding IL-4 in human T-helper cells and its relation to DNA methylation after they become transcriptionally competent. The structure and DNA methylation status of IL-13 gene was shown as an independent control.
Antigenically naive CD4+ TH lymphocytes differentiate into at least two phenotypes after encountering different pathogens, and express different subsets of cytokines (Riviere et al. 1998; Kuo and Leiden 1999; Ansel et al. 2006). TH1 cells are involved in cell-mediated immunity and specifically express IFN-γ. TH2 cells are responsible for humoral immunity and eosinophil differentiation and express IL-4, IL-5 and IL-13. The genes encoding these TH2 cytokines are clustered on human chromosome 5 (see Fig. 1a).
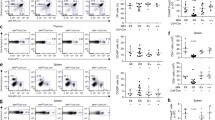
Cytokine expression of TH1 and TH2 cell populations after 28 days’ differentiation. (a) Map of IL-4/IL-13 locus on human chromosome 5. (b) Fluorescent activated cell sorting (FACS) analysis of cytokine expression in the differentiated TH1 and TH2 populations. Activated cells were assessed for competence to express IL-4, IL-13 and IFN-γ by intracellular cytokine staining. Cytokine-positive cells were determined by comparison with control antibodies and resting cells; quadrants were set so that 98% of events in the control sample were in the lower left quadrants. (c) RT-PCR analysis of transcription in resting and activated (by PMA and ionomycin) TH1 and TH2 cells
IL-4 expression plays an important autocrine role in TH2 differentiation as TH2 cells also express the IL-4 receptor. IL-4 has been reported to be expressed either mono- or biallelically and a recent study has shown that there is a random selection of the active allele (Bix et al. 1998; Riviere et al. 1998; Guo et al. 2005). Several trans- and cis-acting factors and/or alternative epigenetic mechanisms of regulation of IL-4 expression must therefore exist. Indeed, TH2 differentiation is accompanied by the appearance of DHSs in both the human and murine IL-4 and IL-13 genes (Agarwal and Rao 1998; Santangelo et al. 2002), by the demethylation of specific CpG dinucleotides (Santangelo et al. 2002), by intergenic transcription (Rogan et al. 1999, 2004) and by core histone modifications (Avni et al. 2002). A locus control region for the TH2 cytokine genes has also been reported in the mouse (Fields et al. 2004). This model of studying transcriptional control has an advantage over others in that the chromatin structure and DNA methylation are studied in competent cells before they are activated for expression, which provides a distinction between genes that are competent for transcription and those that are being actively transcribed.
In this study, after TH cells had been differentiated for 28 days into TH1 or TH2 cells, but were not currently activated (i.e. in resting T cells), their nuclei were digested with MNase and DNase I in parallel and the region around the IL-4 gene 5'-end was examined using indirect end-labelling. The methylation status of CpG dinucleotides in this region was also studied.
Materials and methods
In vitro differentiation of TH1/TH2 cells
Naive CD4+ CD45RA+ T lymphocytes were isolated from venous blood from healthy human volunteers as described (Cousins et al. 2002; Santangelo et al. 2002). These cells were cultured in RPMI 1640 (Life Technologies, UK) supplemented with 10% fetal calf serum, 2 mM l-glutamine, 100 U/ml penicillin (Life Technologies) and 100 μg/ml streptomycin (Life Technologies) and differentiated towards either the TH1 or TH2 phenotype for 28 days as described previously (Cousins et al. 2002). To confirm that the starting population had differentiated into the TH1 and TH2 phenotypes after this period, cytokine expression was analysed by RT-PCR and fluorescence activated cell sorting (FACS), as described previously (Cousins et al. 2002).
Bisulfite modification of DNA and sequencing
DNA was isolated from day-28 differentiated TH1 and TH2 cells and bisulfite-modified as described (Santangelo et al. 2002). The methylation status of CpG sites in the IL-13 promoter, the IL-4 promoter and the IL-4 second intron was determined by amplifying the modified DNA in these regions using PCR, and cloning and sequencing the resultant DNA fragments.
DNase I and MNase digestion analyses
Differentiated TH cell nuclei were permeabilized and digested with DNase I as described (Santangelo et al. 2002). Aliquots of the same populations of permeabilized nuclei containing 50 μg nucleic acid were digested with 0.5-14 units MNase (Worthington Biochemical Corporation, USA) at 25°C for 5 minutes in MNase digestion buffer (20 mM Tris pH 7.5, 50 mM NaCl, 2 mM CaCl2, 20% glycerol, 0.1 mM PMSF and complete EDTA-free protease inhibitor (Roche Diagnostics Ltd., UK), or with 10-26 units of DNase I (Sigma-Aldrich, UK) at 25°C for 10 min in 20 mM Tris pH 7.5, 15 mM NaCl, 5 mM MgCl2, 1 mM CaCl2, 0.1 mM EDTA, 0.1 mM EGTA, 20% glycerol, containing 0.1 mM PMSF and complete EDTA-free protease inhibitor (Roche Diagnostics). DNA was isolated using the DNEasy Tissue Kit (Qiagen, UK).
DNase I- and MNase-digested DNA was digested with BclI (New England Biolabs (NEB), USA) and precipitated, and 5 μg of DNA from each digestion point was separated on a 0.7% agarose TAE gel. The agarose gel showed that the bulk chromatin had been digested to the same degree by DNase I and by MNase in both TH1 and TH2 nuclei (not shown). The DNA fragments were indirectly end-labelled as described (Santangelo et al. 2002).
To analyse the bands produced by nuclease digestion, one digestion point on the autoradiograph from each cell type was digitized using NIH Image software and analysed further using Kaleidagraph (Synergy Software, USA). Nucleosome positioning and repeat lengths were analysed by linear regression. The traces were then expanded into the sum of Gaussian curves using the formula
where A is the amplitude of the peak in arbitrary units, X 0 is its position in base pairs from the restriction site end of the fragment, and Δ is its standard deviation, as described (Staynov and Proykova 1998).
Results
TH cell differentiation
Naive TH lymphocytes were obtained from venous blood collected from healthy volunteers and differentiated, in parallel, into either TH1 or TH2 cells. Previous work has shown that the differentiation of human cells requires many cell cycles (Cousins et al. 2002; Santangelo et al. 2002). Because nuclease digestion and DNA methylation patterns reflect the average of a population of individual cells, the 28-day time point was selected to obtain as homogeneous a population as possible. Differentiation into the two cell types was confirmed by analysis of the cytokine transcription and expression patterns using FACS analysis (Fig. 1b) and RT-PCR (Fig. 1c).
Demethylation of specific CpG sites in TH2 cells
Although IL-4 and IL-13 have a common evolutionary origin and have similar, partially overlapping functions, they show different regulation characteristics at the level of chromatin. There is a TH2-specific DHS at the transcriptional start of IL-13 and the seven closely spaced CpGs around it are strongly demethylated after TH2 (Fig. 2a) but not after TH1 differentiation (Santangelo et al. 2002).
CpG methylation status in TH2 cells after 28 days’ differentiation after cloning and sequencing a number of clones, numbered according to Santangelo et al. (2002). Black boxes denote a methylated CpG site and white boxes an unmethylated site .(a) Methylation status of a group of seven CpG sites in promoter and exon 1 of IL-13. (b) Methylation status of the seven CpG sites in promoter and exon 1 of IL-4. (c) Methylation status of the nine CpG sites in intron 2 of IL-4. Horizontal arrows indicate direction of gene transcription. Base pair coordinates refer to numbers used previously (Santangelo et al. 2002). Vertical arrows indicate previously reported DHS (Santangelo et al., 2002)
By contrast, there is no DHS at the transcriptional start of IL-4 (Santangelo et al. 2002; discussed further below) and the seven CpGs found in an interval of 224 bp (from -77 to +147 bp) around the start of transcription remained almost fully methylated, even after 28 days of differentiation (Fig. 2b). Our previous studies with methylation-sensitive restriction enzymes have shown that the sparsely distributed CpGs downstream in the gene are methylated in both TH1 and TH2 cells (Santangelo et al. 2002). Only one cluster of nine CpGs within a 174 bp stretch around a TH2-specific DHS in the second intron, 1 kb from the start of transcription (from 966 to 1142), was found to become considerably demethylated after 14 days’ differentiation (Santangelo et al. 2002), and after 28 days of differentiation, the demethylation at sites 1 and 9 was almost complete (Fig. 2c). The demethylation at these sites suggests that both alleles of IL-4 are in identical functional states and thus available for random allelic choice of transcription, in agreement with other findings on allelic expression of IL-4 in terminally differentiated cells (Guo et al. 2005).
Changes of the chromatin structure in IL-4 gene caused by TH2 cell differentiation
We investigated the chromatin structure of IL-4 using nuclease digestion. The DNase I and MNase digestion profiles around the 5'-end of IL-4 in resting TH1 and TH2 cells are shown in Fig. 3c. Although naked human DNA from Jurkat T cells shows some faint sites of preferential sequence-specific digestion by MNase, they are at different positions to the cuts observed in chromatin (Fig. 3b). DNase I digestion revealed no DNase I hypersensitive sites here in TH1 cells, but there are six closely spaced cuts over 1.4 kb in the 5'-end of IL-4 in TH2 cells, which resemble a MNase digestion profile (Fig. 3c). Indeed, MNase digestion shows a very similar profile to DNase I in TH2 cells, except that it is better resolved. The bands are labelled alphabetically to aid clarity during discussion.
Analysis of chromatin structure in IL-4 by MNase and DNase I. (a) Map of IL-4 showing the positions of the BclI sites and the radioactively labelled probe used. (b) Autoradiograph of Southern blot of MNase I-digested naked human (Jurkat) DNA used as a control to detect sequence-specific digestion, cut with BclI and hybridized to the same probe as used in (C). (c) Autoradiograph of Southern blot of BclI-digested MNase and DNase I digestion series of TH1 and TH2 cells. DNA marker sizes are shown in kilobases. Bands/peaks have been designated c to j; asterisks indicate linkers protected from MNase digestion. (d) Ethidium bromide-stained agarose gel that was blotted and hybridized in (C). Note that similar amounts of DNA are loaded and that the extents of digestion are equivalent
Cuts are seen at six out of eight intervening linkers between nine positioned nucleosomes (Fig. 3c): one nucleosome in the proximal promoter, one at the start of transcription and five extending into the second intron of the gene. The difference in the DNase I digestion profiles of TH1 and TH2 cells is not caused by different extents of digestion or different amounts of total DNA in the two samples (see Fig. 3d), but by different extents of digestion of that particular gene in TH1 versus TH2 cells. These data show that the chromatin in this region has a more open structure in TH2 cells. Although the same membrane was used for hybridization with probes that were specific for different regions of that gene cluster and MNase digestion showed positioned nucleosomes elsewhere in both TH1 and TH2 cells, this unusual profile was observed only between the second intron and the proximal promoter of IL-4 and thus it does not reflect an experimental artefact (for those in IL-13; see Fig. S1, supplementary material online).
The densitometric traces of the autoradiographs of the MNase digests (Fig. 3c) were expanded into the sum of Gaussian functions (Fig. 4a) as described in Staynov (2000). The traces from both TH1 and TH2 cells are very similar. In both cell types, the MNase cut that would have produced band f is prevented; the linker between bands h and j is also protected (asterisks in Figs 3b, 4a). These linkers are probably protected by a trans-acting factor(s) that is present in both TH1 and TH2 quiescent cells. Band f, however, is not protected from MNase digestion in human primary skin fibroblasts, which also have similarly positioned nucleosomes (Fig. S2, supplementary material online).
Analysis of MNase digestion patterns. (a) Expansion of MNase digestion profiles shown in Fig. 3c (thick lines) into the sum of Gaussian curves (narrow lines). Band designations c to j are shown above the curves. CpG sites and their positions relative to the bands are shown as solid circles, with their methylation status indicated by the scale to the right of the graph. (b) Linear regression analysis of bands produced by MNase digestion of TH1 and TH2 chromatin. Band designations c to j are shown above or below the corresponding points. (c) Linear regression analysis of the standard deviation of Gaussian curves resolved from digitized traces of MNase-generated bands
In TH1 cells, all the bands can be expanded as single Gaussian functions. However, band e in the TH2 cell digest is clearly seen in Fig. 3c as a doublet, while band d shows a trailing smear. In Fig. 4a they both appear as asymmetrical unresolved doublets that can be resolved into two Gaussian functions, each with minor components of 40-50 bp to the right and to the left of the main bands, respectively. These doublets do not result from contamination by a small fraction of chromatin containing an alternatively positioned nucleosome, as this would produce minor bands shifted in the same direction. Most likely, the nucleosome located at the transcriptional start site between bands e and f has two internal sites, about 40-50 bp from the dyad axis, which are accessible to MNase. This suggests that it is a modified nucleosome.
The positions of the cuts from the BclI digestion site versus their numbers are plotted in Fig. 4b. Linear regression analysis shows a nucleosome repeat length of 201 base pairs. In TH2 cells, band j deviates from the regression line by about 60-70 bp, which suggests that the adjacent nucleosome is shifted.
The standard deviations (Δ) of the Gaussian curves can give an idea of the length of the exposed DNA regions, since after cutting, MNase digests exonucleolytically along the naked DNA. However, several factors contribute to the increase in the standard deviation of the Gaussian functions (e.g. the strength, thickness and length of the gel, the type of radioactive label and the film used) (Staynov and Crane-Robinson 1988; Staynov and Proykova 1998). Therefore, Δ indicates only an upper limit of the actual window of accessibility to the enzyme. Because the electrophoretic mobility is not a linear function of the size of a fragment, widening of the bands increases with the molecular weight of the DNA. This can be expressed as a power function of the molecular weight, which in native agarose gels is very close to a linear function, but is slightly different for each gel. Thus, if Δ plotted against the molecular weight of consecutive cuts produces a smooth line, the sizes of the exposed DNA regions must be identical. The standard deviation Δ of the band of smallest molecular weight gives an upper limit to the actual size of the exposed DNA.
Linear regression analysis of the standard deviations of the Gaussian functions in Fig. 4a is shown in Fig. 4c. Most bands have the same widths in both TH1 and TH2 cells and fit a straight line. The standard deviation of the first band corresponds to approximately 40 bp, which is in good agreement with the expected sizes of DNA linkers in chromatin with a repeat length of about 200 bp. Band g is wider than the others in both TH1 and TH2 populations (Fig. 4c, Δ = 70-90 bp); band j is wider only in TH2 cells (Δ = 50-60 bp). The upper limits of the widths of bands g and j are too small to reflect missing nucleosomes. Most probably, they reflect a shift of the nucleosomes along the adjacent linkers and partial overlapping with the nucleosomes, which have one of their linkers protected. However, we cannot rule out the possibility that the widening of band j in the TH2 cells is also caused by minor cuts within the adjacent nucleosome (like those between bands d and e). The resolution of the gel beyond band e is insufficient for more accurate analysis. The size of this ‘open’ structure of about 1.4 kb is a conservative estimate.
Discussion
TH1 and TH2 nuclei were digested with MNase and DNase I in parallel and the full demethylation of several particular CpG dinucleotides was taken as the criterion that both alleles are in the same structural/functional state. After 28 days of differentiation of primary human TH cells towards the TH2 phenotype, individual CpG sites in IL-4 and IL-13 became 100% demethylated. This suggested that both IL-4 and IL-13 alleles were in the same functional environment and that they most likely have identical chromatin structures.
DNase I revealed six hypersensitive sites, which appeared only in TH2 cells, in a 1.4 kb region around the first two introns of the IL-4 gene. This area of interest extends from around the start of transcription approximately 1.4 kb into the IL-4 gene. The digestion patterns have shown that the cuts by both DNase I and MNase are at almost identical sites and revealed nine positioned nucleosomes with six out of the eight intervening linkers exposed to both enzymes. These nucleosomes are positioned similarly in TH1 cells, but their linkers are not accessible to DNase I. There are no missing nucleosomes and this suggests a major perturbation of the chromatin structure above the level of the nucleosome in TH2 cells. Moreover, this structure occurs in resting cells and therefore does not reflect active transcription. We do not observe this unusual structure at other gene loci such as IL-13, where we see some disruption of the regular nucleosomal array in TH2 cells when analysed by MNase, but no DNase I sensitivity (see Fig. S1). Hence, we report a single-copy gene in higher eukaryotes in which the linkers of a set of consecutive nucleosomes are exposed to DNase I under physiological conditions. This region corresponds to that in the second intron of the mouse IL-4 gene where histone H3 AcK9/14 levels were found to be substantially increased in TH2 cells (Baguet and Bix 2004).
CpG methylation status
The almost full demethylation of several selected cytosines in both IL-4 and IL-13 after 28 days of TH2 differentiation contradicts the previously reported broad demethylation across this locus in the murine genes (Agarwal and Rao 1998) and strongly supports a non-random mechanism of demethylation.
The active model of demethylation proposes catalytic removal of the methyl group by enzymatic activity, such as that observed in the IL-2 promoter upon T cell activation (Bruniquel and Schwartz 2003) and in the RAD50 hypersensitive site 7 (RHS7) of the murine TH2 LCR (Kim et al. 2007). However, active demethylation clearly does not take place at these sites, as the demethylation occurs over many cell cycles. After 28 days of differentiation, 78% of the cells express IL-13 and the seven CpGs around the DHS at the transcription start site are 86% demethylated. One can speculate that a longer period of differentiation or sorting of the IL-13-expressing cells would show their full demethylation. Because their demethylation requires a large number of cell cycles, it is most likely passive. This is supported by the fact that all demethylated cytosines in the regions we examined are found in DNA that is accessible to DNase I, which suggests that the opening of the chromatin structure allows binding of specific trans-acting factors that in turn prevent de novo methylation. During passive demethylation, Dnmt1 recruitment is inhibited, most likely by steric hindrance of other DNA-binding factors (Wilson et al. 2005). By contrast, the CpGs that are inside nucleosomes or in internucleosomal DNA that is not accessible to DNase I are not protected from maintenance methylation after replication.
An alternative explanation is that the gene is relocated to a different nuclear domain (environment) where de novo methylation is not efficient (for reviews see Carmo-Fonseca (2002), Sexton et al. (2007)). Interestingly, some CpGs around DNase I cuts do remain methylated, such as the cluster around the IL-4 transcription start site.
Chromatin structure of the IL-4 gene
The MNase digestion profiles of IL-4 show that there are nine similarly positioned nucleosomes with six out of the eight intervening linkers cut in both TH1 and TH2 cells. One nucleosome is in the proximal promoter, one is at the transcription start site, and the following seven extend well into the second intron. The size of this ‘open’ structure of 1.4 kb, shown in the autoradiograph and the plot (Figs 3, 4) is a conservative estimate.
DNase I accesses these linkers in TH2 cells only. This shows that the IL-4 gene is in a fibre (or higher than fibre) structure in the TH1 cells but is in a more open structure in the TH2 cells. DNase I digests were carried out in presence of 6 mM MgCl2 and 1 mM CaCl2 and under these conditions the 30 nm chromatin fibre is very stable. Thus, an additional factor or factors must be responsible for the opening of the chromatin. At this stage we do not know the actual size of this ‘more open’ structure. If it is a long stretch of open chromatin, like the one illustrated in Fig. 5a, it may be caused by a lack of histone H1 and/or of core-histone modifications and nucleosome remodelling. The two additional cuts in the nucleosome at the start of IL-4 transcription might be caused by lack of histone H2a-H2b pairs as was reported for the nucleosome at the start of transcription of the mouse mammary tumour virus (Vicent et al. 2004). If the open structure consists only of these nine nucleosomes, it could be caused by binding of a bulky factor or a multiprotein complex, which causes a kink in the fibre (normally configured as in Fig. 5b), exposing several linkers on each side of the kink (Fig. 5c).
Schematic representations of possible chromatin conformations in the 5(-end of IL-4 in TH2 cells. Red octagons represent nucleosomal DNA wrapped around histone octamers (blue). The thick black lines represent the linker DNA. (a) Non-interacting nucleosomes in a ‘beads on a string’ conformation, with linkers exposed. (b) A 30 nm fibre with linkers criss-crossing inside and unavailable for DNase I digestion. (c) A fibre that has been distorted by a bulky protein complex, which causes a kink, exposing several linkers on each side of it
However, we cannot rule out an alternative explanation, namely that the difference between digestion patterns of the 14- and 28-day-differentiated cells does not reflect different proportions of cells with open and closed chromatin structure, but that chromatin unfolds gradually during many cell cycles. It might start with the appearance of the cut j (previously described as DHS I) around the regulatory element in the second intron, caused by the binding of a trans-acting factor(s), which prevents maintenance methylation of the corresponding CpGs during cell replications. With progression of their demethylation, the opening spreads upstream in the promoter. If this is the case, the order and the causal hierarchy of events should be: (i) local disruption of the chromatin structure caused by the binding of a trans-acting factor to a specific DNA sequence; (ii) prevention of the maintenance methylation by this factor and gradual increase of the number of demethylated CpGs in that cluster; (iii) binding of more trans-acting factors to the demethylated sequence; and (iv) spreading of the unfolded chromatin upstream of the original disruption. To answer this question, single-cell experiments would be required. The cut j, which is the strongest and was clearly observed after 14 days’ differentiation, has been reported in mouse mast and T-cells to be an enhancer (Lee et al. 2001).
A nucleosome ladder produced by DNase I is not observed in single-copy genes in higher eukaryotes. In our experiments DNase I also did not recognize positioned nucleosomes in other regions of this gene cluster in the TH2 cells (Fig. S1 and Santangelo 2002). Thus this ‘more open’ structure, which appears even in unactivated cells, is not a result of transcription and is a prerequisite for transcriptional competence. Its novel structure suggests that it contains an unidentified regulatory element in the human IL-4/IL-13 cluster. Its location corresponds with that of the intronic enhancer located within the second intron of murine IL-4: transgenic reporter assays showed that this region enhanced murine IL-4 promoter activity and, in combination with CNS-1/HSS, conferred GATA-3-dependent enhancement of IL-4 promoter activity (Lee et al. 2001). Further work with higher-resolution gels may provide better understanding of the structure of nucleosomes and their mutual interactions.
In conclusion, the accessibility of the linker DNA to DNase I can provide insight into the higher-order structure of particular genes. How this structure relates to the transcriptional competence or activity of a gene requires further investigation. It may provide additional information about the order of epigenetic events that lead to transcriptional competence of genes or gene clusters in the eukaryotic nucleus. In general, these results support the notion that most of the active genes are in a fibre or a modified fibre conformation that would be unfolded by the transcriptional machinery for transcription and that refolds immediately after transcription.
Abbreviations
- CpG:
-
cysteine-guanine dinucleotide
- DHS:
-
DNase I hypersensitive site
- DNase I:
-
deoxyribonuclease I
- FACS:
-
fluorescence activated cell sorting
- IL-4:
-
interleukin-4
- MNase:
-
micrococcal nuclease
- TAE:
-
Tris, acetate, EDTA buffer
- TH1 cells:
-
differentiated T-helper lymphocytes that express interferon gamma
- TH2 cells:
-
differentiated T-helper lymphocytes that express interleukins-4, -5 and -13
References
Agarwal S, Rao A (1998) Modulation of chromatin structure regulates cytokine gene expression during T cell differentiation. Immunity 9:765–775
Ansel KM, Djuretic I, Tanasa B, Rao A (2006) Regulation of Th2 differentiation and Il4 locus accessibility. Annu Rev Immunol 24:607–656
Ashe HL, Monks J, Wijgerde M, Fraser P, Proudfoot NJ (1997) Intergenic transcription and transinduction of the human beta-globin locus. Genes Dev 11:2494–2509
Avni O, Lee D, Macian F et al (2002) TH cell differentiation is accompanied by dynamic changes in histone acetylation of cytokine genes. Nat Immunol 3:643–651
Baguet A, Bix M (2004) Chromatin landscape dynamics of the Il4-Il13 locus during T helper 1 and 2 development. Proc Natl Acad Sci U S A 101:11410–11415
Bert AG, Johnson BV, Baxter EW, Cockerill PN (2007) A modular enhancer is differentially regulated by GATA and NFAT elements that direct different tissue-specific patterns of nucleosome positioning and inducible chromatin remodeling. Mol Cell Biol 27:2870–2885
Bix M, Wang ZE, Thiel B, Schork NJ, Locksley RM (1998) Genetic regulation of commitment to interleukin 4 production by a CD4+ T cell-intrinsic mechanism. J Exp Med 188:2289–2299
Boeger H, Griesenbeck J, Strattan JS, Kornberg RD (2003) Nucleosomes unfold completely at a transcriptionally active promoter. Mol Cell 11:1587–1598
Bruniquel D, Schwartz RH (2003) Selective, stable demethylation of the interleukin-2 gene enhances transcription by an active process. Nat Immunol 4:235–240
Bulger M, Groudine M (1999) Looping versus linking: toward a model for long-distance gene activation. Genes Dev 13:2465–2477
Carmo-Fonseca M (2002) The contribution of nuclear compartmentalization to gene regulation. Cell 108:513–521
Cirillo LA, Lin FR, Cuesta I et al (2002) Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell 9:279–289
Cousins DJ, Lee TH, Staynov DZ (2002) Cytokine coexpression during human Th1/Th2 cell differentiation: direct evidence for coordinated expression of Th2 cytokines. J Immunol 169:2498–2506
Elgin SCR, Workman JL (2000) Chromatin structure and gene expression. Oxford University Press, Oxford
Felsenfeld G, Groudine M (2003) Controlling the double helix. Nature 421:448–453
Fields PE, Lee GR, Kim ST, Bartsevich VV, Flavell RA (2004) Th2-specific chromatin remodeling and enhancer activity in the Th2 cytokine locus control region. Immunity 21:865–876
Gilbert N, Boyle S, Fiegler H et al (2004) Chromatin architecture of the human genome: gene-rich domains are enriched in open chromatin fibers. Cell 118:555–566
Gribnau J, Diderich K, Pruzina S, Calzolari R, Fraser P (2000) Intergenic transcription and developmental remodeling of chromatin subdomains in the human beta-globin locus. Mol Cell 5:377–386
Gross DS, Garrard WT (1988) Nuclease hypersensitive sites in chromatin. Annu Rev Biochem 57:159–197
Guo L, Hu-Li J, Paul WE (2005) Probabilistic regulation in TH2 cells accounts for monoallelic expression of IL-4 and IL-13. Immunity 23:89–99
Hebbes TR, Clayton AL, Thorne AW, Crane-Robinson C (1994) Core histone hyperacetylation co-maps with generalized DNase I sensitivity in the chicken beta-globin chromosomal domain. EMBO J 13:1823–1830
Higgs DR (1998) Do LCRs open chromatin domains? Cell 95:299–302
Kim ST, Fields PE, Flavell RA (2007) Demethylation of a specific hypersensitive site in the Th2 locus control region. Proc Natl Acad Sci U S A 104:17052–17057
Kimura T, Mills FC, Allan J, Gould H (1983) Selective unfolding of erythroid chromatin in the region of the active beta-globin gene. Nature 306:709–712
Kuo CT, Leiden JM (1999) Transcriptional regulation of T lymphocyte development and function. Annu Rev Immunol 17:149–187
Lee GR, Fields PE, Flavell RA (2001) Regulation of IL-4 gene expression by distal regulatory elements and GATA-3 at the chromatin level. Immunity 14:447–459
Meehan RR, Stancheva I (2001) DNA methylation and control of gene expression in vertebrate development. Essays Biochem 37:59–70
Ravindra A, Weiss K, Simpson RT (1999) High-resolution structural analysis of chromatin at specific loci: Saccharomyces cerevisiae silent mating-type locus HMRa. Mol Cell Biol 19:7944–7950
Reitman M, Felsenfeld G (1990) Developmental regulation of topoisomerase II sites and DNase I-hypersensitive sites in the chicken beta-globin locus. Mol Cell Biol 10:2774–2786
Riviere I, Sunshine MJ, Littman DR (1998) Regulation of IL-4 expression by activation of individual alleles. Immunity 9:217–228
Rogan DF, Cousins DJ, Staynov DZ (1999) Intergenic transcription occurs throughout the human IL-4/IL-13 gene cluster. Biochem Biophys Res Commun 255:556–561
Rogan DF, Cousins DJ, Santangelo S et al (2004) Analysis of intergenic transcription in the human IL-4/IL-13 gene cluster. Proc Natl Acad Sci U S A 101:2446–2451
Santangelo S (2002) Studies on the chromatin structure of the IL-4/IL-13 gene cluster in human T lymphocytes. PhD Thesis, University of London
Santangelo S, Cousins DJ, Winkelmann NE, Staynov DZ (2002) DNA methylation changes at human Th2 cytokine genes coincide with DNase I hypersensitive site formation during CD4+ T cell differentiation. J Immunol 169:1893–1903
Sapojnikova N, Thorne A, Myers F, Staynov D, Crane-Robinson C (2008) The chromatin of active genes is not in a permanently open conformation. J Mol Biol 386:290–299
Sexton T, Schober H, Fraser P, Gasser SM (2007) Gene regulation through nuclear organization. Nat Struct Mol Biol 14:1049–1055
Staynov DZ (1983) Possible nucleosome arrangements in the higher-order structure of chromatin. Int J Biol Macromol 5:3–9
Staynov DZ (2000) DNase I digestion reveals alternating asymmetrical protection of the nucleosome by the higher order chromatin structure. Nucleic Acids Res 28:3092–3099
Staynov DZ, Crane-Robinson C (1988) Footprinting of linker histones H5 and H1 on the nucleosome. EMBO J 7:3685–3691
Staynov DZ, Proykova YG (1998) Quantitative analysis of DNase I digestion patterns of oligo- and polynucleosomes. J Mol Biol 279:59–71
Staynov DZ, Proykova YG (2007) Topological constraints on the possible structures of the 30 nm chromatin fibre. Chromosoma 117:67–76
Trojer P, Reinberg D (2007) Facultative heterochromatin: is there a distinctive molecular signature? Mol Cell 28:1–13
van Holde KE (1988) Chromatin. Springer-Verlag, New York
Vicent GP, Nacht AS, Smith CL et al (2004) DNA instructed displacement of histones H2A and H2B at an inducible promoter. Mol Cell 16:439–452
Wallrath L, Elgin SCR (1995) Position effect variegation in Drosophila is associated with an altered chromatin structure. Genes Dev 9:1263–1277
Wang X, Simpson RT (2001) Chromatin structure mapping in Saccharomyces cerevisiae in vivo with DNase I. Nucleic Acids Res 29:1943–1950
Weintraub H, Groudine M (1976) Chromosomal subunits in active genes have an altered conformation. Science 193:848–856
Wilson CB, Makar KW, Shnyreva M, Fitzpatrick DR (2005) DNA methylation and the expanding epigenetics of T cell lineage commitment. Semin Immunol 17:105–119
Wolffe AP (1998) Chromatin : structure and function. Academic Press, San Diego, London
Woodcock CL, Dimitrov S (2001) Higher-order structure of chromatin and chromosomes. Curr Opin Genet Dev 11:130–135
Workman JL, Kingston RE (1998) Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem 67:545–579
Acknowledgements
This paper is dedicated to the memory of Nicole Winkelmann, a promising young scientist who died aged 25. The work was supported by a MRC grant number G9536930.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Wendy Bickmore.
Nicole Winkelmann passed away in 2004.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1
Analysis of chromatin structure in IL-13 by MNase and DNase I. (A) Map of IL-13 and upstream CpG-rich region showing the positions of the BclI sites and the radioactively labelled probe used. (B) Autoradiograph of Southern blot of BclI-digested MNase and DNase I digestion series of TH1 and TH2 cells. DNA marker sizes are shown in kilobases. The position of the TH2-specific DHS III is shown. The pattern of bands produced by MNase in lanes 11 and 20 were digitized and are shown as densitometric traces to the right of the MNase digestion panels (GIF 139 kb)
Fig. S2
Analysis of chromatin structure in IL-4 by MNase. Autoradiograph of Southern blot of BclI-digested MNase digestion series of fibroblasts. DNA marker sizes are shown in kilobases. The pattern of bands produced by MNase in lane 4 was digitized and is shown as a densitometric trace to the right of the digestion panel (GIF 120 kb)
Rights and permissions
About this article
Cite this article
Santangelo, S., Cousins, D.J., Triantaphyllopoulos, K. et al. Chromatin structure and DNA methylation of the IL-4 gene in human TH2 cells. Chromosome Res 17, 485–496 (2009). https://doi.org/10.1007/s10577-009-9040-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10577-009-9040-3