Abstract
Background
Germline mutations in BRCA1 and BRCA2 (BRCA1/2) account for the majority of hereditary breast and/or ovarian cancers. Pakistan has one of the highest rates of breast cancer incidence in Asia, where BRCA1/2 small-range mutations account for 17% of early-onset and familial breast/ovarian cancer patients. We report the first study from Pakistan evaluating the prevalence of BRCA1/2 large genomic rearrangements (LGRs) in breast and/or ovarian cancer patients who do not harbor small-range BRCA1/2 mutations.
Materials and methods
Both BRCA1/2 genes were comprehensively screened for LGRs using multiplex ligation-dependent probe amplification in 120 BRCA1/2 small-range mutations negative early-onset or familial breast/ovarian cancer patients from Pakistan (Group 1). The breakpoints were characterized by long-range PCR- and DNA-sequencing analyses. An additional cohort of 445 BRCA1/2 negative high-risk patients (Group 2) was analyzed for the presence of LGRs identified in Group 1.
Results
Three different BRCA1 LGRs were identified in Group 1 (4/120; 3.3%), two of these were novel. Exon 1–2 deletion was observed in two unrelated patients: an early-onset breast cancer patient and another bilateral breast cancer patient from a hereditary breast cancer (HBC) family. Novel exon 20–21 deletion was detected in a 29-year-old breast cancer patient from a HBC family. Another novel exon 21–24 deletion was identified in a breast-ovarian cancer patient from a hereditary breast and ovarian cancer family. The breakpoints of all deletions were characterized. Screening of the 445 patients in Group 2 for the three LGRs revealed ten additional patients harboring exon 1–2 deletion or exon 21–24 deletion (10/445; 2.2%). No BRCA2 LGRs were identified.
Conclusions
LGRs in BRCA1 are found with a considerable frequency in Pakistani breast/ovarian cancer cases. Our findings suggest that BRCA1 exons 1–2 deletion and exons 21–24 deletion should be included in the recurrent BRCA1/2 mutations panel for genetic testing of high-risk Pakistani breast/ovarian cancer patients.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Approximately 5–10% of all breast and ovarian cancers are the result of an inherited predisposition due to germline alterations (including small-range mutations and large genomic rearrangements (LGRs)) in the two major high-penetrance breast cancer susceptibility genes, BRCA1 and BRCA2 (BRCA1/2) [1, 2]. Women with pathogenic BRCA1/2 mutations have increased lifetime risk of developing breast and ovarian cancer as well as various other malignancies [3]. Identification of individuals harboring BRCA1/2 mutations is clinically relevant and has significant impact on surveillance and management [4].
The presence of alterations in these genes is variable in different populations and may therefore account for a varying fraction of breast and ovarian cancer cases. In Pakistan, currently with one of the highest rates of breast cancer in Asia, BRCA1/2 small-range mutations account for 17% of hereditary breast/ovarian cancer and early-onset breast and ovarian cancer cases [5]. The mutation frequency reported in Pakistani population may be underestimated as only PCR-based mutation screening methods have previously been applied that would have missed LGRs. Detection of LGRs in BRCA1/2 is important because these alterations have been shown to significantly contribute to hereditary breast and/or ovarian cancer in some populations. At present, more than 100 different LGRs in BRCA1/2 have been described among breast/ovarian cancer cases negative for BRCA1/2 small-range mutations worldwide [6].
Among high-risk breast/ovarian cancer patients, the prevalence and spectrum of LGRs in BRCA1/2 vary significantly with the geographic distribution, ethnicity, and selection criteria of study population. LGRs in BRCA1/2 have been identified in studies (considering those with more than 100 high-risk cases) with frequencies varying from 0 to 5% in Europe [7–16], 3.3 to 4.4% in Australia/New Zealand [17, 18], and 4.8 to 11.7% in US [19–21]. However, increased frequency of LGRs in BRCA1 was reported in those populations where founder effects or recurrent mutations had been identified [8, 10, 16, 22–25]. Contrary to BRCA1, LGRs in BRCA2 were absent in several populations [8, 12, 14–16, 26, 27] or reported rarely [7, 10, 17, 19–21, 28–30].
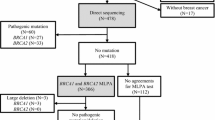
The LGRs in BRCA1/2 have been reported with frequencies varying from 0.4 to 3.0% in Asia [26–30]. Studies from Asia assessing the contribution of BRCA1/2 LGRs to breast/ovarian cancer cases have been small in size [31–34] or analyzed only BRCA1 [31, 34–36]. We report the results of a large study from Pakistan which evaluated the prevalence of LGRs in the BRCA1/2 genes in 565 high-risk breast/ovarian cancer patients who did not harbor small-range BRCA1/2 mutations. Initially, 120 patients (Group 1) were comprehensively screened by multiplex ligation-dependent probe amplification (MLPA) for all exons of BRCA1 and BRCA2. An additional cohort of 445 high-risk cases (Group 2) was analyzed for those LGRs identified in Group 1.
Materials and methods
Study population
Breast and/or ovarian cancer families were identified at the Shaukat Khanum Memorial Cancer Hospital and Research Centre (SKMCH & RC) in Lahore, Pakistan, from June 2001 to August 2015. Index patients diagnosed with invasive breast cancer or epithelial ovarian cancer from 565 unrelated breast and/or ovarian cancer families were included in the current study based on following criteria: (A1) families with one female breast cancer diagnosed ≤30 years of age, (A2) families with two first- or second-degree (through a male) female relatives diagnosed with breast cancer; at least one diagnosed ≤50 years of age, (A3) families with at least three cases of breast cancer; at least one diagnosed ≤50 years of age, (A4) families with one male breast cancer case diagnosed at any age, (B) families with at least one female breast cancer and one ovarian cancer at any age, (C1) families with one ovarian cancer diagnosed ≤45 years of age, and (C2) families with at least two ovarian cancers; at least one diagnosed ≤45 years of age. Bilateral breast cancer or breast and ovarian cancer in the same patient were counted as two independent diagnoses. A detailed description of the 565 index cases is shown in Table 1. All study participants signed informed written consent. The study was approved by the institutional review board of the SKMCH & RC.
Genomic DNA was extracted as described elsewhere [37]. One hundred and twenty cases underwent comprehensive screening for BRCA1/2 using protein-truncation test, single-strand conformational polymorphism analysis and denaturing high-performance liquid chromatography (DHPLC) analysis followed by DNA sequencing of variant fragments and confirmed to be negative for BRCA1/2 small-range mutations as described previously were assigned to Group 1 [5]. Small-range mutations affecting one or a few nucleotides included frameshift deletions or insertions, nonsense mutations, or splice junction alterations. 445 cases additionally enrolled were subsequently analyzed for BRCA1/2 using DHPLC- and DNA-sequencing analyses and found negative for small-range mutations were assigned to Group 2. Ninety-eight patients from Group 1 and 311 cases from Group 2 have previously been included in a Pakistani breast cancer study [38].
After screening for LGRs in the complete BRCA1/2 coding regions in Group 1 (n = 120), Group 2 (n = 445) was screened for the three identified LGRs.
MLPA analyses
For Group 1 cases, comprehensive MLPA analyses was performed for BRCA1 (using probe mix P002 for primary screening and probe mix P087 for confirmatory screening) and BRCA2 (using probe mix P045), as described by manufacturer (MRC Holland, Amsterdam, The Netharlands). Separation and relative quantification of the amplified product was obtained using the Beckman CEQ 8000XL DNA analysis system (Beckman Coulter, Fullerton, USA). A positive control with a known LGR in BRCA1 (exon 1–2 deletion, exon 1–7 deletion, exon 3–16 deletion, exon 9–12 deletion, exon 13–15 deletion, exon 13 duplication, exon 14 deletion, exon 14–20 deletion, exon 17–19 deletion, exon 20 deletion, exon 21–24 deletion, exon 22 deletion, exon 23–24 deletion and exon 24 deletion) or BRCA2 (exon 1–2 deletion) was included in the MLPA analyses. For quality control of each experiment, visual peak pattern evaluation was performed as described by manufacturer (MRC Holland, Amsterdam, The Netharlands). The proportion of each peak relative to the height of all peaks was calculated for every single patient sample and then compared to proportions for the corresponding peak averaged for a set of healthy control samples using a commercially available SeqPilot software (JSI medical systems GmbH, Ettenheim, Germany). The ratio relative peak area (RPA) defined as the RPA of the patient result file divided by the RPA of the control result file was calculated. If this value was around 100%, the RPA of the patient was about the RPA of the control indicative of no copy number change. Samples revealing RPA ratios of ≤50% were considered as deletions and RPA ratios of ≥150% as duplications. Each positive result was independently confirmed in a second experiment. DNA-sequencing analyses was performed in samples showing single exon deletion or duplication to exclude the presence of polymorphisms at the probe ligation sites.
Identification of genomic breakpoints and confirmation
DNA samples with a positive MLPA result indicating exon 1–2 deletion, exon 20–21 deletion, and exon 21–24 deletion in the BRCA1 gene were subjected to further analysis.
For characterization of exon 1–2 deletion, long-range PCR was performed using long template enzyme mix (Fermentas, Vilnius, Lithuania) with the primer pair as previously reported [39].
For characterization of exon 20–21 deletion and exon 21–24 deletion, a combined approach of long-range PCR and restriction fragment length polymorphism (RFLP) was used to narrow down the breakpoint regions with restriction endonucleases PstI or PstI and XmnI, respectively (New England Biolabs, Ipswich MA, USA). The primer pairs (sequences are available upon request) that eventually allowed for characterization of exon 20–21 deletion and exon 21–24 deletion were P1 and P2 (located in intron 19 and 21) and P3 and P4 (located in intron 20 and ~25 kb downstream of the BRCA1 stop codon), respectively (Fig. 1a). Mutation-specific PCR products of 580 and 536 bp containing breakpoints of exon 20–21 deletion and exon 21–24 deletion were obtained, respectively (Fig. 1b, c). PCR products containing the breakpoint regions were purified using QIAquick® PCR purification kit (Qiagen, Hilden, Germany) and bidirectionally sequenced with BigDye Terminator v3.1 Cycle Sequencing kit on the ABI 3130 genetic analyzer (Applied Biosystem, Foster City, CA, USA).
Diagram of the BRCA1 exon 20–21 deletion and exon 21–24 deletion assays. a Genomic structure of breakpoint region with numbered exon. Solid blocks, BRCA1 exon 20–24; dotted bars, intron 19–23; crossed-pattern bar, 25 kb genomic region downstream from BRCA1 stop codon; Brackets above and below the diagram, positions of the respective breakpoints. Primer positions for the specific rearrangement assay (P1, P2 and P3, P4) are indicated along with respective orientation. Photographs of ethidium bromide-stained gels of exon 20–21 deletion specific PCR (b), and exon 21–24 deletion specific PCR (c). M, DNA 100 bp marker; Lanes 1, 2, predicted wild-type allele products of 7.8 kb (b) and 33.6 kb (c) that were not detectable in assays; Lane 3, mutant alleles with a deletion resulting in 580 bp (b) and 536 bp (c) fragments; Lane 4, no template control. DNA sequence analysis of the fragments with breakpoints for exon 20–21 deletion (d) and exon 21–24 deletion (e). Boxes shaded in gray, the 21-bp core identical sequences for both the AluY and AluSc present in intron 19 and 21, respectively (d), and 16-bp core identical sequences for the AluSp present in intron 20 of BRCA1 and intron 1 of VAT1 (e), where the recombination events occur
Sequence traces were aligned to the February 2009 assembly of the Human Genome Browser (available at: http://genome.ucsc.edu/). Mutations were described according to the recommended nomenclature system described by the Human Genome Variation Society (HGVS) [40]. Designation of the exon 1–2 deletion and exon 20–21 deletion in BRCA1 was based on the reference sequence NG_005905.2. Designation of the BRCA1 exon 21–24 deletion was based on the chromosome 17 reference sequence from February 2009 assembly of the Genome Reference Consortium Human genome build 37 (GRCh37/hg19) (available at: https://genome.ucsc.edu). The sequences at breakpoints were also analyzed by RepeatMasker program (http://www.repeatmasker.org). A second blood sample was taken from patients identified with LGR. LGRs were confirmed by PCR-based assays as described above.
The BRCA1 LGRs identified in Group 1 were subsequently analyzed in the 445 cases in Group 2 by deletion specific PCR-based assays using primer pairs flanking the deletion breakpoints. A positive control with a known BRCA1 exon 1–2 deletion, exon 20–21 deletion, or exon 21–24 deletion was included in each experiment. PCR products containing the breakpoint regions were bidirectionally sequenced to confirm the deletions.
Results
Characteristics of the study participants
Characteristics of the 565 unrelated index patients from Pakistani breast and/or ovarian cancer families are shown in Table 1. Of the index cases, 530 patients had a diagnosis of invasive breast cancer—497 females and 33 males. Forty-eight patients had a diagnosis of epithelial ovarian cancer. Majority of the index patients belonged to the Punjabi (68.5%) or Pathan (14.7%) ethnic group. Of the index patients who presented with invasive breast cancer, 140 (28.6%) had triple-negative breast cancer (TNBC), i.e., lack of estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER-2) expression, while 349 (71.4%) were diagnosed with non-TNBC phenotype. In Group 1, the median age of disease onset was 30.4 years (range 21–70) for female breast cancer (n = 108), 48.0 years (range 30–73) for male breast cancer (n = 11), and 31.8 years (range 29–34) for ovarian cancer (n = 5). Seventeen of 120 women (14.2%) were diagnosed with bilateral breast cancer. In Group 2, the median age of disease onset was 35.8 years (range 18–78) for female breast cancer (n = 389), 52.5 years (range 27–69) for male breast cancer (n = 22), and 38.8 years (range 22–66) for ovarian cancer (n = 43). Twenty-nine of 445 (6.5%) women were diagnosed with bilateral breast cancer. All index patients were previously tested and found to be negative for germline small-range mutations in the BRCA1/2 genes [5, 38].
LGRs identified in Group 1
All index patients of Group 1 (n = 120) were comprehensively screened for BRCA1/2 LGRs using MLPA analyses. Genomic breakpoints were identified by the joint approach of long-range PCR-, RFLP-, and DNA-sequencing analyses. Three LGRs were identified in four patients (4/120, 3.3%) (Table 2). All of them were deletions in BRCA1. The frequency of BRCA1 LGRs by family phenotype was 1.4% (1/72) for early-onset breast cancer (EOBC), 6.7% (2/30) for hereditary breast cancer (HBC), and 14.3% (1/7) for hereditary breast and ovarian cancer (HBOC) families (Table 2). All deletion carriers were diagnosed with invasive ductal carcinoma, grade 3 tumor, and displayed TNBC features. No LGRs were detected in the BRCA2 gene.
BRCA1 exon 1–2 deletion
A recurrent and previously reported LGR, BRCA1 exon 1–2 deletion [39] was identified in two patients of Punjabi ethnicity: one with EOBC at 25 years of age (II:3) (Supplementary Fig. 1, FP 89) and another patient with bilateral breast cancer at age 28 and 33 (III:5) and also reported a family history of HBC (Supplementary Fig. 1, FP 147). Characterization of the genomic breakpoints of this LGR revealed a 36,934 bp deletion, which is similar to the deletion previously described [39].
BRCA1 exon 20–21 deletion
A novel LGR, exon 20–21 deletion was identified in a 29-year-old breast cancer patient (III:1) of Punjabi background. The mother (II:4) and maternal uncle (II:2) of the index patient were diagnosed with breast cancer or intestinal cancer at age 47 and 61, respectively (Supplementary Fig. 1, FP 187).
Sequence analysis revealed a deletion of 7223 bp (chr17:41,202,523–41,209,745) (Fig. 1a, b, d). Sequence analysis at the junction point showed that the exon 20–21 deletion fused the AluY sequence of intron 19 with the AluSc of intron 21 (Fig. 1d). The deletion involved an unequal homologous recombination between these two Alu sequences which shared 98% homology. The sites of crossover event were present within a 21-bp sequence.
BRCA1 exon 21–24 deletion
Another novel LGR, exon 21–24 deletion, was identified in a patient (IV:6) from Balochistan, who was diagnosed with breast and ovarian cancer at age 29 and 31, respectively (Supplementary Fig. 1, FP 193). The mutation co-segregated with the disease in two sisters of the index patient, one diagnosed with breast cancer (IV:8) at age 44 and the other diagnosed with ovarian cancer (IV:5) at age 30. Another sister (IV:1) was diagnosed with breast cancer at age 37. Three other breast cancers (II:3, III:6, III:7) and one uterine cancer (II:1) were also reported in this family.
Sequence analysis revealed a deletion of 33,092 bp (chr17:41,172,653–41,205,744) (Fig. 1a, c, e). Sequence analysis at the junction point showed that the exon 21–24 deletion fused the AluSp sequence of intron 20 with AluSp of VAT intron 1, present ~25 kb downstream of stop codon of BRCA1 (Fig. 1e). The deletion involved an unequal homologous recombination between these two Alu sequences which shared 100% homology. The sites of crossover event were present within a 16-bp sequence.
LGRs identified in Group 2
Screening for patients in Group 2 for the presence of the three BRCA1 LGRs identified in Group 1 revealed ten deletions. Exon 1–2 deletion was detected in seven patients, all of Punjabi ethnic origin (Supplementary Fig. 1, FP 229, FP 498, FP 379, FP 406, FP 549, FP 314, and FP 291). Exon 21–24 deletion was detected in three patients of Punjabi background (Supplementary Fig. 1, FP 261, FP 719, and FP 191). The frequency of BRCA1 LGRs by family phenotype was 1.1% (2/182) for EOBC, 2.7% (5/183) for HBC, 6.2% (2/32) for HBOC, and 33.3% (1/3) for hereditary ovarian cancer (HOC) families (Table 2).
Several other cancers were also reported among families harboring LGRs from Group 1 and Group 2 (Table 3).
Discussion
In the current study, we investigated the frequency of BRCA1/2 LGRs in 565 breast and/or ovarian cancer cases, who tested negative for BRCA1/2 small-range mutations. Initally, index patients from Group 1 (n = 120) were comprehensively screened and characterized for LGRs in BRCA1/2. The LGRs identified in Group 1 were next screened in Group 2 (n = 445). Three different LGRs in BRCA1 were identified and characterized in Pakistani high-risk cases, with an overall frequency of 3.3% (4/120) in Group 1 and 2.2% (10/445) in Group 2. No LGRs were detected in the BRCA2 gene.
The exon 1–2 deletion is the most commonly reported LGR in BRCA1. This deletion is pathogenic as it predicts for disruption of the BRCA1 promoter and abolishes the transcription from the mutant allele [41, 42]. In the present study, it was identified in nine Punjabi families from Group 1 and Group 2 including one HBOC, two EOBC, and six HBC families. This deletion comprised 36,934 bp including BRCA1 exon 1–2 and the intergenic region between ΨBRCA1 and BRCA1. It was previously identified in several other European and North-American studies [7, 8, 12, 16, 19, 21, 39, 41–51] and was associated with different breakpoints. Breakpoints similar to those detected in the Pakistani families were observed in one HBOC family from USA [39] and subsequently detected in families from Italy [42], Germany [8], The Netherlands [50], and Czech Republic [16]. Different [12, 21, 47, 51] or uncharacterized breakpoints were reported in several other studies from Europe and Turkey [7, 19, 41, 43–46, 48, 49].
BRCA1 exon 21–24 deletion is another recurrent LGR detected in our study. This deletion is disease-causative as it is predicts to remove the polyA tail and 3′-UTR regions of BRCA1, hence abolishing the transcription from the mutant allele [6]. It was detected in four Pakistani families including one HBC, one HBOC, one HOC family from Punjab, and one HBOC family from Balochistan. This novel large deletion co-segregated with the disease in the Balochi family FP 193 as it was identified in the index patient´s two sisters affected by breast and ovarian cancer at age 44 and 30, respectively, suggesting that this mutation predisposes to breast and ovarian cancer. Her niece, who was unaffected, tested negative for this mutation. The deletion comprised a 33,092 bp fragment, with breakpoints within highly similar duplicated sequences, i.e., Alu elements (AluSp). These findings support the notion that the BRCA1 exon 21–24 deletion is a result of Alu repeat-mediated recombination event. The BRCA1 sequence is composed of about 42% Alu sequences, thereby suggesting that majority of LGRs in this gene are due to Alu repeats [52]. BRCA1 exon 21–24 deletion has previously been reported with the different breakpoints in one HBOC family from Ireland [21] and one HBC family from Czech Republic [16]. It has also been described with uncharacterized breakpoints [19, 20, 45, 46]. The results of these studies indicate that different hotspots are present in intron 20 and downstream of the BRCA1 gene, which result in deletion fragments of different sizes.
A BRCA1 exon 20–21 deletion was identified in one HBC family of Punjabi background, suggesting it is a rare alteration in Pakistan. This deletion is novel and has not been previously reported. It comprised a 7223 bp fragment with breakpoints within Alu elements, AluY and AluSc. This finding supports the notion that the BRCA1 exon 20–21 deletion is because of Alu repeat-mediated recombination event.
In the current study, BRCA1 LGRs were identified in 3.3% of Pakistani patients with EOBC or family history of breast/ovarian cancer. Lower BRCA1 LGR frequencies ranging from 0.4 to 2.1% were observed in other Asian studies from China, Malaysia, Indonesia, Singapore, or Korea, considering those with more than 100 EOBC or familial patients [26–30]. Frequencies ranging from 0 to 10.7% have been reported by other investigators in studies conducted in Finland, Spain, Italy, Germany, Denmark, Czech Republic, the Netherlands, USA (including Hispanics), and Australia [7, 8, 10, 12, 15–17, 19, 21, 24, 53, 54]. These discrepant results may be explained by differences in inclusion criteria, sample size, the sensitivity of the LGR detection assay used, different ethnic or geographic origins of study participants, and founder effects.
The BRCA1-associated breast tumors are reported to have significant numbers of TNBC phenotype [55]. In the current study, the breast tumors linked with BRCA1 LGRs in majority of Pakistani index patients exhibited TNBC features (9/12; 75%). This observation is in agreement with previous findings that breast tumors associated with LGRs in BRCA1 predominantly present with TNBC phenotype [29].
In the present study, no LGR in BRCA2 was detected. Similar findings have been reported in Asian studies from Indonesia and Korea [26, 27] or European studies performed in Germany, Poland, and Czech Republic [8, 12, 14, 16]. LGRs in BRCA2 were identified at low frequencies ranging from 0.2 to 2.0% in some Asian studies from China, Singapore, and Malaysia [28–30], European studies conducted in Spain, Denmark [7, 9, 10, 13], or studies from the USA [19, 21] and Australia [17, 18], suggesting a minimal contribution of BRCA2 LGRs to breast and/or ovarian cancer. The low contribution or lack of LGRs in the BRCA2 gene may be a reflection of the fact that it is composed of few Alu sequences.
No BRCA1/2 LGRs were identified in 33 male breast cancer families included in our study. In agreement with our findings, no LGRs in BRCA1/2 have previously been reported among male breast cancer patients from Italy [56], Finland [57], USA [58], and Turkey [59]. However, LGRs in BRCA2 only have been identified in small subset of male breast cancer patients (7.7%; 3/39) from France [60]. Taken together, the results suggest a marginal contribution of BRCA2 LGRs to hereditary male breast cancer.
In summary, this is the first comprehensive study conducted among Pakistani high-risk breast/ovarian cancer patients to assess the prevalence and spectrum of LGRs in the BRCA1/2 genes. We have not observed any LGR in BRCA2 indicating that such type of alterations may be rare or absent in our population. However, LGRs in BRCA1 are found with a considerable frequency in Pakistani breast/ovarian cancer cases. Two recurrent alterations, BRCA1 exon 1–2 deletion and exon 21–24 deletion, were identified in multiple high-risk breast/ovarian cancer patients. Potential founder effects of the recurrent mutations will be investigated in future haplotype analyses. Our findings suggest that these recurrent LGRs in BRCA1 should be included in the panel of recurrent point mutations in BRCA1/2 for further early-onset and familial-based genetic testing. Our data may improve genetic counseling and may help effective detection strategy in Pakistani early-onset and familial breast/ovarian cancer patients.
Abbreviations
- DHPLC:
-
Denaturing high-performance liquid chromatography
- EOBC:
-
Early-onset breast cancer
- ER:
-
Estrogen receptor
- HBC:
-
Hereditary breast cancer
- HBOC:
-
Hereditary breast and ovarian cancer
- HER-2:
-
Human epidermal growth factor receptor 2
- HGVS:
-
Human Genome Variation Society
- LGRs:
-
Large genomic rearrangements
- MLPA:
-
Multiplex ligation-dependent probe amplification
- PR:
-
Progesterone receptor
- RFLP:
-
Restriction fragment length polymorphism
- RPA:
-
Ratio relative peak area
- TNBC:
-
Triple-negative breast cancer
References
Wooster R, Bignell G, Lancaster J et al (1995) Identification of the breast cancer susceptibility gene BRCA2. Nature 378:789–792
Miki Y, Swensen J, Shattuck-Eidens D et al (1994) A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 266:66–71
Ford D, Easton DF, Stratton M et al (1998) Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. Am J Hum Genet 62:676–689
Robson M, Offit K (2007) Management of an inherited predisposition to breast cancer. N Engl J Med 357:154–162
Rashid MU, Zaidi A, Torres D et al (2006) Prevalence of BRCA1 and BRCA2 mutations in Pakistani breast and ovarian cancer patients. Int J Cancer 119:2832–2839
Sluiter MD, van Rensburg EJ (2011) Large genomic rearrangements of the BRCA1 and BRCA2 genes: review of the literature and report of a novel BRCA1 mutation. Breast Cancer Res Treat 125:325–349
del Valle J, Feliubadalo L, Nadal M et al (2010) Identification and comprehensive characterization of large genomic rearrangements in the BRCA1 and BRCA2 genes. Breast Cancer Res Treat 122:733–743
Engert S, Wappenschmidt B, Betz B et al (2008) MLPA screening in the BRCA1 gene from 1,506 German hereditary breast cancer cases: novel deletions, frequent involvement of exon 17, and occurrence in single early-onset cases. Hum Mutat 29:948–958
Fachal L, Blanco A, Santamarina M, Carracedo A, Vega A (2014) Large genomic rearrangements of BRCA1 and BRCA2 among patients referred for genetic analysis in Galicia (NW Spain): delimitation and mechanism of three novel BRCA1 rearrangements. PLoS ONE 9:e93306
Hansen T, Jonson L, Albrechtsen A, Andersen MK, Ejlertsen B, Nielsen FC (2009) Large BRCA1 and BRCA2 genomic rearrangements in Danish high risk breast-ovarian cancer families. Breast Cancer Res Treat 115:315–323
Moisan AM, Fortin J, Dumont M et al (2006) No Evidence of BRCA1/2 genomic rearrangements in high-risk French-Canadian breast/ovarian cancer families. Genet Test 10:104–115
Preisler-Adams S, Schonbuchner I, Fiebig B, Welling B, Dworniczak B, Weber BH (2006) Gross rearrangements in BRCA1 but not BRCA2 play a notable role in predisposition to breast and ovarian cancer in high-risk families of German origin. Cancer Genet Cytogenet 168:44–49
Rodriguez M, Torres A, Borras J, Salvat M, Guma J (2010) Large genomic rearrangements in mutation-negative BRCA families: a population-based study. Clin Genet 78:405–407
Rudnicka H, Debniak T, Cybulski C et al (2013) Large BRCA1 and BRCA2 genomic rearrangements in Polish high-risk breast and ovarian cancer families. Mol Biol Rep 40:6619–6623
Thomassen M, Gerdes AM, Cruger D, Jensen PK, Kruse TA (2006) Low frequency of large genomic rearrangements of BRCA1 and BRCA2 in western Denmark. Cancer Genet Cytogenet 168:168–171
Ticha I, Kleibl Z, Stribrna J et al (2010) Screening for genomic rearrangements in BRCA1 and BRCA2 genes in Czech high-risk breast/ovarian cancer patients: high proportion of population specific alterations in BRCA1 gene. Breast Cancer Res Treat 124:337–347
James PA, Sawyer S, Boyle S et al (2015) Large genomic rearrangements in the familial breast and ovarian cancer gene BRCA1 are associated with an increased frequency of high risk features. Fam Cancer 14:287–295
Woodward AM, Davis TA, Silva AG, Kirk JA, Leary JA (2005) Large genomic rearrangements of both BRCA2 and BRCA1 are a feature of the inherited breast/ovarian cancer phenotype in selected families. J Med Genet 42:e31
Palma MD, Domchek SM, Stopfer J et al (2008) The relative contribution of point mutations and genomic rearrangements in BRCA1 and BRCA2 in high-risk breast cancer families. Cancer Res 68:7006–7014
Ramus SJ, Harrington PA, Pye C et al (2007) Contribution of BRCA1 and BRCA2 mutations to inherited ovarian cancer. Hum Mutat 28:1207–1215
Walsh T, Casadei S, Coats KH et al (2006) Spectrum of mutations in BRCA1, BRCA2, CHEK2, and TP53 in families at high risk of breast cancer. JAMA 295:1379–1388
Armaou S, Pertesi M, Fostira F et al (2009) Contribution of BRCA1 germ-line mutations to breast cancer in Greece: a hospital-based study of 987 unselected breast cancer cases. Br J Cancer 101:32–37
Hogervorst FB, Nederlof PM, Gille JJ et al (2003) Large genomic deletions and duplications in the BRCA1 gene identified by a novel quantitative method. Cancer Res 63:1449–1453
Petrij-Bosch A, Peelen T, van Vliet M et al (1997) BRCA1 genomic deletions are major founder mutations in Dutch breast cancer patients. Nat Genet 17:341–345
Villarreal-Garza C, varez-Gomez RM, Perez-Plasencia C et al (2015) Significant clinical impact of recurrent BRCA1 and BRCA2 mutations in Mexico. Cancer 121:372–378
Cho JY, Cho DY, Ahn SH et al (2014) Large genomic rearrangement of BRCA1 and BRCA2 genes in familial breast cancer patients in Korea. Fam Cancer 13:205–211
Purnomosari D, Pals G, Wahyono A et al (2007) BRCA1 and BRCA2 germline mutation analysis in the Indonesian population. Breast Cancer Res Treat 106:297–304
Kang P, Mariapun S, Phuah SY et al (2010) Large BRCA1 and BRCA2 genomic rearrangements in Malaysian high risk breast-ovarian cancer families. Breast Cancer Res Treat 124:579–584
Kwong A, Chen J, Shin VY et al (2015) The importance of analysis of long-range rearrangement of BRCA1 and BRCA2 in genetic diagnosis of familial breast cancer. Cancer Genet 208:448–454
Lim YK, Lau PT, Ali AB et al (2007) Identification of novel BRCA large genomic rearrangements in Singapore Asian breast and ovarian patients with cancer. Clin Genet 71:331–342
Al-Moundhri MS, Al-Ansari A, Al-Mawali K, Al-Bahrani B (2013) BRCA1 gene molecular alterations in Omani breast cancer patients. Gulf J Oncolog 1:45–51
Ang P, Lim IH, Lee TC et al (2007) BRCA1 and BRCA2 mutations in an Asian clinic-based population detected using a comprehensive strategy. Cancer Epidemiol Biomark Prev 16:2276–2284
De SS, Tennekoon KH, Karunanayake EH, Amarasinghe I, Angunawela P (2014) Analysis of BRCA1and BRCA2 large genomic rearrangements in Sri Lankan familial breast cancer patients and at risk individuals. BMC Res Notes 7:344
Pietschmann A, Mehdipour P, Mehdipour P et al (2005) Mutation analysis of BRCA1 and BRCA2 genes in Iranian high risk breast cancer families. J Cancer Res Clin Oncol 131:552–558
Seong MW, Cho SI, Kim KH et al (2014) A multi-institutional study of the prevalence of BRCA1 and BRCA2 large genomic rearrangements in familial breast cancer patients. BMC Cancer 14:645
Yap KP, Ang P, Lim IH, Ho GH, Lee AS (2006) Detection of a novel Alu-mediated BRCA1 exon 13 duplication in Chinese breast cancer patients and implications for genetic testing. Clin Genet 70:80–82
Rashid MU, Muzaffar M, Khan FA et al (2015) Association between the Bsm I polymorphism in the vitamin D receptor gene and breast cancer risk: results from a Pakistani case–control study. PLoS ONE 10:e0141562
Rashid MU, Muhammad N, Bajwa S et al (2016) High prevalence and predominance of BRCA1 germline mutations in Pakistani triple-negative breast cancer patients. BMC Cancer 16:673
Puget N, Gad S, Perrin-Vidoz L et al (2002) Distinct BRCA1 rearrangements involving the BRCA1 pseudogene suggest the existence of a recombination hot spot. Am J Hum Genet 70:858–865
den Dunnen JT, Antonarakis SE (2000) Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Hum Mutat 15:7–12
Hartmann C, John AL, Klaes R et al (2004) Large BRCA1 gene deletions are found in 3% of German high-risk breast cancer families. Hum Mutat 24:534
Montagna M, Dalla PM, Menin C et al (2003) Genomic rearrangements account for more than one-third of the BRCA1 mutations in northern Italian breast/ovarian cancer families. Hum Mol Genet 12:1055–1061
Agata S, Viel A, Della PL et al (2006) Prevalence of BRCA1 genomic rearrangements in a large cohort of Italian breast and breast/ovarian cancer families without detectable BRCA1 and BRCA2 point mutations. Genes Chromosom Cancer 45:791–797
Aktas D, Gultekin M, Kabacam S et al (2010) Identification of point mutations and large rearrangements in the BRCA1 gene in 667 Turkish unselected ovarian cancer patients. Gynecol Oncol 119:131–135
Arnold AG, Otegbeye E, Fleischut MH et al (2014) Assessment of individuals with BRCA1 and BRCA2 large rearrangements in high-risk breast and ovarian cancer families. Breast Cancer Res Treat 145:625–634
Jackson SA, Davis AA, Li J et al (2014) Characteristics of individuals with breast cancer rearrangements in BRCA1 and BRCA2. Cancer 120:1557–1564
Palanca SS, Esteban CE, Barragan GE et al (2008) Identification of a novel BRCA1 large genomic rearrangement in a Spanish breast/ovarian cancer family. Breast Cancer Res Treat 112:63–67
Ratajska M, Brozek I, Senkus-Konefka E et al (2008) BRCA1 and BRCA2 point mutations and large rearrangements in breast and ovarian cancer families in Northern Poland. Oncol Rep 19:263–268
Unger MA, Nathanson KL, Calzone K et al (2000) Screening for genomic rearrangements in families with breast and ovarian cancer identifies BRCA1 mutations previously missed by conformation-sensitive gel electrophoresis or sequencing. Am J Hum Genet 67:841–850
van den Ouweland AM, Dinjens WN, Dorssers LC et al (2009) Deletion of exons 1a–2 of BRCA1: a rather frequent pathogenic abnormality. Genet Test Mol Biomark 13:399–406
Vasickova P, Machackova E, Lukesova M et al (2007) High occurrence of BRCA1 intragenic rearrangements in hereditary breast and ovarian cancer syndrome in the Czech Republic. BMC Med Genet 8:32
Smith TM, Lee MK, Szabo CI et al (1996) Complete genomic sequence and analysis of 117 kb of human DNA containing the gene BRCA1. Genome Res 6:1029–1049
Laurila E, Syrjakoski K, Holli K, Kallioniemi A, Karhu R (2005) Search for large genomic alterations of the BRCA1 gene in a Finnish population. Cancer Genet Cytogenet 163:57–61
Weitzel JN, Clague J, Martir-Negron A et al (2013) Prevalence and type of BRCA mutations in Hispanics undergoing genetic cancer risk assessment in the southwestern United States: a report from the clinical cancer genetics community research network. J Clin Oncol 31(2):210–216
Reis-Filho JS, Tutt AN (2008) Triple negative tumours: a critical review. Histopathology 52:108–118
Falchetti M, Lupi R, Rizzolo P et al (2008) BRCA1/BRCA2 rearrangements and CHEK2 common mutations are infrequent in Italian male breast cancer cases. Breast Cancer Res Treat 110:161–167
Karhu R, Laurila E, Kallioniemi A, Syrjakoski K (2006) Large genomic BRCA2 rearrangements and male breast cancer. Cancer Detect Prev 30:530–534
Tchou J, Ward MR, Volpe P et al (2007) Large genomic rearrangement in BRCA1 and BRCA2 and clinical characteristics of men with breast cancer in the United States. Clin Breast Cancer 7:627–633
Manguoglu E, Guran S, Yamac D et al (2011) Genomic large rearrangement screening of BRCA1 and BRCA2 genes in high-risk Turkish breast/ovarian cancer patients by using multiplex ligation-dependent probe amplification assay. Cancer Investig 29:73–77
Tournier I, Paillerets BB, Sobol H et al (2004) Significant contribution of germline BRCA2 rearrangements in male breast cancer families. Cancer Res 64:8143–8147
Acknowledgements
We thank Britta Fiebig, Irene Kostantopoulou, Jeffery N. Weitzel, Jenny Leary, Mads Thomassen, and Trinidad Caldes for providing DNA samples of BRCA1/2 LGR controls. This study was supported by the Shaukat Khanum Memorial Cancer Hospital and Research Centre and the German Cancer Research Center. Dr. Rashid’s visit to the German Cancer Research Center has been supported by a Union for International Cancer Control (UICC) Yamagiwa-Yoshida Memorial International Cancer Study Grant.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary material 1 (DOCX 385 kb)
Pedigrees of Pakistani breast and/or ovarian cancer families carrying BRCA1 LGRs. Circles are females, squares are males, and a diagonal slash indicates a deceased individual. Symbols with filled left upper quadrant: unilateral breast cancer. Symbols with filled upper half: bilateral breast cancer. Symbols with filled left lower quadrant: ovarian cancer. Symbols with filled right lower quadrant: cancer other than breast or ovarian cancer, the name of which is mentioned. Identification numbers of individuals are shown below the symbols. The index patient is indicated by an arrow. BC breast cancer, OC ovarian cancer. The numbers following these abbreviations indicate age at cancer diagnosis. M+ mutation positive. M− mutation negative
Rights and permissions
About this article
Cite this article
Rashid, M.U., Muhammad, N., Amin, A. et al. Contribution of BRCA1 large genomic rearrangements to early-onset and familial breast/ovarian cancer in Pakistan. Breast Cancer Res Treat 161, 191–201 (2017). https://doi.org/10.1007/s10549-016-4044-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-016-4044-0