Abstract
The association between mutations of key driver genes and colorectal cancer (CRC) metastasis has been investigated by many studies. However, the results of these studies have been contradictory. Here, we perform a comprehensive analysis to screen key driver genes from the TCGA database and validate the roles of these mutations in CRC metastasis. Using bioinformatics analysis, we identified six key driver genes, namely APC, KRAS, BRAF, PIK3CA, SMAD4 and p53. Through a systematic search, 120 articles published by November 30, 2017, were included, which all showed roles for these gene mutations in CRC metastasis. A meta-analysis showed that KRAS mutations (combined OR 1.18, 95% CI 1.05–1.33) and p53 mutations (combined OR 1.49, 95% CI 1.23–1.80) were associated with CRC metastasis, including lymphatic and distant metastases. Moreover, CRC patients with a KRAS mutation (combined OR 1.29, 95% CI 1.13–1.47), p53 mutation (combined OR 1.35, 95% CI 1.06–1.72) or SMAD4 mutation (combined OR 2.04, 95% CI 1.41–2.95) were at a higher risk of distant metastasis. Subgroup analysis stratified by ethnic populations indicated that the BRAF mutation was related to CRC metastasis (combined OR 1.42, 95% CI 1.18–1.71) and distant metastasis (combined OR 1.51, 95% CI 1.20–1.91) in an Asian population. No significant association was found between mutations of APC or PIK3CA and CRC metastasis. In conclusion, mutations of KRAS, p53, SMAD4 and BRAF play significant roles in CRC metastasis and may be both potential biomarkers of CRC metastasis as well as therapeutic targets.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Colorectal cancer (CRC) remains one of the leading types of malignancy and causes of deaths worldwide [1]. Despite early detection and therapeutic advances, metastasis, including lymphatic and distant metastases, remains the major cause of death in newly diagnosed CRC patients, and the overall survival of advanced CRC remains unsatisfactory.
Multiple alternative genetic pathways exist in CRC development. APC, as a tumour suppressor gene, plays a principal role in CRC development. Contributing to tumour progression, deletion of the APC protein triggers accumulation of beta-catenin and transcriptional activation of TCF-responsive genes [2]. Additionally, KRAS and BRAF encode a protein that belongs to the Ras-Raf-MEK-ERK signalling pathway. Activation of this pathway is considered to be a molecular switch that leads to cellular growth and proliferation [3]. Occurring late in the pathogenesis of CRC, loss of p53 function is proposed to be one of the major events in the development of CRC [4].
Additionally, as CRC evolves from benign to malignant lesions, some key driver genes acquire a series of mutations over time [5]. Among these key driver genes is APC, and its mutation regulates growth advantages in epithelial cells and results in the formation of a small adenoma. Subsequently, KRAS and BRAF mutations provide a second round of expansion for cells, involving transformation to a large adenoma. Eventually, mutations of PIK3CA, SMAD4 and p53 generate a malignant tumour that has potential for invasion and metastasis. In this genetic model of CRC, mutations of APC, KRAS and BRAF are significant early events in the transition from normal colonic epithelium to adenoma. In addition, mutations of PIK3CA, SMAD4 and p53 are related to the late stage, the transition from adenoma to carcinoma. However, whether early or late genetic events contribute to CRC metastasis remains unknown. It also remains unclear which mutations of these key driver genes are involved in the metastasis of CRC.
Thus far, many studies have estimated the association between mutations of the above-mentioned key driver genes and CRC metastasis, but the results of these studies have been contradictory. For example, the study performed by Josephine Mun-Yee Ko et al. showed that no essential correlations were found between KRAS mutations and Dukes staging, metastasis or recurrence [6]. By contrast, Bazan et al. reported that KRAS mutations were significantly associated with advanced Dukes staging and lymphatic metastasis [7]. Consequently, in the present study, we screened several promising key driver genes by analysing the CRC mutation rank combined with a literature search and investigated the association between mutations of these key driver genes and CRC metastasis. This study may provide further insight into more effective preventive strategies and adjuvant therapies against CRC.
2 Methods
2.1 Gene screening, literature search and study selection
We defined key driver genes in CRC progression and metastasis using the mutation frequency and number of publications. To determine the most well-studied gene mutations in CRC, literature search was applied. Specifically, the search query “(‘colon cancer’ or ‘colon carcinoma’ or ‘colon tumour’ or ‘colon neoplasm’ or ‘rectal cancer’ or ‘rectal carcinoma’ or ‘rectal tumour’ or ‘rectal neoplasm’ or ‘colorectal cancer’ or ‘colorectal carcinoma’ or ‘colorectal tumour’ or ‘colorectal neoplasm’ or ‘CRC’) AND (‘mutation’ or ‘mutations’) AND (‘metastasis’)” was used to retrieve available studies from PubMed that were published by November 30, 2017. The R package, pubmed.mineR, was used to fetch the genes (HGNC proved Symbol) from the abstracts of the articles and tally the number of articles in which a gene was discussed. Furthermore, the most frequently mutated genes in the TCGA Colorectal Adenocarcinoma cohort were retrieved from cBioPortal (TCGA Colorectal Adenocarcinoma datasets, http://www.cbioportal.org/study? id=coadread_tcga# mutations).
To validate the role of mutations of the screened genes in CRC metastasis, we performed systematic literature searches of the PubMed, Embase, Cochrane Library and TCGA databases of articles published by November 30, 2017. The corresponding search term combinations were (‘colon cancer’ or ‘colon carcinoma’ or ‘colon tumour’ or ‘colon neoplasm’ or ‘rectal cancer’ or ‘rectal carcinoma’ or ‘rectal tumour’ or ‘rectal neoplasm’ or ‘colorectal cancer’ or ‘colorectal carcinoma’ or ‘colorectal tumour’ or ‘colorectal neoplasm’ or ‘CRC’) AND (‘mutation’ or ‘mutations’) AND (‘APC’ or ‘p53’ or ‘SMAD4’ or ‘KRAS’ or ‘BRAF’ or ‘PIK3CA’). After excluding potential duplicates, we subsequently manually reviewed the titles, abstracts and full papers for potential eligible studies.
The studies were selected if they met the predefined selection criteria: (1) the outcome of interest was metastasis of CRC, (2) study of interest was mutations of key driver genes, (3) OR estimates with 95% CIs were available and (4) studies published in English or Chinese were included. Of note, only the most recent or detailed information was extracted from overlapping publications.
2.2 Data extraction
Two authors independently extracted all of the datasets from the selected studies, and any disagreements were resolved by consensus or by consulting with a third reviewer. The following information was collected from each study: first author, publication, year of publication and title, study design and participants’ characteristics, including number, age, gender, race and metastasis status as well as tumour stage. The principal author or corresponding author was contacted to acquire additional or unavailable information. The Newcastle-Ottawa Scale was used to estimate the methodological and reporting quality of the included studies ranked in order of scores (8 to 9 points represented high quality; 5 to 7 points represented medium quality; fewer than 5 points represented low quality) [8].
2.3 Statistical analysis
All analyses were conducted using the R software (version 3.4.1). The I2 test was applied to assess the heterogeneity of each study [9]. Displayed in the forest plot, pooled ORs and 95% CIs were calculated by applying a random-effects model for the study with substantial heterogeneity (I2 > 50%); otherwise, a fixed-effects model was applied (I2 < 50%). We conducted primary meta-analyses to investigate the association between mutations of key driver genes and CRC metastasis and further performed subgroup analyses according to ethnic populations. Funnel plots as well as Begg’s and Egger’s tests were performed to identify the risk of publication bias.
3 Results
3.1 Screening analysis and description of the included studies
The key driver genes were identified by combining the literature search results with the mutation rank of the TCGA Colorectal Adenocarcinoma cohort as well as biological relevance. The results of the literature search and mutation rank (top 100 genes) are given in Table S1. As shown in Fig. 1a, the intersection of the two lists resulted in a list of 11 genes, including APC, KRAS, BRAF, PIK3CA, SMAD4, p53, NRAS, FBXW7, PDGFRA, ARID1A and BRCA2. Among these genes, APC, KRAS, PIK3CA and p53 were well studied and the most frequently mutated in CRC. Given that BRAF regulates the Ras-Raf-MEK-ERK pathway and is associated with a poor prognosis in many CRC studies, it was selected as one of the key driver genes, although its mutation rate is slightly lower than those of the other genes. SMAD4 was also selected because of its significant role as a critical regulator of the TGF-beta pathway in the metastasis process. Consequently, APC, KRAS, BRAF, PIK3CA, SMAD4 and p53 were selected for further investigation and comprehensive analysis.
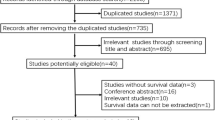
The process of gene screening, literature search and study selection. a Bubble diagram of gene screening. Blue represents an oncogene, orange represents a tumour suppressor gene and grey represents an unselected gene. b The flow chart for the study selection. This figure provides detailed information for study inclusion and exclusion
Additionally, a total of 12,822 records were retrieved from the PubMed, Embase, Cochrane Library and TCGA databases. After exclusion of duplicated and irrelevant articles by data analysis and title and abstract scanning, 173 citations remained for the full-text review. Finally, a total of 120 relevant articles were included in the analysis, with 20 studies for APC, 76 studies for KRAS, 52 studies for BRAF, 24 studies for PIK3CA, 9 studies for SMAD4 and 21 studies for p53, and some studies reported mutations of two or more genes in the same article [3, 4, 6, 10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121,122,123,124,125]. The details for selection are presented in Fig. 1b. All of the included studies were from Asia, Europe and America. Among them, 16 studies enrolled only colon cancer patients and three studies included only rectal cancer patients, whereas the other studies included patients with both colon and rectal malignancy. Most of the studies investigated CRC patients at all TNM stages (I–IV), whereas three studies enrolled only stage I–III patients and another four studies reported II–IV stage patients. The quality of the analysed studies was generally moderate to good.
3.2 Association between mutations of driver genes and CRC metastasis
A total of 39,313 patients were evaluated for the association between these key driver gene mutations and CRC metastasis. There was a significant statistical association between KRAS mutations and CRC metastasis, including lymphatic and distant metastases (combined OR 1.18, 95% CI 1.05–1.33; Fig. 2). Additionally, p53 mutations were also associated with metastasis in CRC patients (combined OR 1.49, 95% CI 1.24–1.80; Fig. 4a).
Moreover, KRAS mutations were significantly related to CRC distant metastasis in 61 studies that included 24,262 patients (combined OR 1.29, 95% CI 1.13–1.47; Fig. 3). Similarly, p53 mutations resulted in a higher risk of distant metastasis (combined OR 1.35, 95% CI 1.06–1.72; Fig. 4b). Furthermore, there was a trend that SMAD4 mutations correlated with distant metastasis (combined OR 2.04, 95% CI 1.41–2.95; Fig. 5b). Of note, no significant association was found between mutations of APC, BRAF, and PIK3CA and CRC metastasis (Figs. S1, S2, S3 and S4).
Forest plots of p53. a Forest plots of the association between p53 mutations and CRC metastasis, including lymphatic and distant metastases. p53 mutations were associated with metastasis in CRC patients. b Forest plots of the association between p53 mutations and CRC distant metastasis. p53 mutations yielded a higher risk of distant metastasis
3.3 Subgroup analyses according to population
Because there was a significant heterogeneity in some gene mutations, subgroup analyses stratified by ethnic populations were further performed for KRAS, BRAF and p53 (Tables 1 and 2). KRAS mutations (combined OR 1.12, 95% CI 1.03–1.22), BRAF mutations (combined OR 1.42, 95% CI 1.18–1.71) and p53 mutations (combined OR 1.55, 95% CI 1.16–2.06) were all significantly related to CRC metastasis, including lymphatic and distant metastases in an Asian population; however, p53 mutations (combined OR 1.44, 95% CI 1.11–1.85) were also associated with CRC metastasis in European and American populations.
In addition, significant associations between gene mutations and CRC distant metastasis were observed in an Asian population for BRAF (combined OR 1.51, 95% CI 1.20–1.91). Furthermore, p53 mutations (combined OR 1.72, 95% CI 1.23–2.40) were correlated with CRC distant metastasis in European and American populations. Interestingly, the CRC distant metastasis risk was enhanced by KRAS mutations both in an Asian population (combined OR 1.23, 95% CI 1.11–1.37) or European and American populations (combined OR 1.35, 95% CI 1.10–1.65). However, CRC metastasis was not influenced by other gene mutations in different ethnic populations according to the present results.
3.4 Publication bias
Funnel plots are displayed in Fig. S4, and the results of Begg’s test are shown in Table S2. No publication bias was found for the included studies for the key driver gene mutations.
4 Discussion
It is well established that many mutations of oncogenes and tumour suppressor genes are involved in the process of tissue transformation from normal epithelial cells to carcinomas in CRC [29]. Indeed, the accumulation of molecular alterations of these key driver genes plays a crucial role in the tumourigenesis and progression of CRC. However, it remains controversial whether mutations of these key driver genes can influence CRC metastasis.
In the present study, we performed a comprehensive analysis to screen several promising key driver genes, APC, KRAS, BRAF, PIK3CA, SMAD4 and p53, and to evaluate the association between mutations of these key driver genes and CRC metastasis (Fig. 6). The results indicated that the frequencies of KRAS as well as p53 mutations were significantly enhanced in patients with CRC metastasis. In addition, SMAD4 mutations were associated with CRC distant metastasis. Of note, the results of the subgroup analysis stratified by ethnic populations indicated that KRAS, BRAF and p53 mutations were associated with CRC metastasis, including lymphatic and distant metastases in an Asian population, whereas only p53 mutations promoted CRC metastasis, including lymphatic and distant metastases, in European and American populations. In addition, the CRC distant metastasis risk was facilitated by KRAS and BRAF mutations in an Asian population and by KRAS and p53 mutations in European and American populations.
Summary of our study for the association between key driver gene mutations and CRC metastasis. KRAS mutations and p53 mutations were associated with CRC metastasis, including lymphatic and distant metastases in Asian, European and American populations. Moreover, CRC patients with KRAS mutations, p53 mutations or SMAD4 mutations were at a higher risk of distant metastasis in Asian, European and American populations. No significant associations were found between mutations of APC, BRAF, PIK3CA and CRC metastasis in Asian, European and American populations. However, KRAS, BRAF and p53 mutations were associated with CRC metastasis, including lymphatic and distant metastases, in an Asian population, whereas only p53 mutations promoted CRC metastasis, including lymphatic and distant metastases, in European and American populations. In addition, the CRC distant metastasis risk was facilitated by KRAS and BRAF mutations in an Asian population and by KRAS and p53 mutations in European and American populations
KRAS mutations occur early during the progression from colorectal adenoma to malignant carcinoma, accumulating a sequential tumour growth advantage [97]. Large cohort studies, such as the collaborative RASCAL study, indicated that KRAS mutations increased the risk of recurrence and death [64]. Of note, the Ras-Raf-MEK-ERK and PI3K/AKT pathways are strongly interconnected, forming a signalling network. Activation of the different partners in this network by mutation deregulates survival, mobility and proliferation of cells [91]. In the present study, we found that KRAS mutations had a significant impact on both lymphatic metastasis and distant metastasis.
It is well known that p53 mutations are relatively late events of CRC progression. p53, as a tumour suppressor gene, causes apoptosis, releasing oncogenic stress and terminating DNA damage [86]. By repressing epithelial marker expression and increasing mesenchymal marker expression, the absence of p53 led to EMT transition and provided potential for the invasion and metastasis of CRC cells [126]. In the present study, p53 mutations were also observed to have significant associations with both lymphatic metastasis and distant metastasis in CRC.
TGF-beta signalling regulates tumourigenesis and tumour progression and is also involved in metastasis, particularly as an important EMT inducer [123]. As the pivotal factor of the TGF-beta pathway, in the present study, SMAD4 mutations tend to promote distant metastasis rather than lymphatic metastasis of CRC, because of different molecular mechanism between lymphatic and distant metastases or lymphatic and distant metastases originated from independent subclones in colorectal cancer [127]. In addition, as a downstream molecule and mutual exclusion mutation of KRAS, BRAF mutations were only shown to promote CRC metastasis including lymphatic or distant metastases in an Asian population, but not in European and American populations, in the current study. The reason for this difference is that the mutation rate of BRAF in the Asian population included in our study was 5.7%, which was consistent with a previous report that found a mutation rate of 5.0–10.0% for BRAF mutations in CRC patients [128]. Nevertheless, the BRAF mutation rate in the European and American populations included in the current study reached 14.8%, which was higher than a previous study [20].
Our study comprehensively analysed the association between mutations of several key driver genes and CRC metastasis, and the results suggested that some CRC metastasis-related driver genes, such as KRAS and p53, may be more important for clinical practice and diagnosis of CRC than other metastasis-related driver genes. However, the current analysis has several limitations. First, subgroup analysis for certain genes, such as SMAD4, could not be performed due to a lack of sufficient studies. Second, the heterogeneity of the studies for some gene analyses could not be fully explained. In the current study, ethnic populations were focused on to explain the heterogeneity. However, the effects of genetic factors on metastasis risk may be confounded by gender, age and mutational site. Therefore, further individual participant data analysis is warranted to avoid these confounding factors.
In conclusion, the results from the current study demonstrated that mutations of key driver genes, particularly KRAS and p53, promote CRC metastasis. This study highlights that KRAS, p53, SMAD4 and BRAF are potential markers to estimate prognosis, including the status of metastasis for patients with CRC. Future studies are needed to confirm the mechanisms and effects and to provide deeper insight into the role of mutations of these key driver genes in CRC metastasis.
References
Siegel, R. L., Miller, K. D., & Jemal, A. (2017). Cancer statistics, 2017. CA: a Cancer Journal for Clinicians, 67(1), 7–30.
Dow, L. E., O’Rourke, K. P., Simon, J., Tschaharganeh, D. F., van Es, J. H., Clevers, H., & Lowe, S. W. (2015). Apc restoration promotes cellular differentiation and reestablishes crypt homeostasis in colorectal cancer. Cell, 161(7), 1539–1552.
Eklöf, V., Wikberg, M. L., Edin, S., Dahlin, A. M., Jonsson, B.-A., Öberg, Å., Rutegård, J., & Palmqvist, R. (2013). The prognostic role of KRAS, BRAF, PIK3CA and PTEN in colorectal cancer. British Journal of Cancer, 108(10), 2153–2163.
Rechsteiner, M., Von Teichman, A., Rüschoff, J. H., Fankhauser, N., Pestalozzi, B., Schraml, P., Weber, A., Wild, P., Zimmermann, D., & Moch, H. (2013). KRAS, BRAF, and TP53 deep sequencing for colorectal carcinoma patient diagnostics. The Journal of Molecular Diagnostics, 15(3), 299–311.
Vogelstein, B., Papadopoulos, N., Velculescu, V. E., Zhou, S., Diaz, L. A., & Kinzler, K. W. (2013). Cancer genome landscapes. Science (New York, N.Y.), 339, (6127), 1546–1558.
Ko, J. M.-Y., Cheung, M. H.-Y., Wong, C.-M., Lau, K.-W., Tang, C. M.-C., Kwan, M. W., & Lung, M. L. (1998). Ki-ras codon 12 point mutational activation in Hong Kong colorectal carcinoma patients. Cancer Letters, 134(2), 169–176.
Bazan, V., Migliavacca, M., Zanna, I., Tubiolo, C., Corsale, S., Calò, V., Russo, A., et al. (2002). DNA ploidy and S-phase fraction, but not p53 or NM23-H1 expression, predict outcome in colorectal cancer patients. Result of a 5-year prospective study. Journal of Cancer Research and Clinical Oncology, 128(12), 650–658.
Stang, A. (2010). Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. European Journal of Epidemiology, 25(9), 603–605.
Higgins, J. P. T., & Thompson, S. G. (2002). Quantifying heterogeneity in a meta-analysis. Statistics in Medicine, 21(11), 1539–1558.
Sameer, A. S., Shah, Z. A., Abdullah, S., Chowdri, N. A., & Siddiqi, M. A. (2011). Analysis of molecular aberrations of Wnt pathway gladiators in colorectal cancer in the Kashmiri population. Human Genomics, 5(5), 441–452.
Lin, J.-K., Lin, P.-C., Lin, C.-H., Jiang, J.-K., Yang, S.-H., Liang, W.-Y., Chang, S.-C., et al. (2014). Clinical relevance of alterations in quantity and quality of plasma DNA in colorectal cancer patients: based on the mutation spectra detected in primary tumors. Annals of Surgical Oncology, 21(4), 680–686.
Syngal, S., Stoffel, E., Chung, D., Willett, C., Schoetz, D., Schroy, P., Ross, M., et al. (2006). Detection of stool DNA mutations before and after treatment of colorectal neoplasia. Cancer, 106(2), 277–283.
Frattini, M., Balestra, D., Suardi, S., Oggionni, M., Alberici, P., Radice, P., Pierotti, M. A., et al. (2004). Different genetic features associated with colon and rectal carcinogenesis. Clinical Cancer Research, 10(12), 4015–4021.
Calistri, D., Rengucci, C., Bocchini, R., Saragoni, L., Zoli, W., & Amadori, D. (2003). Fecal multiple molecular tests to detect colorectal cancer in stool. Clinical Gastroenterology and Hepatology, 1(5), 377–383.
Ko, J. M.-Y., Cheung, M. H.-Y., Kwan, M.-W., Wong, C.-M., Lau, K.-W., Tang, C. M.-C., & Lung, M. L. (1999). Genomic instability and alterations in Apc, Mcc and Dcc in Hong Kong patients with colorectal carcinoma. International Journal of Cancer, 84(4), 404–409.
Chiang, J.-M., Chou, Y.-H. W., Ma, S.-C., & Chen, J.-R. (2004). Influence of age on adenomatous polyposis coli and p53 mutation frequency in sporadic colorectal cancer—rarity of co-occurrence of mutations in APC, K-ras, and p53 genes. Virchows Archiv, 445(5), 465–471.
Liu, X., Shan, X., Friedl, W., Uhlhaas, S., Propping, P., Li, J., & Wang, Y. (2007). May the APC gene somatic mutations in tumor tissues influence the clinical features of Chinese sporadic colorectal cancers? Acta Oncologica, 46(6), 757–762.
Sánchez-de-Abajo, A., de la Hoya, M., van Puijenbroek, M., Tosar, A., López-Asenjo, J. A., Díaz-Rubio, E., Caldes, T., et al. (2007). Molecular analysis of colorectal cancer tumors from patients with mismatch repair-proficient hereditary nonpolyposis colorectal cancer suggests novel carcinogenic pathways. Clinical Cancer Research, 13(19), 5729–5735.
Vasovcak, P., Pavlikova, K., Sedlacek, Z., Skapa, P., Kouda, M., Hoch, J., & Krepelova, A. (2011). Molecular genetic analysis of 103 sporadic colorectal tumours in Czech patients. PLoS One, 6(8), e24114.
Al-Shamsi, H. O., Jones, J., Fahmawi, Y., Dahbour, I., Tabash, A., Abdel-Wahab, R., Wolff, R. A., et al. (2016). Molecular spectrum of KRAS, NRAS, BRAF, PIK3CA, TP53, and APC somatic gene mutations in Arab patients with colorectal cancer: determination of frequency and distribution pattern. Journal of Gastrointestinal Oncology, 7(6), 882–902.
Russo, A. L., Borger, D. R., Szymonifka, J., Ryan, D. P., Wo, J. Y., Blaszkowsky, L. S., Hong, T. S., et al. (2014). Mutational analysis and clinical correlation of metastatic colorectal cancer. Cancer, 120(10), 1482–1490.
Chang, Y. C., Chang, J.-G., Liu, T.-C., Lin, C.-Y., Yang, S.-F., Ho, C.-M., Chang, Y.-S., et al. (2016). Mutation analysis of 13 driver genes of colorectal cancer-related pathways in Taiwanese patients. World Journal of Gastroenterology, 22(7), 2314–2325.
Lüchtenborg, M., Weijenberg, M. P., Wark, P. A., Saritas, A. M., Roemen, G. M., van Muijen, G. N., de Goeij, A. F., et al. (2005). Mutations in APC, CTNNB1 and K-ras genes and expression of hMLH1 in sporadic colorectal carcinomas from the Netherlands Cohort Study. BMC Cancer, 5, 160.
Leslie, A., Pratt, N. R., Gillespie, K., Sales, M., Kernohan, N. M., Smith, G., Steele, R. J. C., et al. (2003). Mutations of APC, K-ras, and p53 are associated with specific chromosomal aberrations in colorectal adenocarcinomas. Cancer Research, 63(15), 4656–4661.
Chen, T.-H., Chang, S.-W., Huang, C.-C., Wang, K.-L., Yeh, K.-T., Liu, C.-N., Cheng, Y.-W., et al. (2013). The prognostic significance of APC gene mutation and miR-21 expression in advanced-stage colorectal cancer. Colorectal Disease, 15(11), 1367–1374.
Jorissen, R. N., Christie, M., Mouradov, D., Sakthianandeswaren, A., Li, S., Love, C., Sieber, O. M., et al. (2015). Wild-type APC predicts poor prognosis in microsatellite-stable proximal colon cancer. British Journal of Cancer, 113(6), 979–988.
Chang, P.-Y., Chen, J.-S., Chang, S.-C., Wang, M.-C., Chang, N.-C., Wen, Y.-H., Lu, J.-J., et al. (2017). Acquired somatic TP53 or PIK3CA mutations are potential predictors of when polyps evolve into colorectal cancer. Oncotarget, 8(42), 72352–72362.
Jauhri, M., Bhatnagar, A., Gupta, S., BP, M., Minhas, S., Shokeen, Y., & Aggarwal, S. (2017). Prevalence and coexistence of KRAS, BRAF, PIK3CA, NRAS, TP53, and APC mutations in Indian colorectal cancer patients: next-generation sequencing–based cohort study. Tumor Biology, 39(2), 1–11.
Neumann, J., Wehweck, L., Maatz, S., Engel, J., Kirchner, T., & Jung, A. (2013). Alterations in the EGFR pathway coincide in colorectal cancer and impact on prognosis. Virchows Archiv, 463(4), 509–523.
Feng, Q., Liang, L., Ren, L., Chen, J., Wei, Y., Chang, W., Xu, J., et al. (2015). A specific KRAS codon 13 mutation is an independent predictor for colorectal cancer metachronous distant metastases. American Journal of Cancer Research, 5(2), 674–688.
Siraj, A. K., Bu, R., Prabhakaran, S., Bavi, P., Beg, S., Al Hazmi, M., Al-Kuraya, K. S., et al. (2014). A very low incidence of BRAF mutations in Middle Eastern colorectal carcinoma. Molecular Cancer, 13, 168.
Pai, R. K., Jayachandran, P., Koong, A. C., Chang, D. T., Kwok, S., Ma, L., Pai, R. K., et al. (2012). BRAF-mutated, microsatellite-stable adenocarcinoma of the proximal colon: an aggressive adenocarcinoma with poor survival, mucinous differentiation, and adverse morphologic features. The American Journal of Surgical Pathology, 36(5), 744–752.
Landau, M. S., Kuan, S.-F., Chiosea, S., & Pai, R. K. (2014). BRAF-mutated microsatellite stable colorectal carcinoma: an aggressive adenocarcinoma with reduced CDX2 and increased cytokeratin 7 immunohistochemical expression. Human Pathology, 45(8), 1704–1712.
Tanaka, H., Deng, G., Matsuzaki, K., Kakar, S., Kim, G. E., Miura, S., Kim, Y. S., et al. (2006). BRAF mutation, CpG island methylator phenotype and microsatellite instability occur more frequently and concordantly in mucinous than non-mucinous colorectal cancer. International Journal of Cancer, 118(11), 2765–2771.
Yaeger, R., Cercek, A., Chou, J. F., Sylvester, B. E., Kemeny, N. E., Hechtman, J. F., Saltz, L. B., et al. (2014). BRAF mutation predicts for poor outcomes after metastasectomy in patients with metastatic colorectal cancer. Cancer, 120(15), 2316–2324.
Kalady, M. F., DeJulius, K. L., Sanchez, J. A., Jarrar, A., Liu, X., Manilich, E., Church, J. M., et al. (2012). BRAF mutations in colorectal cancer are associated with distinct clinical characteristics and worse prognosis. Diseases of the Colon & Rectum, 55(2), 128–133.
Nam, S. K., Yun, S., Koh, J., Kwak, Y., Seo, A. N., Park, K. U., Lee, H. S., et al. (2016). BRAF, PIK3CA, and HER2 oncogenic alterations according to KRAS mutation status in advanced colorectal cancers with distant metastasis. PLoS One, 11(3), e0151865.
Boulagnon, C., Dudez, O., Beaudoux, O., Dalstein, V., Kianmanesh, R., Bouché, O., & Diebold, M.-D. (2016). BRAFV600E gene mutation in colonic adenocarcinomas. Immunohistochemical detection using tissue microarray and clinicopathologic characteristics: an 86 case series. Applied Immunohistochemistry & Molecular Morphology, 24(2), 88–96.
Chen, J., Guo, F., Shi, X., Zhang, L., Zhang, A., Jin, H., & He, Y. (2014). BRAF V600E mutation and KRAS codon 13 mutations predict poor survival in Chinese colorectal cancer patients. BMC Cancer, 14, 802.
Aldiab, A., Khayal, K. A. A., Obaid, O. A. A., Alsheikh, A., Alsaleh, K., Shahid, M., & Alkharji, H. (2017). Clinicopathological features and predictive factors for colorectal cancer outcome in the Kingdom of Saudi Arabia. Oncology, 92(2), 75–86.
Hanna, M. C., Go, C., Roden, C., Jones, R. T., Pochanard, P., Javed, A. Y., MacConaill, L. E., et al. (2013). Colorectal cancers from distinct ancestral populations show variations in BRAF mutation frequency. PLoS One, 8(9), e74950.
Amaki-Takao, M., Yamaguchi, T., Natsume, S., Iijima, T., Wakaume, R., Takahashi, K., Miyaki, M., et al. (2016). Colorectal cancer with BRAF D594G mutation is not associated with microsatellite instability or poor prognosis. Oncology, 91(3), 162–170.
Li, W., Qiu, T., Zhi, W., Shi, S., Zou, S., Ling, Y., Lu, N., et al. (2015). Colorectal carcinomas with KRAS codon 12 mutation are associated with more advanced tumor stages. BMC Cancer, 15, 340.
Seppälä, T. T., Böhm, J. P., Friman, M., Lahtinen, L., Väyrynen, V. M. J., Liipo, T. K. E., Mecklin, J.-P., et al. (2015). Combination of microsatellite instability and BRAF mutation status for subtyping colorectal cancer. British Journal of Cancer, 112(12), 1966–1975.
Korphaisarn, K., Pongpaibul, A., Limwongse, C., Roothumnong, E., Klaisuban, W., Nimmannit, A., Akewanlop, C., et al. (2015). Deficient DNA mismatch repair is associated with favorable prognosis in Thai patients with sporadic colorectal cancer. World Journal of Gastroenterology: WJG, 21(3), 926–934.
Shen, Y., Wang, J., Han, X., Yang, H., Wang, S., Lin, D., & Shi, Y. (2013). Effectors of epidermal growth factor receptor pathway: the genetic profiling of KRAS, BRAF, PIK3CA, NRAS mutations in colorectal cancer characteristics and personalized medicine. PLoS One, 8(12).
Tsai, J.-H., Liau, J.-Y., Lin, Y.-L., Tseng, L.-H., Lin, L.-I., Yeh, K.-H., & Jeng, Y.-M. (2016). Frequent BRAF mutation in early-onset colorectal cancer in Taiwan: association with distinct clinicopathological and molecular features and poor clinical outcome. Journal of Clinical Pathology, 69(4), 319–325.
Jeantet, M., Tougeron, D., Tachon, G., Cortes, U., Archambaut, C., Fromont, G., & Karayan-Tapon, L. (2016). High intra- and inter-tumoral heterogeneity of RAS mutations in colorectal cancer. International Journal of Molecular Sciences, 17(12), 2015.
Hang, J.-F., Li, A. F.-Y., Chang, S.-C., & Liang, W.-Y. (2016). Immunohistochemical detection of the BRAF V600E mutant protein in colorectal cancers in Taiwan is highly concordant with the molecular test. Histopathology, 69(1), 54–62.
Ye, J.-X., Liu, Y., Qin, Y., Zhong, H.-H., Yi, W.-N., & Shi, X.-Y. (2015). KRAS and BRAF gene mutations and DNA mismatch repair status in Chinese colorectal carcinoma patients. World Journal of Gastroenterology: WJG, 21(5), 1595–1605.
Martinetti, D., Costanzo, R., Kadare, S., Alimehmeti, M., Colarossi, C., Canzonieri, V., Memeo, L., et al. (2014). KRAS and BRAF mutational status in colon cancer from Albanian patients. Diagnostic Pathology, 9, 187.
Bond, C. E., Bettington, M. L., Pearson, S.-A., McKeone, D. M., Leggett, B. A., & Whitehall, V. L. (2015). Methylation and expression of the tumour suppressor, PRDM5, in colorectal cancer and polyp subgroups. BMC Cancer, 15, 20.
Birgisson, H., Edlund, K., Wallin, U., Påhlman, L., Kultima, H. G., Mayrhofer, M., Sundström, M., et al. (2015). Microsatellite instability and mutations in BRAF and KRAS are significant predictors of disseminated disease in colon cancer. BMC Cancer, 15, 125.
Bond, C. E., Nancarrow, D. J., Wockner, L. F., Wallace, L., Montgomery, G. W., Leggett, B. A., & Whitehall, V. L. J. (2014). Microsatellite stable colorectal cancers stratified by the BRAF V600E mutation show distinct patterns of chromosomal instability. PLoS One, 9(3), e91739.
Sylvester, B. E., Huo, D., Khramtsov, A., Zhang, J., Smalling, R. V., Olugbile, S., Olopade, O. I., et al. (2012). Molecular analysis of colorectal tumors within a diverse patient cohort at a single institution. Clinical Cancer Research, 18(2), 350–359.
Zhang, J., Zheng, J., Yang, Y., Lu, J., Gao, J., Lu, T., Liu, T., et al. (2015). Molecular spectrum of KRAS, NRAS, BRAF and PIK3CA mutations in Chinese colorectal cancer patients: analysis of 1,110 cases. Scientific Reports, 5, 18678.
Liou, J.-M., Wu, M.-S., Shun, C.-T., Chiu, H.-M., Chen, M.-J., Chen, C.-C., Liang, J.-T., et al. (2011). Mutations in BRAF correlate with poor survival of colorectal cancers in Chinese population. International Journal of Colorectal Disease, 26(11), 1387–1395.
Gao, J., Sun, Z., Li, Y., & Shen, L. (2012). Mutations of KRAS and BRAF in Chinese patients with colorectal carcinoma: analyses of 966 cases. Chinese Journal of Pathology, 41(9), 579–583.
Zhu, K., Yan, H., Wang, R., Zhu, H., Meng, X., Xu, X., Chen, D., et al. (2014). Mutations of KRAS and PIK3CA as independent predictors of distant metastases in colorectal cancer. Medical Oncology (Northwood, London, England), 31(7), 16.
Palomba, G., Doneddu, V., Cossu, A., Paliogiannis, P., Manca, A., Casula, M., Palmieri, G., et al. (2016). Prognostic impact of KRAS, NRAS, BRAF, and PIK3CA mutations in primary colorectal carcinomas: a population-based study. Journal of Translational Medicine, 14, 292.
Nakanishi, R., Harada, J., Tuul, M., Zhao, Y., Ando, K., Saeki, H., Maehara, Y., et al. (2013). Prognostic relevance of KRAS and BRAF mutations in Japanese patients with colorectal cancer. International Journal of Clinical Oncology, 18(6), 1042–1048.
Nakaji, Y., Oki, E., Nakanishi, R., Ando, K., Sugiyama, M., Nakashima, Y., Maehara, Y., et al. (2017). Prognostic value of BRAF V600E mutation and microsatellite instability in Japanese patients with sporadic colorectal cancer. Journal of Cancer Research and Clinical Oncology, 143(1), 151–160.
Wangefjord, S., Sundström, M., Zendehrokh, N., Lindquist, K. E., Nodin, B., Jirström, K., & Eberhard, J. (2013). Sex differences in the prognostic significance of KRAS codons 12 and 13, and BRAF mutations in colorectal cancer: a cohort study. Biology of Sex Differences, 4, 17.
Ahn, T. S., Jeong, D., Son, M. W., Jung, H., Park, S., Kim, H., Baek, M.-J., et al. (2014). The BRAF mutation is associated with the prognosis in colorectal cancer. Journal of Cancer Research and Clinical Oncology, 140(11), 1863–1871.
Jang, M. H., Kim, S., Hwang, D. Y., Kim, W. Y., Lim, S. D., Kim, W. S., Han, H. S., et al. (2017). BRAF-mutated colorectal cancer exhibits distinct clinicopathological features from wild-type BRAF-expressing cancer independent of the microsatellite instability status. Journal of Korean Medical Science, 32(1), 38–46.
Yalcin, S., & Onguru, O. (2017). BRAF mutation in colorectal carcinomas with signet ring cell component. Cancer Biology & Medicine, 14(3), 287–292.
Takane, K., Akagi, K., Fukuyo, M., Yagi, K., Takayama, T., & Kaneda, A. (2017). DNA methylation epigenotype and clinical features of NRAS-mutation(+) colorectal cancer. Cancer Medicine, 6(5), 1023–1035.
Alvarez, K., Orellana, P., Villarroel, C., Contreras, L., Kawachi, H., Kobayashi, M., López-Köstner, F., et al. (2017). EGFR pathway subgroups in Chilean colorectal cancer patients, detected by mutational and expression profiles, associated to different clinicopathological features. Tumor Biology, 39(9), 1–12.
Mariani, S., Bertero, L., Osella-Abate, S., Di Bello, C., Francia di Celle, P., Coppola, V., Marchiò, C., et al. (2017). Extreme assay sensitivity in molecular diagnostics further unveils intratumour heterogeneity in metastatic colorectal cancer as well as artifactual low-frequency mutations in the KRAS gene. British Journal of Cancer, 117(3), 358–366.
Hao, Y.-X., Li, Y.-M., Ye, M., Guo, Y.-Y., Li, Q.-W., Peng, X.-M., Xiao, W.-H., et al. (2017). KRAS and BRAF mutations in serum exosomes from patients with colorectal cancer in a Chinese population. Oncology Letters, 13(5), 3608–3616.
Le Balc’h, E., Grandin, N., Demattei, M.-V., Guyétant, S., Tallet, A., Pagès, J.-C., Charbonneau, M., et al. (2017). Measurement of telomere length in colorectal cancers for improved molecular diagnosis. International Journal of Molecular Sciences, 18(9).
Fujiyoshi, K., Yamamoto, G., Takenoya, T., Takahashi, A., Arai, Y., Yamada, M., Akagi, K., et al. (2017). Metastatic pattern of stage IV colorectal cancer with high-frequency microsatellite instability as a prognostic factor. Anticancer Research, 37(1), 239–247.
Lee, C.-T., Huang, Y.-C., Hung, L.-Y., Chow, N.-H., Su, P.-F., Ho, C.-L., Lee, J.-C., et al. (2017). Serrated adenocarcinoma morphology in colorectal mucinous adenocarcinoma is associated with improved patient survival. Oncotarget, 8(21), 35165–35175.
Won, D. D., Lee, J. I., Lee, I. K., Oh, S.-T., Jung, E. S., & Lee, S. H. (2017). The prognostic significance of KRAS and BRAF mutation status in Korean colorectal cancer patients. BMC Cancer, 17(1), 403.
Imamura, Y., Lochhead, P., Yamauchi, M., Kuchiba, A., Qian, Z. R., Liao, X., Ogino, S., et al. (2014). Analyses of clinicopathological, molecular, and prognostic associations of KRAS codon 61 and codon 146 mutations in colorectal cancer: cohort study and literature review. Molecular Cancer, 13, 135.
Dolatkhah, R., Somi, M. H., Asvadi Kermani, I., Bonyadi, M., Sepehri, B., Boostani, K., Dastgiri, S., et al. (2016). Association between proto-oncogene mutations and clinicopathologic characteristics and overall survival in colorectal cancer in East Azerbaijan, Iran. OncoTargets and Therapy, 9, 7385–7395.
Kodaz, H., Hacibekiroglu, I., Erdogan, B., Turkmen, E., Tozkir, H., Albayrak, D., Cicin, I., et al. (2015). Association between specific KRAS mutations and the clinicopathological characteristics of colorectal tumors. Molecular and Clinical Oncology, 3(1), 179–184.
Hu, J., Yan, W.-Y., Xie, L., Cheng, L., Yang, M., Li, L., Qian, X.-P., et al. (2016). Coexistence of MSI with KRAS mutation is associated with worse prognosis in colorectal cancer. Medicine, 95(50), e5649.
Geido, E., Sciutto, A., Rubagotti, A., Oliani, C., Monaco, R., Risio, M., & Giaretti, W. (2002). Combined DNA flow cytometry and sorting with k-ras2 mutation spectrum analysis and the prognosis of human sporadic colorectal cancer. Cytometry, 50(4), 216–224.
Pu, X., Pan, Z., Huang, Y., Tian, Y., Guo, H., Wu, L., Lin, T., et al. (2013). Comparison of KRAS/BRAF mutations between primary tumors and serum in colorectal cancer: biological and clinical implications. Oncology Letters, 5(1), 249–254.
Kuo, Y.-B., Chen, J.-S., Fan, C.-W., Li, Y.-S., & Chan, E.-C. (2014). Comparison of KRAS mutation analysis of primary tumors and matched circulating cell-free DNA in plasmas of patients with colorectal cancer. Clinica Chimica Acta, 433, 284–289.
Wang, Q., Zhong, M., Lü, Y., Yuan, J., & Wei, L. (2012). Correlation of KRAS gene mutations and clinicopathologic parameters in colorectal carcinoma. Chinese Journal of Pathology, 41(9), 603–606.
Xu, C., Liu, Y., Huang, J., He, D., Hou, Y., Ji, Y., Tan, Y., et al. (2012). Detection of KRAS gene mutation and its clinical significance in colorectal adenocarcinoma. Chinese Journal of Pathology, 41(10), 667–670.
Hosoya, K., Matsusaka, S., Kashiwada, T., Suzuki, K., Ureshino, N., Sato, A., Sueoka-Aragane, N., et al. (2017). Detection of KRAS mutations in plasma DNA using a fully automated rapid detection system in colorectal cancer patients. Pathology & Oncology Research, 23(4), 737–744.
Schweiger, T., Hegedüs, B., Nikolowsky, C., Hegedüs, Z., Szirtes, I., Mair, R., Hoetzenecker, K., et al. (2014). EGFR, BRAF and KRAS status in patients undergoing pulmonary metastasectomy from primary colorectal carcinoma: a prospective follow-up study. Annals of Surgical Oncology, 21(3), 946–954.
Losi, L., Ponz de Leon, M., Jiricny, J., Di Gregorio, C., Benatti, P., Percesepe, A., Benhattar, J., et al. (1997). K-ras and p53 mutations in hereditary non-polyposis colorectal cancers. International Journal of Cancer, 74(1), 94–96.
Ruiz-Candelaria, Y., Miranda-Diaz, C., & Hunter-Mellado, R. F. (2013). K-RAS mutation profile in Puerto Rican patients with colorectal cancer: trends from April 2009 to January 2011. The International Journal of Biological Markers, 28(4), e393–e397.
Cejas, P., López-Gómez, M., Aguayo, C., Madero, R., de C. Carpeño, J., Belda-Iniesta, C., Feliu, J., et al. (2009). KRAS mutations in primary colorectal cancer tumors and related metastases: a potential role in prediction of lung metastasis. PLoS One, 4(12), e8199.
Garrido-Laguna, I., Hong, D. S., Janku, F., Nguyen, L. M., Falchook, G. S., Fu, S., Kurzrock, R., et al. (2012). KRASness and PIK3CAness in patients with advanced colorectal cancer: outcome after treatment with early-phase trials with targeted pathway inhibitors. PLoS One, 7(5), e38033.
Calistri, D., Rengucci, C., Seymour, I., Lattuneddu, A., Polifemo, A. M., Monti, F., Amadori, D., et al. (2005). Mutation analysis of p53, K-ras, and BRAF genes in colorectal cancer progression. Journal of Cellular Physiology, 204(2), 484–488.
Barault, L., Veyrie, N., Jooste, V., Lecorre, D., Chapusot, C., Ferraz, J.-M., Piard, F., et al. (2008). Mutations in the RAS-MAPK, PI(3)K (phosphatidylinositol-3-OH kinase) signaling network correlate with poor survival in a population-based series of colon cancers. International Journal of Cancer, 122(10), 2255–2259.
Rao, B., Gao, Y., Huang, J., Gao, X., Fu, X., Huang, M., Wang, J., et al. (2011). Mutations of p53 and K-ras correlate TF expression in human colorectal carcinomas: TF downregulation as a marker of poor prognosis. International Journal of Colorectal Disease, 26(5), 593.
Chang, Y.-Y., Lin, P.-C., Lin, H.-H., Lin, J.-K., Chen, W.-S., Jiang, J.-K., Chang, S.-C., et al. (2016). Mutation spectra of RAS gene family in colorectal cancer. The American Journal of Surgery, 212, (3), 537–544.e3.
Corso, G., Pascale, V., Flauti, G., Ferrara, F., Marrelli, D., & Roviello, F. (2013). Oncogenic mutations and microsatellite instability phenotype predict specific anatomical subsite in colorectal cancer patients. European Journal of Human Genetics, 21(12), 1383–1388.
Tortola, S., Marcuello, E., González, I., Reyes, G., Arribas, R., Aiza, G., Capella, G., et al. (1999). p53 and K-ras gene mutations correlate with tumor aggressiveness but are not of routine prognostic value in colorectal cancer. Journal of Clinical Oncology, 17(5), 1375–1375.
Kato, S., Iida, S., Higuchi, T., Ishikawa, T., Takagi, Y., Yasuno, M., Sugihara, K., et al. (2007). PIK3CA mutation is predictive of poor survival in patients with colorectal cancer. International Journal of Cancer, 121(8), 1771–1778.
Palomba, G., Colombino, M., Contu, A., Massidda, B., Baldino, G., Pazzola, A., Cossu, A., et al. (2012). Prevalence of KRAS, BRAF, and PIK3CA somatic mutations in patients with colorectal carcinoma may vary in the same population: clues from Sardinia. Journal of Translational Medicine, 10, 178.
Palomba, G., Cossu, A., Paliogiannis, P., Pazzola, A., Baldino, G., Scartozzi, M., Palmieri, G., et al. (2016). Prognostic role of KRAS mutations in Sardinian patients with colorectal carcinoma. Oncology Letters, 12(2), 1415–1421.
Inoue, Y., Saigusa, S., Iwata, T., Okugawa, Y., Toiyama, Y., Tanaka, K., Kusunoki, M., et al. (2012). The prognostic value of KRAS mutations in patients with colorectal cancer. Oncology Reports, 28(5), 1579–1584.
Hasegawa, S., Goto, S., Matsumoto, T., Hida, K., Kawada, K., Matsusue, R., Sakai, Y., et al. (2017). A multicenter phase 2 study on the feasibility and efficacy of neoadjuvant chemotherapy without radiotherapy for locally advanced rectal cancer. Annals of Surgical Oncology, 24(12), 3587–3595.
Thiebault, Q., Defossez, G., Karayan-Tapon, L., Ingrand, P., Silvain, C., & Tougeron, D. (2017). Analysis of factors influencing molecular testing at diagnostic of colorectal cancer. BMC Cancer, 17, 765.
Pang, X.-L., Li, Q.-X., Ma, Z.-P., Shi, Y., Ma, Y.-Q., Li, X.-X., Zhang, W., et al. (2017). Association between clinicopathological features and survival in patients with primary and paired metastatic colorectal cancer and KRAS mutation. OncoTargets and Therapy, 10, 2645–2654.
Ganesh, K., Shah, R. H., Vakiani, E., Nash, G. M., Skottowe, H. P., Yaeger, R., Stadler, Z. K., et al. (2017). Clinical and genetic determinants of ovarian metastases from colorectal cancer. Cancer, 123(7), 1134–1143.
Cho, A., Jo, K., Hwang, S. H., Lee, N., Jung, M., Yun, M., & Hwang, H. S. (2017). Correlation between KRAS mutation and 18F-FDG uptake in stage IV colorectal cancer. Abdominal Radiology, 42(6), 1621–1626.
Charlton, M. E., Karlitz, J. J., Schlichting, J. A., Chen, V. W., & Lynch, C. F. (2017). Factors associated with guideline-recommended KRAS testing in colorectal cancer patients: a population-based study. American Journal of Clinical Oncology, 40(5), 498–506.
Huang, S.-C., Huang, S.-F., Chen, Y.-T., Chang, Y., Chiu, Y.-T., Chang, I.-C., Chen, J.-S., et al. (2017). Overexpression of MutL homolog 1 and MutS homolog 2 proteins have reversed prognostic implications for stage I–II colon cancer patients. Biomedical Journal, 40(1), 39–48.
Jones, R. P., Sutton, P. A., Evans, J. P., Clifford, R., McAvoy, A., Lewis, J., Malik, H. Z., et al. (2017). Specific mutations in KRAS codon 12 are associated with worse overall survival in patients with advanced and recurrent colorectal cancer. British Journal of Cancer, 116(7), bjc201737.
Peng, J., Huang, D., Poston, G., Ma, X., Wang, R., Sheng, W., Cai, S., et al. (2017). The molecular heterogeneity of sporadic colorectal cancer with different tumor sites in Chinese patients. Oncotarget, 8(30), 49076–49083.
Godai, T., Suda, T., Sugano, N., Tsuchida, K., Shiozawa, M., Sekiguchi, H., Miyagi, Y., et al. (2009). Identification of colorectal cancer patients with tumors carrying the TP53 mutation on the codon 72 proline allele that benefited most from 5-fluorouracil (5-FU) based postoperative chemotherapy. BMC Cancer, 9, 420.
Ferraz, J.-M., Zinzindohoué, F., Lecomte, T., Cugnenc, P.-H., Loriot, M.-A., Beaune, P., Laurent-Puig, P., et al. (2004). Impact of GSTT1, GSTM1, GSTP1 and NAT2 genotypes on KRAS2 and TP53 gene mutations in colorectal cancer. International Journal of Cancer, 110(2), 183–187.
Rengucci, C., Maiolo, P., Saragoni, L., Zoli, W., Amadori, D., & Calistri, D. (2001). Multiple detection of genetic alterations in tumors and stool. Clinical Cancer Research, 7(3), 590–593.
Iniesta, P., Vega, F. J., Caldés, T., Massa, M.-J., de Juan, C., Cerdán, F. J., Benito, M., et al. (1998). p53 Exon 7 mutations as a predictor of poor prognosis in patients with colorectal cancer. Cancer Letters, 130(1–2), 153–160.
Zhao, Y., Oki, E., Ando, K., Morita, M., Kakeji, Y., & Maehara, Y. (2010). The impact of a high-frequency microsatellite instability phenotype on the tumor location-related genetic differences in colorectal cancer. Cancer Genetics and Cytogenetics, 196(2), 133–139.
Naguib, A., Cooke, J. C., Happerfield, L., Kerr, L., Gay, L. J., Luben, R. N., Arends, M. J., et al. (2011). Alterations in PTEN and PIK3CA in colorectal cancers in the EPIC Norfolk study: associations with clinicopathological and dietary factors. BMC Cancer, 11, 123.
Phipps, A. I., Makar, K. W., & Newcomb, P. A. (2013). Descriptive profile of PIK3CA-mutated colorectal cancer in postmenopausal women. International Journal of Colorectal Disease, 28(12).
Day, F. L., Jorissen, R. N., Lipton, L., Mouradov, D., Sakthianandeswaren, A., Christie, M., Sieber, O. M., et al. (2013). PIK3CA and PTEN gene and exon mutation-specific clinicopathologic and molecular associations in colorectal cancer. Clinical Cancer Research, 19(12), 3285–3296.
Iida, S., Kato, S., Ishiguro, M., Matsuyama, T., Ishikawa, T., Kobayashi, H., Sugihara, K., et al. (2012). PIK3CA mutation and methylation influences the outcome of colorectal cancer. Oncology Letters, 3(3), 565–570.
Nosho, K., Kawasaki, T., Ohnishi, M., Suemoto, Y., Kirkner, G. J., Zepf, D., Ogino, S., et al. (2008). PIK3CA mutation in colorectal cancer: relationship with genetic and epigenetic alterations. Neoplasia (New York, N.Y.), 10(6), 534–541.
Herreros-Villanueva, M., Gomez-Manero, N., Muñiz, P., García-Girón, C., & del Corral, M. J. C. (2011). PIK3CA mutations in KRAS and BRAF wild type colorectal cancer patients. A study of Spanish population. Molecular Biology Reports, 38(2), 1347–1351.
Ando, T., Sugai, T., Habano, W., Jiao, Y.-F., & Suzuki, K. (2005). Analysis of SMAD4/DPC4 gene alterations in multiploid colorectal carcinomas. Journal of Gastroenterology, 40(7), 708–715.
Miyaki, M., Iijima, T., Konishi, M., Sakai, K., Ishii, A., Yasuno, M., Hishima, T., Koike, M., Shitara, N., Iwama, T., Utsunomiya, J., et al. (1999). Higher frequency of Smad4 gene mutation in human colorectal cancer with distant metastasis. Oncogene, 18(2), 3098–3103.
Fleming, N. I., Jorissen, R. N., Mouradov, D., Christie, M., Sakthianandeswaren, A., Palmieri, M., Sieber, O. M., et al. (2013). SMAD2, SMAD3 and SMAD4 mutations in colorectal cancer. Cancer Research, 73(2), 725–735.
Sameer, A. S., Chowdri, N. A., Syeed, N., Banday, M. Z., Shah, Z. A., & Siddiqi, M. A. (2010). SMAD4—molecular gladiator of the TGF-β signaling is trampled upon by mutational insufficiency in colorectal carcinoma of Kashmiri population: an analysis with relation to KRAS proto-oncogene. BMC Cancer, 10, 300.
Sarshekeh, A. M., Advani, S., Overman, M. J., Manyam, G., Kee, B. K., Fogelman, D. R., Kopetz, S., et al. (2017). Association of SMAD4 mutation with patient demographics, tumor characteristics, and clinical outcomes in colorectal cancer. PLoS One, 12(3), e0173345.
Jia, X., Shanmugam, C., Paluri, R. K., Jhala, N. C., Behring, M. P., Katkoori, V. R., Manne, U., et al. (2017). Prognostic value of loss of heterozygosity and sub-cellular localization of SMAD4 varies with tumor stage in colorectal cancer. Oncotarget, 8(12), 20198–20212.
Sui, X., Zhu, J., Tang, H., Wang, C., Zhou, J., Han, W., He, C., et al. (2015). p53 controls colorectal cancer cell invasion by inhibiting the NF-κB-mediated activation of Fascin. Oncotarget, 6(26), 22869–22879.
Naxerova, K., Reiter, J. G., Brachtel, E., Lennerz, J. K., van de Wetering, M., Rowan, A., Jain, R. K., et al. (2017). Origins of lymphatic and distant metastases in human colorectal cancer. Science, 357(6346), 55–60.
Mao, C., Zhou, J., Yang, Z., Huang, Y., Wu, X., Shen, H., Chen, Q., et al. (2012). KRAS, BRAF and PIK3CA mutations and the loss of PTEN expression in Chinese patients with colorectal cancer. PLoS One, 7(5), e36653.
Acknowledgments
The authors thank Prof. Pengyuan Liu (Zhejiang University), an expert bio-statistician, for reviewing the analysis portion of the manuscript.
Funding
This work is supported by the National Natural Science Foundation of China (81672730 to H.Z. and 81572716 to M.L.), the Fundamental Research Funds for the Central Universities (2017FZA7005 to H.Z.), the Natural Science Foundation of Zhejiang Province (LY17H160025 to E.X.) and the Department of Science and Technology of Zhejiang Province (2016C33150 to E.X.)
Author information
Authors and Affiliations
Contributions
H.Z. and Y.W. contributed to the conception and design of the study. D.H., W.S. and Y.Z. contributed to the conception, design of the study and editing of the manuscript. D.H., Y.Z., P.L., F.C., H.C. and D.X. contributed to the statistical analysis. E.X. and M.L. contributed to the analysis and interpretation of data. All of the authors commented on drafts of the paper and approved the final draft of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare that they have no conflicts of interest.
Electronic supplementary material
Fig. S1
Forest plots of APC. (A) Forest plots of the association between APC mutations and CRC metastasis, including lymphatic and distant metastases. No association was found between APC mutations and CRC metastasis. (B) Forest plots of the association between APC mutations and CRC distant metastasis. No association was found between APC mutations and CRC distant metastasis. (GIF 168 kb)
Fig. S2
Forest plots of the association between BRAF mutations and CRC metastasis, including lymphatic and distant metastases. No association was found between BRAF mutations and CRC metastasis. (GIF 185 kb)
Fig. S3
Forest plots of the association between BRAF mutations and CRC distant metastasis. No association was found between BRAF mutations and CRC distant metastasis. (GIF 164 kb)
Fig. S4
Forest plots of PIK3CA. (A) Forest plots of the association between PIK3CA mutation and CRC metastasis, including lymphatic and distant metastases. No association was found between PIK3CA mutations and CRC metastasis. (B) Forest plots of the association between PIK3CA mutations and CRC distant metastasis. No association was found between PIK3CA mutations and CRC distant metastasis. (GIF 203 kb)
Fig. S5
Funnel plots for the relationship between 6 genes and CRC metastasis in the current study. (GIF 93 kb)
Table S1
The results of key driver gene screening. (XLSX 9 kb)
Table S2
The results of the Begg’s test for the included studies of each gene. (XLSX 12 kb)
Rights and permissions
About this article
Cite this article
Huang, D., Sun, W., Zhou, Y. et al. Mutations of key driver genes in colorectal cancer progression and metastasis. Cancer Metastasis Rev 37, 173–187 (2018). https://doi.org/10.1007/s10555-017-9726-5
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10555-017-9726-5