Abstract
Cytochrome P450 2D6 (CYP2D6) plays an important role in the formation of endoxifen, the active metabolite of tamoxifen. In this study the association between the most prevalent CYP2D6 null-allele in Caucasians (CYP2D6*4) and breast cancer mortality was examined among all incident users of tamoxifen in a population-based cohort study. Breast cancer mortality was significantly increased in patients with the * 4/*4 genotype (HR = 4.1, CI 95% 1.1–15.9, P = 0.041) compared to wild type patients. The breast cancer mortality increased with a hazard ratio of 2.0 (CI 95% 1.1–3.4, P = 0.015) with each additional variant allele. No increased risk of all-cause mortality or all-cancer mortality was found in tamoxifen users carrying a CYP2D6*4 allele. The risk of breast cancer mortality is increased in tamoxifen users with decreased CYP2D6 activity, consistent with the model in which endoxifen formation is dependent on CYP2D6 activity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Breast cancer is a major public health problem. In the Netherlands the incidence of breast cancer increased from 85 per 100,000 person years in 1974 to almost 130 per 100,000 person years in 2004 [1]. Tamoxifen is one of the most widely used drugs for post-menopausal women with estrogen receptor-positive breast cancer. In metastatic breast cancer, about 30% of the women respond to tamoxifen therapy [2, 3]. Women with estrogen receptor-positive breast cancer taking adjuvant tamoxifen for 5 years have a decreased risk of breast cancer recurrence and significantly lower mortality rates compared to women not using tamoxifen [2, 3] .
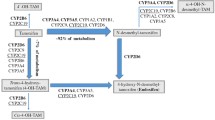
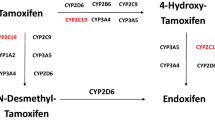
Tamoxifen undergoes extensive hepatic metabolism to more potent metabolites, including 4-hydroxy-tamoxifen and endoxifen (4-hydroxy-N-desmethyl-tamoxfen). Endoxifen and 4-hydroxy-tamoxifen have a 50-fold higher affinity for the estrogen receptor than tamoxifen. Plasma concentrations of endoxifen are on average 5–10 times higher than those of 4-hydroxy-tamoxifen, making endoxifen the most active substance [4, 5]. Cytochrome P450 2D6 (CYP2D6) plays an important role in the formation of endoxifen from tamoxifen [6]. The activity of CYP2D6 is mainly determined by the presence of genetic polymorphisms, rather than by induction or inhibition of expression, giving rise to a 1,000-fold difference in CYP2D6 metabolic capacity [7]. Individuals carrying two non-functional alleles of the CYP2D6 gene lack CYP2D6 enzyme activity and are therefore classified as poor metabolizer (PM), whereas extensive metabolizers (EMs) have 2 functional alleles and exhibit normal enzyme activity. Carriers of one functional and one non-functional allele are usually classified as intermediate metabolizers (IMs) [8, 9]. For CYP2D6, the CYP2D6*4 allele is the most common variant allele in Caucasians leading to the PM phenotype [9, 10].
Homozygosity for CYP2D6 non-functional alleles has been associated with lower plasma concentrations of endoxifen [11, 12]. Goetz et al. were the first to describe that women using adjuvant tamoxifen had a higher risk of breast cancer recurrence and a lower incidence of hot flashes when they had the CYP2D6 *4/*4 genotype [13]. In another study patients with decreased CYP2D6 activity due to co-administration of CYP2D6 inhibitors or *4 carriership had a significantly shorter time to breast cancer recurrence and shorter disease free survival, again illustrating the important role of CYP2D6 in tamoxifen therapy [14]. Recently, Schroth et al. confirmed that patients with impaired CYP2D6 metabolism had an increased risk of breast cancer recurrence and worse event free survival rates [15]. In contrast, two other studies showed no association or a tendency towards a decreased recurrence rate in users of tamoxifen with the CYP2D6*4 variant allele [16–18]. In fact, Wegman et al. surprisingly found that patients carrying a CYP2D6*4 allele surprisingly had a significantly better prognosis than wild type individuals [17, 18]. A comparable, but not statistically significant, result was found by Nowell et al. in which a hazard ratio of 0.67 (95% CI 0.33–1.35) on progression free survival was found in CYP2D6*4 carriers [16].
The differences in the effect of the CYP2D6*4 polymorphism on breast cancer survival found among the different studies are as yet unresolved, and therefore additional studies are needed, as indicated by Lash et al. [19].
The current study investigated the association between the CYP2D6*4 polymorphism and breast cancer mortality in incident tamoxifen users in a population-based cohort study.
Methods
Setting
Data were obtained from the Rotterdam Study, a population-based cohort study among 7,983 persons aged 55 years and older [20, 21]. The Medical Ethics Committee of the Erasmus Medical Center approved the study and written informed consent was obtained from all participants. Baseline examination took place between 1990 and 1993 and consisted of a home interview followed by two visits to the research center. Blood samples were obtained from which DNA was isolated. Since the start of the study, participants have been re-examined periodically. To identify all mortality cases, the vital status of the participants was obtained regularly from the municipal population registry. The cause of death was established by information from the general practitioner, including medical history, and in case of hospitalization, discharge reports from medical specialists were obtained. Two research physicians coded all events independently according to the International Classification of Diseases-10th edition. Information on medication use for all participants was available since January 1991. The seven computerized pharmacies cover the research area and provide information on the drug dispensed (Anatomical Therapeutical Chemical (ATC)-code), dispensing date, the total amount of drug units per prescription and the prescribed daily number of units of the drugs.
Study design and outcomes
The study cohort consisted of all women in the Rotterdam Study, who received a first prescription of tamoxifen between April 1st 1991 and July 1st 2005. Subjects in whom no CYP2D6 genotype was available and who received tamoxifen in the first 3 months of available pharmacy data were excluded from the analysis in order to have a complete medication survey and to include only incident users. Participants should at least have a follow-up of 180 days. Subjects were followed from their first tamoxifen prescription until death or the end of the study period whichever came first. Cancer mortality and breast cancer mortality were independently assessed by two medical doctors on the basis of the medical record and pathology data according to the International Classification of Diseases (ICD-10). In case of discrepancy, a cancer epidemiologist decided.
Genotyping
All participants in the Rotterdam Study were genotyped for the CYP2D6*4 polymorphism (1846G>A) as described earlier [22]. Briefly, 1 ng of genomic DNA was amplified in 40 cycles of denaturation at 92°C for 15 s and annealing and extension at 60°C for 1 min using Taqman assays (Applied Biosystems, Foster City, USA).
Individuals were classified as homozygous *4/*4 (PM), heterozygous *1/*4 (IM) or, in the absence of the 1846G>A SNP, as *1/*1 (EM).
Statistical Analysis
The association between CYP2D6 genotype and mortality due to any cause, cancer mortality, or mortality due to breast cancer was examined using Cox proportional hazard models with drug exposure as time-dependent variable. In this model the mortality date was taken as the index date. To each mortality case, all persons using tamoxifen who were still alive on the index date of the case were matched on duration of use of tamoxifen. Analyses were adjusted for age at the index date, total tamoxifen duration, average tamoxifen dose and calendar time. Co-administration of strong CYP2D6 inhibitors (fluoxetine, paroxetine, bupropion, quinidine) or weak inhibitors (sertraline, duloxetine, cimetidine, terbinafine, amiodaron) was considered as potential confounder or effect modifier. Confounders were adjusted for in the analyses if they caused a change in the point estimate of more than 10 percent. The association between CYP2D6 and breast cancer mortality was studied with an allele-effect model (gene-dose effect), with a genotype-effect model (CYP2D6*1/*1, *1/*4, *4/*4 separately), and with a dominant/recessive model (*1/*4 and *4/*4 vs. *1/*1 or *4/*4 vs. *1/*4 and *1/*1).
In addition, we used all women in the Rotterdam Study, in whom information on CYP2D6 genotype was available, to examine the association between CYP2D6 and breast cancer mortality in the whole population including non-users.
Genotype frequencies were tested for Hardy Weinberg equilibrium using a Chi-square test. All analyses were performed using SPSS software (version 11.0, Chicago, USA).
Results
Of the 4,878 women in the Rotterdam Study 108 patients used tamoxifen at any time during the study period. CYP2D6 genotype was known in 85 of these patients. In the other 23 patients there was no DNA sample available for genotyping. The characteristics of the study population are given in Table 1. The allele frequency of the CYP2D6*4 allele was 21.8%. Genotype frequencies were in Hardy-Weinberg Equilibrium (χ2 = 0.0003; P = 0.987).
The association between CYP2D6 genotype and mortality is shown in Table 2. The risk of all-cause mortality did not significantly differ between the different CYP2D6 genotypes in tamoxifen users. There was also no increased risk of cancer mortality in PMs compared to EMs. However, in an allele-effect model there was a significantly increased breast cancer mortality risk with a hazard ratio of 2.0 per additional variant allele (CI 95% 1.1–3.4, P = 0.015). In a genotype-effect analysis, the risk of death due to breast cancer was significantly increased in tamoxifen users with the CYP2D6 *4/*4 genotype (HR = 4.1, CI 95% 1.1–15.9, P = 0.041) compared to EMs. In IMs using tamoxifen there was a non-significantly increased risk of breast cancer mortality (HR = 1.9, CI 95% 0.9–3.9, P = 0.075).
Taking homozygous and heterozygous *4 individuals together in a dominant model, the breast cancer mortality risk was 2.1 (CI 95% 1.1–4.2, P = 0.031). These results are graphically represented in Fig. 1. There was no increased mortality due to breast cancer among women with the CYP2D6 *4/*4 genotype in the whole population (HR = 1.1; CI 95% 0.3–3.6, P = 0,88).
Co-administration of CYP2D6 inhibitors (fluoxetine, paroxetine, sertraline, cimetidine, amiodaron) occurred in 11 subjects (13%) taking tamoxifen. The duration of use varied from 15 days to 1.7 year. Co-administration of any CYP2D6 inhibitor did not influence our model as a confounder or effect modifier. In order to assess the potential effect of CYP2D6 co-medication, we analyzed a CYP2D6*1/*1 person concurrently prescribed a CYP2D6 inhibitor as an intermediate metabolizer. Likewise, a heterozygous CYP2D6*1/*4 was analyzed as a poor metabolizer when taking CYP2D6 co-medication. In this way, the risk of breast cancer mortality in poor metabolizers was 4.0 instead of 4.1 (P = 0.025).
Discussion
Our population-based study showed that tamoxifen users with a decreased CYP2D6 enzyme activity are at increased risk of breast cancer mortality. Per additional variant allele the hazard ratio of breast cancer mortality increased with a factor 2. These results support previous findings of Goetz et al. and Schroth et al. [13–15], and are consistent with the model in which endoxifen formation, the most active substance of tamoxifen therapy, is dependent on CYP2D6 activity. Interestingly, we found a statistical significant effect despite the small group (n = 4) of CYP2D6 PMs, indicating that the impact of this phenotype on breast cancer survival can be impressive. In our study intermediate metabolizers tended to have an increased risk of breast cancer mortality, suggesting an allele dose effect rather than a recessive model where the risk increase would be limited to *4/*4 homozygous individuals. Goetz et al. included the heterozygous *4 carriers (with or without potent CYP2D6 inhibitors) in the decreased enzyme activity group [14]. They found an increased risk of breast cancer recurrence in tamoxifen users with decreased CYP2D6 activity (HR = 1.91) and worse relapse free survival (HR = 1.74). IMs in that study did not have a shorter time to breast cancer recurrence, but tended to have worse relapse-free survival, albeit marginally non-significant (P = 0.07).
Co-administration of CYP2D6 inhibitors leads to lower endoxifen plasma concentrations [11, 12] and Goetz et al. showed that in subjects taking CYP2D6 inhibitors tamoxifen presumably was less effective. In our study, the use of CYP2D6 co-medication was limited to approximately 10% of the patients. When we took CYP2D6 co-medication into account, we found a comparable risk of breast cancer mortality in CYP2D6 PMs as to analysis with genotype alone, but concurrent use of CYP2D6 inhibitors was scarce and differed in duration of use.
Potential biases of population-based studies are selection bias, information bias and confounding. In our study selection bias probably did not occur, since all tamoxifen users were selected independently of CYP2D6 metabolizer status in a large cohort study. Patients for whom no blood sample was available were slightly younger (mean age of 74.7) and probably more diseased, but missing blood samples were not likely to be related to CYP2D6 genotype. Information bias is unlikely as both information on exposure and disease were gathered prospectively and without knowledge of the research hypothesis and genotype status. Some random misclassification may have occurred, because only the *4 variant allele of the CYP2D6 gene was determined in our study. This variant is by far the most common polymorphism in Caucasians and >75% of PMs can be identified by genotyping this polymorphism [9, 10]. Other less frequent CYP2D6 non-functional alleles like *3 and *6 were not determined, based on their low allele frequencies in Caucasians [10]. The CYP2D6*5 allele (gene deletion), which has an allele frequency of 5% was incorporated in our assay, because *4/*5 individuals will be scored as *4/*4, with the correct PM phenotype. In the heterozygous CYP2D6*1/*4 patients, no *5 allele can be present. Only in the 52 individuals of the CYP2D6*1/*1 group, 5% (2–3 individuals) may have had a *1/*5 genotype which was missed, misclassifying these patients as EM instead of IM. However, this misclassification would lead to a conservative estimation of the currently described association. We adjusted for age, total tamoxifen use, average tamoxifen dose and calendar time. Although we had no complete information on breast cancer stage, tumor size, nodal stage or estrogen receptor (ER) status, CYP2D6 genotype is not known as being associated with these parameters. Schroth et al. did not find a correlation between genotype and tumor size, nodal stage, histologic grade or ER status [15]. From the information available, we deduced that approximately 75% of the women in our study received tamoxifen for metastatic breast cancer. Most studies on tamoxifen efficacy and CYP2D6 are performed in patients taking adjuvant therapy.
Improved efficacy of the aromatase inhibitor anastrozole compared to tamoxifen in metastatic breast cancer and as adjuvant treatment for breast cancer was reported in other studies [23, 24]. The difference could be explained by a worse outcome in CYP2D6 PMs using tamoxifen. Modeling suggested that breast cancer survival outcomes in tamoxifen EMs are indeed similar or even superior to those in aromatase inhibitors [25].
Our study demonstrated that the risk of breast cancer mortality is increased in patients carrying the CYP2D6*4 allele. These patients probably benefit more from aromatase inhibitors, which activity is independent of CYP2D6 enzyme activity. Another option for these patients is to increase the dose of tamoxifen. In our study all *4/*4 individuals received tamoxifen 40 mg once daily and still had an increased risk of breast cancer mortality (after adjustment of dose).
Genotyping of the CYP2D6 gene before start of endocrine treatment in breast cancer could identify PMs who will better respond to aromatase inhibitors than to tamoxifen therapy. CYP2D6 EMs could be prescribed tamoxifen, since aromatase inhibitors are more expensive. Cost-effectiveness studies should be done in order to support the implementation of CYP2D6 genotyping in clinical practice.
In conclusion, our population-based study does confirm that tamoxifen users with decreased CYP2D6 activity have an increased risk of breast cancer mortality. Other drugs may have to be considered in these patients.
References
Louwman WJ, Voogd AC, van Dijck JA et al (2008) On the rising trends of incidence and prognosis for breast cancer patients diagnosed 1975–2004: a long-term population-based study in southeastern Netherlands. Cancer Causes Control 19(1):97–106. doi:10.1007/s10552-007-9075-8
Osborne CK (1998) Tamoxifen in the treatment of breast cancer. N Engl J Med 339(22):1609–1618. doi:10.1056/NEJM199811263392207
Early Breast Cancer Trialists’ Collaborative Group (2005) Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet 365(9472):1687–1717. doi:10.1016/S0140-6736(05)66544-0
Jordan VC, Collins MM, Rowsby L, Prestwich G (1977) A monohydroxylated metabolite of tamoxifen with potent antioestrogenic activity. J Endocrinol 75(2):305–316
Stearns V, Johnson MD, Rae JM et al (2003) Active tamoxifen metabolite plasma concentrations after coadministration of tamoxifen and the selective serotonin reuptake inhibitor paroxetine. J Natl Cancer Inst 95(23):1758–1764
Desta Z, Ward BA, Soukhova NV, Flockhart DA (2004) Comprehensive evaluation of tamoxifen sequential biotransformation by the human cytochrome P450 system in vitro: prominent roles for CYP3A and CYP2D6. J Pharmacol Exp Ther 310(3):1062–1075. doi:10.1124/jpet.104.065607
Ingelman-Sundberg M, Sim SC, Gomez A, Rodriguez-Antona C (2007) Influence of cytochrome P450 polymorphisms on drug therapies: pharmacogenetic, pharmacoepigenetic and clinical aspects. Pharmacol Ther 116(3):496–526. doi:10.1016/j.pharmthera.2007.09.004
Sachse C, Brockmoller J, Bauer S, Roots I (1997) Cytochrome P450 2D6 variants in a Caucasian population: allele frequencies and phenotypic consequences. Am J Hum Genet 60(2):284–295
Bradford LD (2002) CYP2D6 allele frequency in European Caucasians, Asians, Africans and their descendants. Pharmacogenomics 3(2):229–243. doi:10.1517/14622416.3.2.229
Gaedigk A, Gotschall RR, Forbes NS, Simon SD, Kearns GL, Leeder JS (1999) Optimization of cytochrome P4502D6 (CYP2D6) phenotype assignment using a genotyping algorithm based on allele frequency data. Pharmacogenetics 9(6):669–682. doi:10.1097/00008571-199912000-00002
Jin Y, Desta Z, Stearns V et al (2005) CYP2D6 genotype, antidepressant use, and tamoxifen metabolism during adjuvant breast cancer treatment. J Natl Cancer Inst 97(1):30–39
Borges S, Desta Z, Li L et al (2006) Quantitative effect of CYP2D6 genotype and inhibitors on tamoxifen metabolism: implication for optimization of breast cancer treatment. Clin Pharmacol Ther 80(1):61–74. doi:10.1016/j.clpt.2006.03.013
Goetz MP, Rae JM, Suman VJ et al (2005) Pharmacogenetics of tamoxifen biotransformation is associated with clinical outcomes of efficacy and hot flashes. J Clin Oncol 23(36):9312–9318. doi:10.1200/JCO.2005.03.3266
Goetz MP, Knox SK, Suman VJ et al (2007) The impact of cytochrome P450 2D6 metabolism in women receiving adjuvant tamoxifen. Breast Cancer Res Treat 101(1):113–121. doi:10.1007/s10549-006-9428-0
Schroth W, Antoniadou L, Fritz P et al (2007) Breast cancer treatment outcome with adjuvant tamoxifen relative to patient CYP2D6 and CYP2C19 genotypes. J Clin Oncol 25(33):5187–5193. doi:10.1200/JCO.2007.12.2705
Nowell SA, Ahn J, Rae JM et al (2005) Association of genetic variation in tamoxifen-metabolizing enzymes with overall survival and recurrence of disease in breast cancer patients. Breast Cancer Res Treat 91(3):249–258. doi:10.1007/s10549-004-7751-x
Wegman P, Vainikka L, Stal O et al (2005) Genotype of metabolic enzymes and the benefit of tamoxifen in postmenopausal breast cancer patients. Breast Cancer Res 7(3):R284–R290. doi:10.1186/bcr993
Wegman P, Elingarami S, Carstensen J, Stal O, Nordenskjold B, Wingren S (2007) Genetic variants of CYP3A5, CYP2D6, SULT1A1, UGT2B15 and tamoxifen response in postmenopausal patients with breast cancer. Breast Cancer Res 9(1):R7. doi:10.1186/bcr1640
Lash TL, Ahern TP, Cronin-Fenton D et al (2008) Modification of tamoxifen response: what have we learned? J Clin Oncol 26(10):1764–1765. doi:10.1200/JCO.2007.15.5432
Hofman A, Grobbee DE, de Jong PT, van den Ouweland FA (1991) Determinants of disease and disability in the elderly: the Rotterdam Elderly Study. Eur J Epidemiol 7(4):403–422. doi:10.1007/BF00145007
Hofman A, Breteler MM, van Duijn CM et al (2007) The Rotterdam study: objectives and design update. Eur J Epidemiol 22(11):819–829. doi:10.1007/s10654-007-9199-x
Bijl MJ, Visser LE, Hofman A et al (2008) Influence of the CYP2D6*4 polymorphism on dose, switching and discontinuation of antidepressants. Br J Clin Pharmacol 65(4):558–564. doi:10.1111/j.1365-2125.2007.03052.x
Bonneterre J, Buzdar A, Nabholtz JM et al (2001) Anastrozole is superior to tamoxifen as first-line therapy in hormone receptor positive advanced breast carcinoma. Cancer 92(9):2247–2258. doi:10.1002/1097-0142(20011101)92:9<2247::AID-CNCR1570>3.0.CO;2-Y
Forbes JF, Cuzick J, Buzdar A, Howell A, Tobias JS, Baum M (2008) Effect of anastrozole and tamoxifen as adjuvant treatment for early-stage breast cancer: 100-month analysis of the ATAC trial. Lancet Oncol 9(1):45–53. doi:10.1016/S1470-2045(07)70385-6
Punglia RS, Burstein HJ, Winer EP, Weeks JC (2008) Pharmacogenomic variation of CYP2D6 and the choice of optimal adjuvant endocrine therapy for postmenopausal breast cancer: a modeling analysis. J Natl Cancer Inst 100(9):642–648. doi:10.1093/jnci/djn100
Acknowledgments
This work is supported by the Netherlands Genomics Initiative (NGI)/Netherlands Organisation for Scientific Research (NWO) [050.060.810]. The Rotterdam Study is supported by the Erasmus Medical Center and Erasmus University Rotterdam; the Netherlands Organization for Scientific Research; the Netherlands Organization for Health Research and Development (ZonMw); the Research Institute for Diseases in the Elderly; the Ministry of Education, Culture and Science; the Ministry of Health Welfare and Sports; the European Commission; and the Municipality of Rotterdam.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bijl, M.J., van Schaik, R.H.N., Lammers, L.A. et al. The CYP2D6*4 polymorphism affects breast cancer survival in tamoxifen users. Breast Cancer Res Treat 118, 125–130 (2009). https://doi.org/10.1007/s10549-008-0272-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-008-0272-2