Abstract
Purpose
Breast cancer (BC) is a heterogeneous disorder, with variable response to systemic chemotherapy. Likewise, BC shows highly complex immune activation patterns, only in part reflecting classical histopathological subtyping. Schlafen-11 (SLFN11) is a nuclear protein we independently described as causal factor of sensitivity to DNA damaging agents (DDA) in cancer cell line models. SLFN11 has been reported as a predictive biomarker for DDA and PARP inhibitors in human neoplasms. SLFN11 has been implicated in several immune processes such as thymocyte maturation and antiviral response through the activation of interferon signaling pathway, suggesting its potential relevance as a link between immunity and cancer. In the present work, we investigated the transcriptional landscape of SLFN11, its potential prognostic value, and the clinico-pathological associations with its variability in BC.
Methods
We assessed SLFN11 determinants in a gene expression meta-set of 5061 breast cancer patients annotated with clinical data and multigene signatures.
Results
We found that 537 transcripts are highly correlated with SLFN11, identifying “immune response”, “lymphocyte activation”, and “T cell activation” as top Gene Ontology processes. We established a strong association of SLFN11 with stromal signatures of basal-like phenotype and response to chemotherapy in estrogen receptor negative (ER-) BC. We identified a distinct subgroup of patients, characterized by high SLFN11 levels, ER- status, basal-like phenotype, immune activation, and younger age. Finally, we observed an independent positive predictive role for SLFN11 in BC.
Conclusions
Our findings are suggestive of a relevant role for SLFN11 in BC and its immune and molecular variability.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Breast cancer (BC) is the second most common cancer in the world and, by far, the most frequent neoplasm among women [1].
BC is a clinically and molecularly heterogeneous disease and genomic microarray analyses have corroborated the presence of at least four distinct intrinsic molecular subtypes: luminal A, luminal B, basal-like, and HER2 enriched subsets [2, 3]. These subtypes display varying degrees of sensitivity to treatment and highlight the molecular heterogeneity of BC [4].
We and an independent group [5] recently discovered the role of a putative DNA/RNA helicase, Schlafen-11 (SLFN11), for its causal association with sensitivity to DNA damaging agents (DDA), such as platinum salts, topoisomerase I and II inhibitors, and other alkylators in the NCI-60 panel of cancer cell lines [6].
SLFN11 belongs to the Schlafen protein family, which has been implicated in the regulation of important mammalian biological functions, such as control of cell proliferation [7], induction of immune responses [8], and regulation of viral replication [9].
Schlafen genes were originally identified during screening for growth regulatory genes, and they are differentially expressed during lymphocyte development [10,11,12,13]. Later, SLFN11 was described as an early interferon response gene, in association with HIV infection [9]. Furthermore, Murai et al. described molecular mechanisms detailing how SLFN11 is a dominant determinant of sensitivity to DNA-targeted therapies [14]. In particular, SLFN11 inhibits checkpoint maintenance and homologous recombination by removing Replication Protein A from single-stranded DNA [15]. Tang et al. demonstrated that the use of histone deacetylase inhibitors can be used to sensitize SLFN11-inactivated cancers to DDA [16]. Recently, the importance of SLFN11 as a predictor of sensitivity to DDA has been proven in Ewing’s sarcomas, ovarian cancer, and colorectal cancer [17,18,19,20]. SLFN11 has also been confirmed as a predictive biomarker of PARP inhibitor sensitivity in small cell lung cancer [21].
The aims of our study were to investigate the transcriptional landscape of SLFN11 expression in invasive BC and to identify clinical and pathological parameters that could help explain SLFN11 modulation in BC. In addition, we set up to determine whether SLFN11 expression could be associated with prognosis or response to treatment in this neoplasm.
Materials and methods
Datasets retrieval, pre-processing, and data normalization
Thirty-five gene expression datasets of expression profiles from 7737 tumors were retrieved from public databases or authors’ websites [32 sets previously described in [22] and another three: PNC, METABRIC and TCGA [23,24,25]. Immune phenotypes for TCGA BC cases and leucocyte infiltration were obtained as described in Hendrickx et al. [26].
To ensure comparability of expression values across multiple datasets and microarray platforms (Agilent, Affymetrix or Illumina), we performed 0.95 quantile normalization (using the R/Bioconductor package genefu [27]).
SLFN 11 expression analysis and gene signature enrichment
Whole transcriptome correlation of SLFN11 was performed using Spearman’s rank correlation. We selected the top 5th percentile of transcripts that better correlated with SLFN11 expression. Functional annotation of correlators was further performed using DAVID (Database for Annotation, Visualization and Integrated Discovery) v6.7 [28] in order to identify significantly enriched pathways [false discovery rate (FDR) < 0.05], particularly Gene Ontology (GO) terms (The Gene Ontology Consortium). DAVID identifies GO categories to which genes belong, determining the statistical significance of non-random representation. To provide an independent assessment of enrichment analysis, we classified patients in molecular subtypes, extracting relative genomic signatures from the genefu package [27]. Patients labeled as “normal” PAM50 phenotype were removed, upon concerns of low cancer cellularity and possible ensuing contamination by normal breast tissue [29]. The most significant gene signatures were extracted using a feature selection machine learning approach, called LASSO regression (glmnet package).
Multiple correspondence analysis
We investigated the modulation of SLNF11 in breast cancer through the study of the mutual distribution of clinical and pathological categorical data. First, we removed T1a samples, due to their small relative number and size, Tx and Nx tumor patients and all those patients with unknown age information, estrogen receptor or HER2 status. For this analysis, SLFN11 expression was subdivided in tertiles of expression (low, intermediate, and high). Exploratory assessment and inter-dependencies relations of data, combined with the extracted gene signatures, were accomplished by multiple correspondence analysis with the FactoMineR package.
Survival analysis and time dependency correlation
Survival analyses were performed in order to determine the association of SLFN11 with prognosis in BC. We defined, by univariable statistical analysis, the association between disease-free survival (DFS) and SLFN11 expression (“low” if in the lower two tertiles and “high” if in the top tertile). The DFS curves were generated using Kaplan–Meier estimators (survcomp package), and p values were obtained with the log-rank test. For what concerns the analysis of more than one covariates, we employed a stepwise backward-forward Cox proportional hazards regression model. The Akaike Information Criterion allowed the estimation of the best set of clinical and pathological variables described above (MASS package).
To explore time dependency of SLFN11 modulation, we tested the proportional hazards assumption for a Cox regression model as described previously [30]. We tested a two-sided hypothesis, rejecting the null ones with a p < 0.05 and applied multiple corrections of resulting p values using the Benjamini–Hochberg method.
Results
SLFN11 expression correlates with BC immune-related transcripts
To investigate the transcriptional landscape of SLFN11 in BC, we conducted a gene expression microarray meta-analysis of 7737 cases from 35 publicly available datasets.
Of 7737 cases, we assessed 5061 patients with SLFN11 expression values. Then, we performed a whole transcriptome correlation analysis with SLFN11 and identified 537 genes in the top 5th percentile of correlation. The list of these 537 transcripts was analyzed for gene ontology (GO) enrichment. Strikingly, immune function processes represented most of the GO terms resulting from such analysis. The overrepresented terms in our sample set are listed in Table 1.
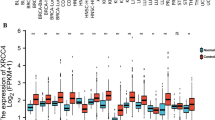
In agreement with such finding, we observed a strong positive association between well-established markers of tumor lymphocytic infiltration with SLFN11 expression such as CD3 and CD8 (Spearman’s ρ = 0.527, FDR < 0.0001 with the expression of CD3, and ρ = 0.514 with the expression of CD8—FDR < 0.0001, see Fig. 1).
Overall, this data purports an association of SLFN11 with immune modulation in BC.
SLFN11 expression correlates with BC immune gene signatures
Next, to validate our observations from an independent perspective, we inferred gene expression signatures from 4740 patients after removing the “normal-like” intrinsic phenotype cases (in light of their low cellularity) and exploited LASSO penalized regression to extract the most relevant signatures associated with SLFN11 expression. In harmony with our previous observations, we observed an independent, strict association with immune-related signatures, in particular with two publicly described signatures ‘Immune2’ [31] (ρ = 0.508, FDR < 0.0001) and ‘Stroma1’ [31] (ρ = 0.377, FDR < 0.0001, see Fig. 2).
a Bar plot shows the LASSO regression coefficient weights related to the gene signatures of interest: the highest weighted signatures are highlighted in a red contoured box. b The upper and lower scatterplots show the correlation between SLNF11 and the most relevant gene signatures resulting from previous variable selection analysis
High expression of SLNF11 is linked with aggressive BC
To better understand the role of SLFN11 in BC modulation, we performed multiple correspondence analysis (MCA) including clinical and pathological parameters, as well as SLFN11 levels ranked by tertiles of expression.
2581 patients from 7 datasets, presenting with all clinical and pathological features including ER and progesterone receptor immunohistochemistry, HER2 status, grade, T, N, intrinsic subtype, and STAT1 signature as a proxy for immune activation [32] were considered for such analysis.
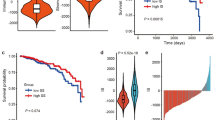
MCA highlighted two clearly separated patient clusters. The “SLFN11-hot” cluster is defined by high SLFN11 expression, ER-negative status, high histological grade, basal-like phenotype, immune activation, and younger age at diagnosis (< 50 years old).
The “SLFN11-cold” cluster is characterized by low/intermediate SLFN11 expression, ER-positive status, lack of HER2 amplification, older age at diagnosis (> 50 years old), and low/intermediate STAT1 expression (see Fig. 3).
MCA showing the relationship patterns between clinico-pathological variables and SLFN11 expression in breast cancer. x- and y-axes represent the first and second dimension (Dim.1 and Dim.2) of the MCA analysis performed on clinical and pathological data, as well as SLFN11 expression, divided in tertiles, from 2581 BC patients. Patients are represented by small grey dots and categorical variables are colored. In particular, patients with high-grade tumors also show high SLFN11 expression levels (highlighted by the red dashed circle), whereas the cluster of patients with SLFN11 low and intermediate expression tumors is also characterized by low and intermediate (Low/Intm) STAT1 expression, HER2-, ER+ cancers (steel-blue dashed circle)
In summary, high SLFN11 expression correlates with aggressive tumors with signs of immune activation (basal-like phenotype, higher histological grade, younger age), whereas lower SLFN11 expression can be observed in luminal, less aggressive neoplasms characterized by low immune activation.
SLFN11 overexpression is independently associated with better prognosis
To evaluate whether SLFN11 expression could be associated with prognosis or response to treatment in BC, we evaluated 2093 patients from 3 different datasets with complete information concerning DFS and type of treatment.
By univariable analysis, SLFN11 was not associated with prognosis (HR = 1.09 for SLFN11-high vs. low expression, 95% CI 0.88–1.36, p = 0.37).
On the other hand, when taking into account clinical and pathological parameters as well as type of treatment and intrinsic subtypes, SLFN11 high expression was independently associated with better prognosis (HR = 0.61, 95% CI 0.41–0.91, p = 0.0153). Moreover, we could define an interaction between SLFN11 expression and hormone treatment (HT), with high-SLFN11 patients undergoing HT being characterized by worse outcome (HR: 1.81, 95% CI 1.11–2.96, p value for interaction = 0.0175, Fig. 4, panel a).
a Forest plot of Cox regression model for DFS in 2093 BC patients with complete anatomopathological and clinical follow-up data. b Plot of scaled Schoenfeld residuals. Red dashed and blue dotted lines represent, respectively, the null effect (null log hazard ratio) and a ± 2-standard-error band around the fit. On the x-axis, time is expressed in years
To better understand this not obvious observation, we investigated SLFN11 expression and HT in relation with possible time dependencies violating the Cox proportional hazards assumption. Indeed, in our analysis high SLFN11 levels subtended a worse prognosis in the first 2 years after diagnosis only in patients undergoing HT (Fig. 4, panel b).
SLFN11 is independent from BC immune activation status in prognosis prediction
Finally, we derived leucocyte infiltration and immune phenotypes in the most extensively analyzed set available to us, TCGA, as previously described [26]. We could indeed confirm that SLFN11 expression is associated with leucocyte infiltration (Spearman’s ρ = 0.61, p < 0.0001, see Supplementary Figure S1). However, in a survival model taking into account the interaction of SLFN11 and the recently described BC “low” and “high” immunological constant of rejection (ICR) phenotypes (N = 318) [26], we could not find a significant interaction in determining prognosis between the two variables. Surprisingly, however, our model suggested that high SLFN11/high ICR cases may have a short-term worse prognosis than other cases (adjusted HR = 2.68, 95% CI 0.28–25.56, p = 0.1483, with a p value for violating the proportional hazards assumption = 0.1114).
Discussion
In the present article, we investigated for the first time how SLFN11 is modulated in BC, analyzing more of 7000 BC cases available from 35 public datasets. Our findings demonstrated a strong correlation of SLNF11 expression with immune system transcriptomic markers, in particular with transcripts involved in immune system processes such as “prolymphocyte activation”, “immune response”, and “T cell activation”. Our findings document the relationship between SLFN11 and immunity in BC, initially suggested by previous works in other settings [9, 33, 34]. In analogy with our findings, Stewart et al. recently published that SLFN11 high expression in small cell lung cancer is positively correlated with immune regulatory pathways, particularly with Type 1 interferon pathway genes [33]. Therefore, SLFN11 appears to have a significant role not only in innate immunity processes such as defense response to virus [9] or DNA damage repair [6], but also in adaptive immune response to cancer.
SLFN11 in addition to its known expression by cancer cells [6] could indeed be expressed by immune cells during anti-tumoral response, potentially behaving as a marker of T cell infiltration in BC as well as in other tumor types. The consistent association of SLNF11 with immunity is exemplified by its strong correlation with tumor infiltrating lymphocyte markers (CD3 and CD8 in our analyses).
Of note, we identified a strong independent correlation of SLFN11 with two immune gene signatures, namely stroma1 and immune2. In the last few years, several prognostic and/or predictive gene expression signatures have been published in BC [35,36,37]. Desmedt et al. in their comprehensive meta-analysis showed as several prognostic gene signatures differ in prognostic abilities according to the BC subtype and as only immune response modules seem to predict prognosis in ER-negative/HER2 negative BC patients [32]. On the other hand, we previously pointed out the prognostic and predictive value of immune gene signatures in primary TNBC underlining the activation of Th1/effector immune response [35]. Our findings show both high expression of SFLN11 in a subgroup of patients with TNBC-like features and a strong correlation with immune signatures, in particular immune2, supporting an involvement of SLFN11 during the effector immune response in BC. In parallel, stroma signatures have also been developed in BC in order to predict clinical outcome and treatment response [38,39,40]. Particularly, Finak et al. developed a 26 gene stroma-derived prognostic predictor in which a good-outcome cluster overexpresses a distinct set of immune-related genes, including T cell and NK cell markers indicative of a Th1-type immune response (GZMA, CD52, CD247, CD8A) [41]. Winslow et al. showed that a specific immune gene signature (C1Q), represented by genes such as DZMH, GZMA, GZMK, CD3D, CD3G, CD247, CD8A, coding for proteins involved in cytotoxic immune response in TNBC, is associated with low risk of recurrence. Finally, their results support that the molecular profile of a Th-1/immune response (CD4+ T cells) is an important prognostic marker in BC [39] as also hypothesized by Gu-Trantien et al. in her work [42]. In good agreement with such independent observations, in our study SFLN11 is highly associated with stromal signatures, in particular stroma1, and expression of T cell markers, supporting the idea of a role of SLFN11 in Th-1/effector immune response in BC.
Through our unbiased analysis of SLFN11 expression in relation with clinico-pathological BC variables, we discovered two distinct BC patient subgroups. In the “SLFN11-hot” cluster, we observed a high expression of the signature of STAT1, a key mediator of type I and type II interferon response. Among its many functions, STAT1 promotes Th1 immune response and TCD8+ cell recruitment [43]. This type of immune activation is predominant in TNBC, a subgroup of BC that is considered highly immunogenic. TNBC typically presents a worse prognosis than other BC subtypes, with—however—a very heterogeneous response to current systemic chemotherapies and absence of actionable molecular targets. To overcome this issue, current clinical trials testing a combination of immunotherapy and chemotherapy in TNBC are ongoing [44].
In our analysis, we demonstrated that SLFN11 expression is strictly related to BC-immunity, in particular in TNBC. The “SLFN11-hot” cluster encompasses a distinct BC subgroup with TNBC-like features, strong immune activation, better prognosis, and better response to systemic treatments compared to other BC subtypes. On the other hand, the “SLFN11-cold” cluster might represent a different subgroup of scarcely immunogenic BC with minor response to systemic treatment. Therefore, SLFN11 as immune-related biomarker is an intriguing venue for further translational research.
In our time dependency analysis, we identified a subgroup of high-SLFN11 BC patients treated with HT presenting with worse outcome in the first 2 years of follow-up. This behavior shows similarities with TNBC and suggests that the phenomenon that we observed might be actually due to a subset of hormone receptor-poor patients with a biological behavior analogous to that of TNBC. This is, however, just a hypothesis since we did not have the availability of ER expression level by immunohistochemistry in the evaluated dataset for a precise quantitation of ER by standard methods. Our observation is in agreement with recent literature, since several papers confirmed the analogies between TNBC and Luminal-B BC concerning survival rates [45], response to neoadjuvant chemotherapy [46], high mutational burden, and immunogenic profile characterized by higher expression of TIL [47]. Finally, Luminal-B BC are poorly responsive to HT [48] and could be stratified by immune profile analysis into different prognostic groups [49], so that in future studies on BC, we believe SLFN11 expression should be assessed together with other established parameters for prognostic and predictive purposes.
Our lack of identifying a clear association between SLFN11 levels and immune activation in BC in determining prognosis is somehow puzzling. We may speculate that SLFN11 levels in cancer cells play an independent role in response to DDAs when considered together with immune status in BC. As a consequence, we strongly advocate for future studies to morphologically deconvolute SLFN11 expression in cancer cells and in immune infiltrate in selected BC cohorts to reach a causal understanding of the role of this protein. On the other hand, our inconclusive results in assessing the relation of SLFN11 and immune activation in BC may be due to both the relatively low number of events (N = 42) and the insufficient length of the follow-up time (median 2.5 years) of publicly available BC TCGA data. Moreover, the suggestion of a worse short-term prognosis again favors the idea of high SLFN11 being a characteristic of BC cancer with such behavior, as TNBC is. The negative prognostic effect of SLFN11 in the high ICR BC cases is puzzling, and we should be careful in overinterpreting substantially indecisive results. Our analyses have several limitations. Among them the heterogeneity concerning the origin of data, chip design, and clinical annotation are unavoidable. Moreover, we did not perform preclinical experiments for our findings, which are of associative nature so far—albeit suggestive—, and SLFN11 location in BC is yet to be determined, since the contribution from infiltrating lymphocytes may be determinant in this regard.
Conclusion
In summary, a consistent and evident pattern emerges, highlighting the strong correlation of SLFN11 with the immune system in BC, as well as its meaningful associations with clearly distinct clinico-pathological BC phenotypes and clinical outcome. Further studies will have to focus on biological, well-annotated, and homogeneous specimens from clinical BC cohorts to further unravel SLFN11 role in BC.
Change history
12 July 2019
In the original publication of the article, the funding information was incorrectly published. The corrected funding statement is given in this correction article.
Abbreviations
- BC:
-
Breast cancer
- DDA:
-
DNA damaging agents
- DFS:
-
Disease-free survival
- ER:
-
Estrogen receptor
- HT:
-
Hormone treatment
- ICR:
-
Immunological constant of rejection
- MCA:
-
Multiple correspondence analysis
- SLFN11:
-
Schlafen-11
- TNBC:
-
Triple-negative breast cancer
References
Ferlay J, Soerjomataram I, Dikshit RES, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F (2015) Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 136:E359–E386. https://doi.org/10.1002/ijc.29210
Perou CM, Sørlie T, Eisen MB et al (2000) Molecular portraits of human breast tumours. Nature 406:747–752. https://doi.org/10.1038/35021093
Sotiriou C, Pusztai L (2009) Gene-expression signatures in breast cancer. N Engl J Med 360:790–800. https://doi.org/10.1056/NEJMra0801289
Hart CD, Sanna G, Siclari O, Biganzoli L, Di Leo A (2015) Defining optimal duration and predicting benefit from chemotherapy in patients with luminal-like subtypes. Breast 24(Suppl 2):S136–S142. https://doi.org/10.1016/j.breast.2015.07.033
Barretina J, Caponigro G, Stransky N et al (2012) The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483:603–607. https://doi.org/10.1038/nature11003
Zoppoli G, Regairaz M, Leo E, Reinhold WC, Varma S, Ballestrero A, Doroshow JH, Pommier Y (2012) Putative DNA/RNA helicase Schlafen-11 (SLFN11) sensitizes cancer cells to DNA-damaging agents. Proc Natl Acad Sci USA 109:15030–15035. https://doi.org/10.1073/pnas.1205943109
Katsoulidis E, Carayol N, Woodard J et al (2009) Role of Schlafen 2 (SLFN2) in the generation of interferon alpha-induced growth inhibitory responses. J Biol Chem 284:25051–25064. https://doi.org/10.1074/jbc.M109.030445
Geserick P, Kaiser F, Klemm U, Kaufmann SH, Zerrahn J (2004) Modulation of T cell development and activation by novel members of the Schlafen (slfn) gene family harbouring an RNA helicase-like motif. Int Immunol 16:1535–1548. https://doi.org/10.1093/intimm/dxh155
Li M, Kao E, Gao X et al (2012) Codon-usage-based inhibition of HIV protein synthesis by human schlafen 11. Nature 491:125–128. https://doi.org/10.1038/nature11433
Mavrommatis E, Fish EN, Platanias LC (2013) The Schlafen family of proteins and their regulation by interferons. J Interferon Cytokine Res 33:206–210. https://doi.org/10.1089/jir.2012.0133
Bustos O, Naik S, Ayers G, Casola C, Perez-Lamigueiro MA, Chippindale PT, Pritham EJ, de la Casa-Esperón E (2009) Evolution of the Schlafen genes, a gene family associated with embryonic lethality, meiotic drive, immune processes and orthopoxvirus virulence. Gene 447:1–11. https://doi.org/10.1016/j.gene.2009.07.006
Schwarz DA, Katayama CD, Hedrick SM (1998) Schlafen, a new family of growth regulatory genes that affect thymocyte development. Immunity 9:657–668
Neumann B, Zhao L, Murphy K, Gonda TJ (2008) Subcellular localization of the Schlafen protein family. Biochem Biophys Res Commun 370:62–66. https://doi.org/10.1016/j.bbrc.2008.03.032
Murai J, Tang S-W, Leo E et al (2018) SLFN11 blocks stressed replication forks independently of ATR. Mol Cell 69:371–384. https://doi.org/10.1016/j.molcel.2018.01.012
Mu Y, Lou J, Srivastava M, Srivastava M, Zhao B, Feng XH, Liu T, Chen J, Huang J (2016) SLFN11 inhibits checkpoint maintenance and homologous recombination repair. EMBO Rep 17:94–109. https://doi.org/10.15252/embr.201540964
Tang S-W, Thomas A, Murai J, Trepel JB, Bates SE, Rajapakse VN, Pommier Y (2018) Overcoming resistance to DNA targeted agents by epigenetic activation of Schlafen 11 (SLFN11) expression with class I histone deacetylase inhibitors. Clin Cancer Res. https://doi.org/10.1158/1078-0432.ccr-17-0443
Tang S-W, Bilke S, Cao L et al (2015) SLFN11 is a transcriptional target of EWS-FLI1 and a determinant of drug response in ewing sarcoma. Clin Cancer Res 21:4184–4193. https://doi.org/10.1158/1078-0432.CCR-14-2112
Tian L, Song S, Liu X et al (2014) Schlafen-11 sensitizes colorectal carcinoma cells to irinotecan. Anticancer Drugs 25:1175–1181. https://doi.org/10.1097/CAD.0000000000000151
Nogales V, Reinhold WC, Varma S et al (2016) Epigenetic inactivation of the putative DNA/RNA helicase SLFN11 in human cancer confers resistance to platinum drugs. Oncotarget 7:3084–3097. https://doi.org/10.18632/oncotarget.6413
He T, Zhang M, Zheng R et al (2017) Methylation of SLFN11is a marker of poor prognosis and cisplatin resistance in colorectal cancer. Epigenomics 9:849–862. https://doi.org/10.2217/epi-2017-0019
Pietanza MC, Waqar SN, Krug LM et al (2018) Randomized, double-blind, phase II study of temozolomide in combination with either veliparib or placebo in patients with relapsed-sensitive or refractory small-cell lung cancer. J Clin Oncol. https://doi.org/10.1200/jco.2018.77.7672
Haibe-Kains B, Desmedt C, Loi S et al (2012) A three-gene model to robustly identify breast cancer molecular subtypes. J Natl Cancer Inst 104:311–325. https://doi.org/10.1093/jnci/djr545
Cancer Genome Atlas Network (2012) Comprehensive molecular portraits of human breast tumours. Nature 490:61–70. https://doi.org/10.1038/nature11412
Curtis C, Shah SP, Chin S-F et al (2012) The genomic and transcriptomic architecture of 2000 breast tumours reveals novel subgroups. Nature 486:346–352. https://doi.org/10.1038/nature10983
Dedeurwaerder S, Desmedt C, Calonne E et al (2011) DNA methylation profiling reveals a predominant immune component in breast cancers. EMBO Mol Med 3:726–741. https://doi.org/10.1002/emmm.201100801
Hendrickx W, Simeone I, Anjum S et al (2017) Identification of genetic determinants of breast cancer immune phenotypes by integrative genome-scale analysis. OncoImmunology 6:e1253654. https://doi.org/10.1080/2162402X.2016.1253654
Gendoo DMA, Ratanasirigulchai N, Schröder MS et al (2016) Genefu: an R/bioconductor package for computation of gene expression-based signatures in breast cancer. Bioinformatics 32:1097–1099. https://doi.org/10.1093/bioinformatics/btv693
Huang DW, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57. https://doi.org/10.1038/nprot.2008.211
Prat A, Perou CM (2010) Deconstructing the molecular portraits of breast cancer. Mol Oncol 5:5–23. https://doi.org/10.1016/j.molonc.2010.11.003
Desmedt C, Zoppoli G, Gundem G et al (2016) Genomic characterization of primary invasive lobular breast cancer. J Clin Oncol 34:1872–1881. https://doi.org/10.1200/JCO.2015.64.0334
Ignatiadis M, Singhal SK, Desmedt C et al (2012) Gene modules and response to neoadjuvant chemotherapy in breast cancer subtypes: a pooled analysis. J Clin Oncol 30:1996–2004. https://doi.org/10.1200/JCO.2011.39.5624
Desmedt C, Haibe-Kains B, Wirapati P et al (2008) Biological processes associated with breast cancer clinical outcome depend on the molecular subtypes. Clin Cancer Res 14:5158–5165. https://doi.org/10.1158/1078-0432.CCR-07-4756
Allison Stewart C, Tong P, Cardnell RJ et al (2017) Dynamic variations in epithelial-to-mesenchymal transition (EMT), ATM, and SLFN11 govern response to PARP inhibitors and cisplatin in small cell lung cancer. Oncotarget. 17:28575–28587. https://doi.org/10.18632/oncotarget.15338
Puck A, Aigner R, Modak M et al (2015) Expression and regulation of Schlafen (SLFN) family members in primary human monocytes, monocyte-derived dendritic cells and T cells. Results Immunol 5:23–32. https://doi.org/10.1016/j.rinim.2015.10.001
Bedognetti D, Hendrickx W, Marincola FM, Miller LD (2015) Prognostic and predictive immune gene signatures in breast cancer. Curr Opin Oncol 27:433–444. https://doi.org/10.1097/CCO.0000000000000234
Denkert C, von Minckwitz G, Brase JC et al (2015) Tumor-infiltrating lymphocytes and response to neoadjuvant chemotherapy with or without carboplatin in human epidermal growth factor receptor 2-positive and triple-negative primary breast cancers. J Clin Oncol 33:983–991. https://doi.org/10.1200/JCO.2014.58.1967
Gingras I, Desmedt C, Ignatiadis M, Sotiriou C (2015) CCR 20th anniversary commentary: gene-expression signature in breast cancer-where did it start and where are we now? Clin Cancer Res 21:4743–4746. https://doi.org/10.1158/1078-0432.CCR-14-3127
Cleator SJ, Powles TJ, Dexter T et al (2006) The effect of the stromal component of breast tumours on prediction of clinical outcome using gene expression microarray analysis. Breast Cancer Res 8:R32. https://doi.org/10.1186/bcr1506
Winslow S, Leandersson K, Edsjö A, Larsson C (2015) Prognostic stromal gene signatures in breast cancer. Breast Cancer Res 17:747. https://doi.org/10.1186/s13058-015-0530-2
Farmer P, Bonnefoi H, Anderle P et al (2009) A stroma-related gene signature predicts resistance to neoadjuvant chemotherapy in breast cancer. Nat Med 15:68–74. https://doi.org/10.1038/nm.1908
Finak G, Bertos N, Pepin F et al (2008) Stromal gene expression predicts clinical outcome in breast cancer. Nat Med 14:518–527. https://doi.org/10.1038/nm1764
Gu-Trantien C, Loi S, Garaud S et al (2013) CD4+ follicular helper T cell infiltration predicts breast cancer survival. J Clin Invest 123:2873–2892. https://doi.org/10.1172/JCI67428
Meissl K, Macho-Maschler S, Müller M, Strobl B (2017) The good and the bad faces of STAT1 in solid tumours. Cytokine 89:12–20. https://doi.org/10.1016/j.cyto.2015.11.011
Bianchini G, Balko JM, Mayer IA et al (2016) Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease. Nat Pub Group 13:674–690. https://doi.org/10.1038/nrclinonc.2016.66
Metzger-Filho O, Sun Z, Viale G et al (2013) Patterns of Recurrence and outcome according to breast cancer subtypes in lymph node-negative disease: results from international breast cancer study group trials VIII and IX. J Clin Oncol 31:3083–3090. https://doi.org/10.1200/JCO.2012.46.1574
Bhargava R, Beriwal S, Dabbs DJ et al (2010) Immunohistochemical surrogate markers of breast cancer molecular classes predicts response to neoadjuvant chemotherapy. Cancer 116:1431–1439. https://doi.org/10.1002/cncr.24876
Nelson DJ, Clark B, Munyard K et al (2017) A review of the importance of immune responses in luminal B breast cancer. OncoImmunology 6:e1282590. https://doi.org/10.1080/2162402X.2017.1282590
Yersal O, Barutca S (2014) Biological subtypes of breast cancer: prognostic and therapeutic implications. World J Clin Oncol 5:412–424. https://doi.org/10.5306/wjco.v5.i3.412
Miller LD, Chou JA, Black MA et al (2016) Immunogenic subtypes of breast cancer delineated by gene classifiers of immune responsiveness. Cancer Immunol Res 4:600–610. https://doi.org/10.1158/2326-6066.CIR-15-0149
Acknowledgements
GZ would like to thank Dr. P. Blandini, MD, for his invaluable scientific insights during all the phases of this project.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Availability of data and material
All raw data used for the generation of the expression set we analyzed are available in GEO under their respective publication IDs. Normalized expression data are available upon request to the Corresponding Author.
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Isnaldi, E., Ferraioli, D., Ferrando, L. et al. Schlafen-11 expression is associated with immune signatures and basal-like phenotype in breast cancer. Breast Cancer Res Treat 177, 335–343 (2019). https://doi.org/10.1007/s10549-019-05313-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-019-05313-w