Abstract
Purpose
The purpose of this study is to characterize a novel structural variant, a large duplication involving exons 1–19 of the BRCA1 gene in four independent families, and to provide diagnostically valuable information including the position of the breakpoints as well as clues to its clinical significance.
Methods
The duplication of exons 1–19 of the BRCA1 gene was initially detected by routine laboratory testing including MLPA analysis and next generation sequencing. For detailed characterization we performed array-comparative genome hybridization analysis, fluorescent in situ hybridization, next generation mapping, and long-distance PCR for break-point sequencing.
Results
Our data revealed a tandem duplication on chromosome 17 that encompassed 357 kb and included exons 1–19 of the BRCA1 gene and the genes NBR2, NBR1, TMEM106A, LOC100130581, ARL4D, MIR2117 up to parts of the DHX8 gene. This structural variant appeared as a tandem duplication with breakpoints in intron 19 of the BRCA1 gene and in intron 3 of the DHX8 gene (HGVS:chr17(hg19):g.41210776_41568516dup). Segregation analysis indicated that this structural rearrangement is phased in trans with a known pathogenic exon deletion of the BRCA1 gene in one family.
Conclusions
The copy number variation initially recognized as duplication of exon 1–19 of the BRCA1 gene by MLPA analysis is a structural variation with breakpoints in the BRCA1 and DHX8 genes. Although currently to be classified as a variant of unknown significance, our family data indicates that this duplication may be a benign variation or at least of markedly reduced penetrance since it occurs in trans with another known fully pathogenic variant in the BRCA1 gene.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Accurate molecular diagnosis of hereditary breast and ovarian cancer syndrome is crucial to providing adapted risk estimates for affected patients and healthy but at risk family members. The identification of a pathogenic variant in the BRCA1 gene can have especially great implications on medical care, including increased surveillance for the occurrence of cancers and risk-reducing interventions. The empirical life-time risk of female mutation carriers in the BRCA1 gene is the highest of all genes known so far to be implicated in hereditary breast and ovarian cancer syndrome: 72% for breast cancer and 44% for ovarian cancer in female BRCA1 mutation carriers [1].
However, these risk estimates can only be applied to carriers of variants that are classified as pathogenic or likely pathogenic [2]. In 7% (European ancestry) to 21% (African-American ancestry) of cases a variant of unknown significance in the BRCA1 or BRCA2 gene was reported to the patient by the Myriad laboratory in 2011 [3]. Novel variants continue to be identified and collective efforts are made to gather data on benign and pathogenic variation [4]. Due to the uncertainty of the clinical significance of such a variant, no accurate risk estimate and advice can be given to patients and family members carriers. Therefore, research on collecting families with a given variant, improved data on genetic variation in the general population as well as functional laboratory studies are crucial to improve judgment of the clinical significance of a given variant.
In this study we aimed to characterize a novel genomic duplication that was initially identified by routine MLPA analysis and reported as a variant of unknown significance by the diagnostic laboratory. In-depth characterization by array-CGH analysis, fluorescent in situ hybridization, high molecular weight DNA next generation mapping (NGM), and long-distance PCR revealed that this structural variation is a tandem duplication with breakpoints in the BRCA1 and DHX8 genes. Interestingly, in one of the families this variation is confirmed to be phased in trans to a known pathogenic deletion of BRCA1 exon 15. Although the structural variant is currently classified as a variant of unknown significance, our data indicate that this duplication may be a benign variation or at least a hypomorphic allele. Furthermore our data show that NGM is a useful tool in characterizing genomic structural variation.
Methods
Patients
The patients and, where applicable, their family members were referred to the hereditary cancer risk evaluation clinics at Hanover Medical School (Fam #9349, #10636) or to the Centre of Familial Breast and Ovarian Cancer, University Hospital of Cologne (#59314), and/or had routine diagnostic analysis for hereditary breast and ovarian cancer syndrome performed at the diagnostic laboratory of the Institute of Human Genetics at Hanover Medical School (Fam #9349, #10636), Centre for Hereditary Breast- and Ovarian Cancer Cologne (#59314) or LADR laboratory Recklinghausen (#27689). All patients gave written informed consent to diagnostic analyses as well as for participation in research studies.
Next generation sequencing
Routine diagnostic sequencing for genes implicated in hereditary breast and ovarian cancer syndrome including the BRCA1 gene was performed using TruSight Cancer Panel (Illumina) or TruRisk Panel (German Consortium for Hereditary Breast and Ovarian Cancer) following the manufacturer’s instructions and as published previously [5].
Multiplex ligation-dependent probe amplification (MLPA) analysis
Routine diagnostic MLPA analysis for deletion and duplication analyses was performed according to the manufacturer’s instructions. The kits P002 and P087 were used for dosage analysis at the BRCA1 locus (MRC-Holland).
Array-comparative genome hybridization (aCGH) analysis
High resolution aCGH analysis using of oligo-arrays (2 × 400 k, Agilent Technologies, Waldbronn, Germany) was performed following the manufacturers’ instructions (Oligonucleotide Array-Based CGH for Genomic DNA Analysis v. 4.0). The direct labeling protocol was used with 0.75 µg test DNA (from blood of individual patients) and 0.75 µg control DNA (Kreatech Biotechnology, Amsterdam, The Netherlands). Fluorescence signals were scanned using a Dual Laser Scanner G2565CA (Agilent Technologies). Raw data analysis was performed using Feature extraction version 11.0.1.1 (Agilent Technologies). For further data analysis, Genomic Workbench 7.0.4.0 (Agilent Technologies) was used.
Chromosome preparation and fluorescent in situ hybridization
Metaphase chromosomes were prepared from heparinized blood samples according to standard cytogenetic procedures. Fluorescent in situ hybridization was performed according to the manufacturer’s manual using probe BAC clone RP11-242D8 (BlueFISH /Illumina) targeting the region in question (17q21.31) and control probes BAC clone RP11-143E18 and RP11-94C24 (both BlueFISH /Illumina) upstream (17q12) and downstream (17q21.33) the same chromosome, respectively. Twenty metaphases were analyzed.
Next generation mapping
High molecular weight DNA extraction was performed using the Bionano Prep Blood and Cell Culture DNA Isolation Kit (#80004). Briefly, 400 µl of whole blood was centrifuged at 400 rcf for 2 min. at 4 °C. The supernatant was removed, leaving behind a 2 µl volume at the bottom of the tube (no visible pellet). This was resuspended and embedded into low melting point agarose plugs, using the Bio-Rad CHEF Mammalian Genomic DNA Plug Kit (#1703591). Plugs were processed according to the kit protocol: plugs were treated with Lysis Buffer (Bionano) and Proteinase K (Qiagen #158,920), then washed with Wash Buffer (Bionano) and TE Buffer (Thermo Fisher #AM9849). DNA was then nicked and labeled using the NLRS Kit (Bionano #RE-012-10) and analyzed on the Irys instrument. Single-molecule data were collected and de novo genome map assembly was performed. Structural variants were called against hg19 reference genome.
Long-distance PCR and sanger sequencing
Long-distance PCR was performed using GoTaq® DNA Polymerase Mix according to the manufacturer’s instructions (Promega, Mannheim, Germany). Primer sequences for sequencing the breakpoint in the BRCA1 gene: forward 5′-TTG CTA CAC TCC TCT TTC TGC T-3′, reverse 5′-GGC CCA CTT GCA TAT ACC TTT G-3′. An annealing temperature of 68 °C was used. PCR products were sequenced using a nested reverse primer: 5′-CAG TCA CAA TTG TTT CTG AGC G-3′. Primer sequences for sequencing the DHX8 gene: forward 5′-CAT GAT GGC GTG CCT CCG TAG-3′, reverse 5′-GTA GAC ACT CTT AAA ATG GTA TCA G-3′. Sanger sequencing was performed on an ABI genetic analyzer 3130xl (Applied Biosystems, Darmstadt, Germany).
Results
Patient families and results of diagnostic testing
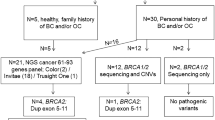
We report four unrelated families with hereditary breast and ovarian cancer, in which we identified a novel duplication at the BRCA1 locus.
In the initial family (Fam #9349), the index patient was diagnosed with unilateral breast cancer at the age of 40 years (Fig. 1a). Histopathologic examinations showed an invasive ductal carcinoma negative for estrogene, progesterone, and Her2 receptors. Later, she developed triple-negative carcinoma in situ in the contralateral breast at the age of 48 years and underwent bilateral mastectomy. In respect to her family history, the patient reported that a maternal aunt was diagnosed with ovarian cancer at the age of 52 years, therefore genetic testing for hereditary breast and ovarian cancer syndrome was indicated [6, 7]. Molecular testing by MLPA analysis in the index patient revealed a duplication of exons 1–19 at the BRCA1 locus, within which the dosage of exon 15 appeared normal due to a deletion in trans on the second allele (Suppl. Figure 1). Segregation analysis showed that the deletion of exon 15 was inherited from the mother, while the duplication of exons 1–19 of the BRCA1 locus was inherited from the father. Regarding the clinical management, the deletion of exon 15 in the BRCA1 gene was classified as the causal variant due to a predicted frame shift followed by a premature stop codon (NM_007294.3:c.(4484+1_4485-1)_(4675+1_4676-1);p.(Ser1496Glyfs*14)), while the duplication of exons 1–19 on the other allele was interpreted as clinically innocuous.
The index patient of the second family (Fam #27689) was diagnosed with triple-negative breast cancer at the age of 44 years (Fig. 1b). She reported that her maternal grandmother was diagnosed with breast cancer in her fourties and deceased in her seventies. Molecular testing in the index patient identified a nonsense variant, c.4327C>T;p.(Arg1443*), in exon 13 of the BRCA1 gene and a duplication of exons 1–19 of the BRCA1 locus. The stop variant is a well-known pathogenic founder mutation in the French-Canadian population [11]. The duplication in the BRCA1 gene was reported by MLPA analysis to affect upstream sequences and to encompass a minimum of aproximately 66.8 kb. Segregation analysis identified both the duplication and the nonsense variant in the father, while the mother did not carry either of these variants. Therefore, both variants are expected to be phased in cis in the index patient. Interestingly, next generation sequencing (NGS) showed that the nonsense variant c.4327C>T was present in 29% of all reads covering that position in the index patient (data not shown), which indicates that only one of three copies of BRCA1 exon 13 is carrying the stop variant. At present, our data cannot discriminate if the nonsense point variant in exon 13 affects the duplicated BRCA1 sequences or the original full-length BRCA1 locus of the paternal chromosome.
Another female patient (Fam #10636) presented with unilateral breast cancer at the age of 38 years (Fig. 1c). Histopathologic examinations showed an invasive ductal carcinoma, positive for hormone receptor expression and negative for Her2 receptor expression. This index patient reported that her maternal grandmother developed breast cancer past the age of 70 years. In additon, she reported that her deceased mother was diagnosed with colon cancer at the age of 66 years and that her father died of prostate cancer at the age of 79 years. Genetic testing of the index patient by NGS identified the same nonsense variant in exon 13 of the BRCA1 gene as in the previous patient, c.4327C>T;p.(Arg1443*). In addition, a duplication in the BRCA1 locus was reported by MLPA analysis, which encompassed exons 1–19 of the BRCA1 gene as well as at least 6.0 kb upstream of the transcriptonal start site in exon 2. Interestingly, we observed the nonsense variant c.4327C>T in 33% of all reads covering that position (data not shown). These data indicate that the nonsense variant has not been duplicated itself and is only present in one of the three alleles of BRCA1 exon 13. In an attempt to validate the genotype, we performed genetic testing on the healthy sister since both parents were deceased and therefore not available for segregation analysis. However, the analysis of the sister remained uninformative since she did not carry either of the BRCA1 variants of the index patient.
The index patient of the fourth family (Fam #59314) was a 49-year-old woman who was diagnosed with hormone receptor postive and Her2 negative breast cancer (Fig. 1d). She reported that her mother was diagnosed with ovarian cancer at the age of 56 years. Other affected family members included a maternal cousin of the mother and a three maternal aunts of the mother, all of them diagnosed with breast cancer in their fourties. Notably, the maternal grandmother of the index patient remained free of cancer until the age of 80 years. Routine MLPA analysis performed on the index patient revealed a duplication of BRCA1 exons 1–19. NGS did not reveal any pathogenic variants in other genes associated with hereditary breast and ovarian cancer syndrome.
Molecular and cytogenetic characterizations
The duplication in the BRCA1 locus that we identified in four unrelated families was revealed by MLPA analysis (Fig. 2). The downstream limit of the duplication was marked by a normal dosage signal in exon 20 of the BRCA1 gene, indicating a break point between exons 19 and 20 of the BRCA1 gene. The upstream border of this copy number variant could not be identified by this method since the most upstream probe set at 6.0 kb upstream of the transcriptional start codon in exon 2 of the BRCA1 locus was already affected by the duplication. Thus, this duplication encompassed at least 66.8 kb of the BRCA1 locus on MLPA analysis and was reported as BRCA1[NM_007294.3]:c.(?_-1)_(5193+1_5194-1) in accordance with HGVS nomenclature.
Schematic view of the duplication at the BRCA1 locus as identified by diagnostic testing by MLPA and array-CGH analyses. MLPA analysis showed a duplication of the BRCA1 locus that ranged from probe “BRCA1-up” 6.0 kb upstream (equals q-terminal) the transcriptonal start site in exon 2 through probe “BRCA1-19” in exon 19 of the BRCA1 gene. The q-terminal limit of the duplication was mapped by array-CGH. The entire duplication encompassed 357 kb. Depiction not to scale. Genomic locations refer to GRCh37/hg19
In order to identify the upstream, i.e., the q-terminal border of this duplication (the BRCA1 gene is on the reverse strand of chromosome 17), we performed array-CGH analysis. As shown in Figs. 3a, b, and 2, array-CGH revealed a duplication of approximately 357 kb, reported as arr[GRCh37] 17q21.31(41,210,812_41,568,343)×3 according to ICSN nomenclature. This duplication ranged from BRCA1 exon 19 at its p-terminal end, up to parts of the DHX8 gene at the q-terminal end, affecting the genes NBR2, NBR1, TMEM106A, LOC100130581, ARL4D and MIR2117.
Molecular cytogenetic mapping of the identified duplication on chromosome 17. a Array-CGH revealed a duplication of 357 kb that included exon 1–19 of the BRCA1 gene as well as the genes NBR2, NBR1, TMEM106A, LOC100130581, ARL4D, MIR2117 and parts of DHX8. (B) Close-up of the duplicated BRCA1 region (same order of samples). Note that the duplication in exon 15 appears as a normal copy number call in the index patient due to a deletion of exon 15 on the second allele. a, b Asterisks mark deletion of BRCA1 exon 15. Arrows mark loci. c, d Fluorescent in situ hybridization analysis on metaphase chromosomes on a sample obtained from the index patient of Fam #59314 showed two adjacent signals for control and duplication probes. c BAC clone RP11_242D8 (chr17:41,316,200-41,322,420) labeled in red (region of interest) co-hybridized with BAC clone RP11-143E18 (chr17:35,985,121-36,129,469) labeled green (upstream control). d BAC clone RP11-242D8 co-hybridized with BAC clone RP11-94C24 (chr17:48,507,413-48,686,813) labeled green (downstream control)
Since array-CGH is limited to copy number evaluation and cannot distinguish the chromosomal location of the duplicated sequences within the genome, we performed fluorescent in situ hybridization analysis on metaphase chromosomes. A probe targeting the duplicated region showed one signal on each chromosome 17 in all 20 analyzed metaphases, each adjacent to a signal of a control probe hybridizing either further upstream (Fig. 3c) or downstream on the same chromosome (Fig. 3d). No signal was observed on other chromosomes. These data indicate that the duplication is limited to the BRCA1 locus on chromosome 17, yet not detectable by cytogenetic resolution.
Next generation mapping and breakpoint identification
In order to characterize the genomic location and orientation of the duplication of exons 1–19 of the BRCA1 gene, we performed de novo genome map assembly of the BRCA1 region of chromosome 17 using Bionano NGM technology. Aligment with hg19 genome reference identified a tandem duplication event that affected genomic sequences from a start point between chromosomal coordinates chr17:41,208,795 and 41,214,425, and an end point between chr17:41,566,390 and 41,570,323 (Fig. 4). The range given for the first breakpoint was 5630 bp and spanned from 925 bp downstream exon 19–274 bp downstream exon 20 of the BRCA1 gene, according to the results of the Bionano analysis. Taking the data obtained by MLPA analysis into account, which showed that exon 20 was not affected by the duplication, the range of the tentative location of the first breakpoint was further reduced to a range from 925 bp downstream exon 19 up to exon 20. The range for the second breakpoint reported by Bionano analysis spanned 3933 bp from 427 bp upstream exon 2 to codon 778 in exon 6 of the DHX8 gene (NM_004941.2). Due to a “fragile site” within the region of interest, resulting from single-stranded nicks (during fluorescent nick-labeling) on opposite strands of the DNA less than 50 bp apart, a complete contiguous assembly of the entire copy of the BRCA1 region could not be achieved. Despite this technical limitation, based on the fusion points captured a model of the tandem duplication event was hypothesized: the complete copy of the BRCA1 gene up to a breakpoint in the DHX8 gene is followed by a duplication in the same orientation that contains the trunctated copy of the BRCA1 gene that starts with BRCA1 exon 19.
De novo genome map assembly of the BRCA1 region. a Next generation mapping revealed a tandem duplication event involving the BRCA1 locus. The breakpoints were localized around chr17:41,211,610 (+/− 2815 bp) and chr17:41,568,356 (+/− 1,966 bp) in hg19. b Hypothesized model of the duplication event: One full-length copy of the BRCA1 gene is followed by a truncated copy BRCA1 gene in the same orientation
In order to define the break points at single nucleotide level, we performed long-range PCR on DNA obtained from all four index patients (Fig. 5a). Sequencing of the PCR products reveald that the last position in intron 3 of the DHX8 gene at chr17:41,568,516 (NM_004941.2: c.308-17) is followed by the first position mapping to chr17:41,210,776 in intron 19 of the BRCA1 gene (NM_007294.3: c.5194-1624) (Fig. 5b). Thus, the duplication encomasses 357,740 bp mapped to the hg19 genome assembly. Sequencing of the second breakpoint in intron 3 of the DHX8 gene showed no sequence alteration, so that one functional DHX8 gene is to be expected (Suppl. Figure 3).
Breakpoint identification by long-range PCR and sequencing. a Long-range PCR amplified the breakpoint region in all index patients and family members that carry the duplication of BRCA1 exons 1–19. b Sequencing results using a reverse primer. Along chromosome 17 (from left to right) the last base of the DHX8 gene at chr17:41,568,516 is followed by the first base of a truncated copy of the BRCA1 gene at chr17:41,210,776. c Schematic model of the duplication event
Discussion
In this study we provide detailed characterization of a large structural variant that appeared as duplication of BRCA1 exons 1–19 on routine MLPA analysis. Our data show that this variation in fact comprises a tandem duplication affecting eight genes from the BRCA1 locus to the DHX8 locus on chromosome 17.
Interestingly, we observed the duplication from BRCA1 intron 19 to DHX9 intron 3 phased in trans with a pathogenic deletion of exon 15 of the BRCA1 gene in a non-syndromic patient with hereditary breast and ovarian cancer syndrome. In contrast to other genes implicated in hereditary breast and ovarian cancer syndrome, the BRCA1 gene is in such remarkable that only two cases of biallelic pathogenic variants have been observed in the BRCA1 gene to date. This observation implies that most combinations of pathogenic variants are embryonically lethal. Both published cases concerned women with a syndromic clinical presentation and missense variants in the C-terminal BRCT repeat of the BRCA1 gene (c.5207T>C;p.(Val1736Ala) and c.5095C>T;p.Arg1699Trp, respectively) in trans with another truncating variant in the BRCA1 gene (c.2457delC;p.(Asp821Ilefs*25) and c.594_597del, respectively) [8, 9]. One of the patients reported developed ovarian cancer at the age of 28 years; the other patient was diagnosed with breast cancer at the age of 23. Another report of biallelic BRCA1 variation concerned a non-syndromic woman diagnosed with breast cancer at the age of 30. The patient carried a deleterious truncating variant (c.2681_2682delAA) in trans with a splice variant (c.594-2A>C) that was predicted to be pathogenic [10]. However, while no full-length BRCA1 transcript was expressed from either allele, isoform analyses showed that the expression of functional isoforms was upregulated. The authors concluded that the expression of isoforms may rescue protein function to an extent that allows the patient to lack a syndromic phenotype. Therefore, given the non-syndromic presentation of our patient it seems very unlikely that both alleles are fully pathogenic. The duplication on chromosome 17 is currently to be classified as variant of unknown significance according to international diagnostic guidelines given the limited amount of data [2]. At this point a clinical significance as hypomorphic allele for hereditary breast and ovarian cancer cannot be excluded; however this structural variant seems unlikely to be a fully penetrant pathogenic allele.
In one of the reported families we observed this structural variant on chromosome 17 in cis with a deleterious nonsense variant in the BRCA1 gene. NGS data indicated that the nonsense variant c.4327C>T;p.(Arg1443*) in exon 13 affected only one of the copies of the BRCA1 gene—either the full-length copy or the duplicated truncated copy on the same chromosome. Given that this particular nonsense variant is a well-known founder variant in the BRCA1 gene [11], it seems more likely that this point mutation arose at an evolutionary date prior to duplication. Instead of being duplicated on the same allele we hypothesize that a truncated BRCA1 locus arose from an insertion of a “wild type” second allele to a chromosome that contained the nonsense variant c.4327C>T. Such structural rearrangement may be linked to the “fragile site” discovered by NGM that prevented the assembly of a complete map of the region. However, it is also possible that the nonsense variant only affects the truncated copy of the BRCA1 gene. In this case, it would be questionable whether this nonsense variant has any clinical significance. To solve this question, continuous sequencing of the breakpoint up to the nonsense variant in exon 13 would be necessary. This encompasses about 23 kb from the breakpoint to exon 13 of the truncated copy and 357 kb from the breakpoint to exon 13 of the original BRCA1 gene. Although technically theoretically manageable by nanopore sequencing, such an endeavor seems currently not feasible since it would require enrichment of unfragmented DNA of high quality.
In spite of the importance of the BRCA1 gene and the identification of this novel structural variation in families with hereditary breast and ovarian cancer syndrome, the clinical significance of this duplication may not be restricted to a potentially increased risk of cancer. The duplication involves seven other genes, including some gene products that seem to be involved in important cellular mechanisms such as the AMPK pathway (NBR2) [12], autophagy (NBR1) [13], GTP-binding proteins (ARL4D) [14] and snRNP regulation (DHX8) [15]; the ARL4D locus has already been identified to be associated with Bardet-Biedl Syndrome [16]. Further research is needed to elucidate if any of the duplicated genes are implicated in disease or phenotypical modulation.
References
Kuchenbaecker KB, Hopper JL, Barnes DR, Phillips KA, Mooij TM, Roos-Blom MJ, Jervis S, van Leeuwen FE, Milne RL, Andrieu N, Goldgar DE, Terry MB, Rookus MA, Easton DF, Antoniou AC, Brca, Consortium BC, McGuffog L, Evans DG, Barrowdale D, Frost D, Adlard J, Ong KR, Izatt L, Tischkowitz M, Eeles R, Davidson R, Hodgson S, Ellis S, Nogues C, Lasset C, Stoppa-Lyonnet D, Fricker JP, Faivre L, Berthet P, Hooning MJ, van der Kolk LE, Kets CM, Adank MA, John EM, Chung WK, Andrulis IL, Southey M, Daly MB, Buys SS, Osorio A, Engel C, Kast K, Schmutzler RK, Caldes T, Jakubowska A, Simard J, Friedlander ML, McLachlan SA, Machackova E, Foretova L, Tan YY, Singer CF, Olah E, Gerdes AM, Arver B, Olsson H (2017) Risks of breast, ovarian, and contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. JAMA 317(23):2402–2416. https://doi.org/10.1001/jama.2017.7112
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL, Committee ALQA (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17(5):405–424. https://doi.org/10.1038/gim.2015.30
Ready K, Gutierrez-Barrera AM, Amos C, Meric-Bernstam F, Lu K, Hortobagyi G, Arun B (2011) Cancer risk management decisions of women with BRCA1 or BRCA2 variants of uncertain significance. Breast J 17(2):210–212. https://doi.org/10.1111/j.1524-4741.2010.01055.x
Cheon JY, Mozersky J, Cook-Deegan R (2014) Variants of uncertain significance in BRCA: a harbinger of ethical and policy issues to come? Genome Med 6(12):121. https://doi.org/10.1186/s13073-014-0121-3
Hauke J, Horvath J, Gross E, Gehrig A, Honisch E, Hackmann K, Schmidt G, Arnold N, Faust U, Sutter C, Hentschel J, Wang-Gohrke S, Smogavec M, Weber BHF, Weber-Lassalle N, Weber-Lassalle K, Borde J, Ernst C, Altmuller J, Volk AE, Thiele H, Hubbel V, Nurnberg P, Keupp K, Versmold B, Pohl E, Kubisch C, Grill S, Paul V, Herold N, Lichey N, Rhiem K, Ditsch N, Ruckert C, Wappenschmidt B, Auber B, Rump A, Niederacher D, Haaf T, Ramser J, Dworniczak B, Engel C, Meindl A, Schmutzler RK, Hahnen E (2018) Gene panel testing of 5589 BRCA1/2-negative index patients with breast cancer in a routine diagnostic setting: results of the German Consortium for Hereditary Breast and Ovarian Cancer. Cancer Med. https://doi.org/10.1002/cam4.1376
Gadzicki D, Evans DG, Harris H, Julian-Reynier C, Nippert I, Schmidtke J, Tibben A, van Asperen CJ, Schlegelberger B (2011) Genetic testing for familial/hereditary breast cancer-comparison of guidelines and recommendations from the UK, France, the Netherlands and Germany. J Community Genet 2:53–69
Petrucelli N, Daly MB, Pal T (2016) BRCA1- and BRCA2-Associated Hereditary Breast and Ovarian Cancer. In: Adam MP, Ardinger HH, Pagon RA et al (eds) GeneReviews® [Internet]. University of Washington, Seattle(WA). Available from: https://www.ncbi.nlm.nih.gov/books/NBK1247/
Domchek SM, Tang J, Stopfer J, Lilli DR, Hamel N, Tischkowitz M, Monteiro AN, Messick TE, Powers J, Yonker A, Couch FJ, Goldgar DE, Davidson HR, Nathanson KL, Foulkes WD, Greenberg RA (2013) Biallelic deleterious BRCA1 mutations in a woman with early-onset ovarian cancer. Cancer Discov 3(4):399–405. https://doi.org/10.1158/2159-8290.CD-12-0421
Sawyer SL, Tian L, Kahkonen M, Schwartzentruber J, Kircher M, University of Washington Centre for Mendelian G, Consortium FC, Majewski J, Dyment DA, Innes AM, Boycott KM, Moreau LA, Moilanen JS, Greenberg RA (2015) Biallelic mutations in BRCA1 cause a new Fanconi anemia subtype. Cancer Discov 5(2):135–142. https://doi.org/10.1158/2159-8290.CD-14-1156
Wong-Brown M, McPhillips M, Gleeson M, Spigelman AD, Meldrum CJ, Dooley S, Scott RJ (2016) When is a mutation not a mutation: the case of the c.594-2A> C splice variant in a woman harbouring another BRCA1 mutation in trans. Hered Cancer Clin Pract 14:6. https://doi.org/10.1186/s13053-015-0045-y
Janavicius R (2010) Founder BRCA1/2 mutations in the Europe: implications for hereditary breast-ovarian cancer prevention and control. EPMA J 1(3):397–412. https://doi.org/10.1007/s13167-010-0037-y
Liu X, Xiao ZD, Han L, Zhang J, Lee SW, Wang W, Lee H, Zhuang L, Chen J, Lin HK, Wang J, Liang H, Gan B (2016) LncRNA NBR2 engages a metabolic checkpoint by regulating AMPK under energy stress. Nat Cell Biol 18(4):431–442. https://doi.org/10.1038/ncb3328
Deosaran E, Larsen KB, Hua R, Sargent G, Wang Y, Kim S, Lamark T, Jauregui M, Law K, Lippincott-Schwartz J, Brech A, Johansen T, Kim PK (2013) NBR1 acts as an autophagy receptor for peroxisomes. J Cell Sci 126(Pt 4):939–952. https://doi.org/10.1242/jcs.114819
Smith SA, Holik PR, Stevens J, Melis R, White R, Albertsen H (1995) Isolation and mapping of a gene encoding a novel human ADP-ribosylation factor on chromosome 17q12-q21. Genomics 28(1):113–115. https://doi.org/10.1006/geno.1995.1115
Ohno M, Shimura Y (1996) A human RNA helicase-like protein, HRH1, facilitates nuclear export of spliced mRNA by releasing the RNA from the spliceosome. Genes Dev 10(8):997–1007
Chiang AP, Nishimura D, Searby C, Elbedour K, Carmi R, Ferguson AL, Secrist J, Braun T, Casavant T, Stone EM, Sheffield VC (2004) Comparative genomic analysis identifies an ADP-ribosylation factor-like gene as the cause of Bardet-Biedl syndrome (BBS3). Am J Hum Genet 75(3):475–484. https://doi.org/10.1086/423903
Acknowledgements
The authors would like to thank the patients and their families for supporting this study and Marcel Tauscher and Hildegard Frye-Boukhriss for excellent technical assistance.
Funding
This work was funded in parts by the German Research Foundation (DFG), Cluster of Excellence REBIRTH to B.S. and D.S.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Disclosures
SC and HC hold stocks of Bionano Genomics, Inc. All other authors have no conflict of interest to declare.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Du, C., Mark, D., Wappenschmidt, B. et al. A tandem duplication of BRCA1 exons 1–19 through DHX8 exon 2 in four families with hereditary breast and ovarian cancer syndrome. Breast Cancer Res Treat 172, 561–569 (2018). https://doi.org/10.1007/s10549-018-4957-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-018-4957-x