Abstract
Enhancer of zeste homolog 2 (EZH2) is a histone methyltransferase polycomb group (PcG) protein, which has been implicated in the process of cellular differentiation and cancer progression for both breast and prostate cancer. Although transcriptional repression by histone modification appears to contribute to the process of cellular differentiation, it is unclear what mediates the specificity of PcG proteins. Since EZH2 requires a binding partner for its histone methyltransferase activity, we surmised that evaluating interacting proteins might shed light on how the activity of EZH2 is regulated. Here we describe the identification of a novel binding partner of EZH2, the repressor of estrogen receptor activity (REA). REA functions as a transcriptional corepressor of the estrogen receptor and can potentiate the effect of anti-estrogens. REA expression levels have also previously been associated with the degree of differentiation of human breast cancers. We show here that EZH2 can also mediate the repression of estrogen-dependent transcription, and that moreover, the ability of both REA and EZH2 to repress estrogen-dependent transcription are mutually dependent. These data suggest that EZH2 may be recruited to specific target genes by its interaction with the estrogen receptor corepressor REA. The identification of a novel interaction between EZH2 and REA, two transcription factors that have been linked to breast cancer carcinogenesis, may lead to further insights into the process of deregulated gene expression in breast cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
A growing body of evidence is accumulating to suggest a relationship between the fields of cancer biology and epigenetics, the study of heritable changes in gene function that occur without a change in DNA sequence [1]. These heritable changes, which are independent of DNA sequence, thus establish cellular identity within a multicellular organism. A major determinant of these heritable changes is modification of chromatin, the term for the structures formed by histone proteins and chromosomal DNA. Transcriptional regulation has been shown to depend in part on the dynamic change between open and compacted chromatin structure. Significantly, a growing number of genes involved in chromatin regulation have been implicated in cancer, including BRCA1, histone deacetylases (HDAC), and members of the polycomb group (PcG) of proteins. Much recent interest has focused on EZH2, a PcG protein. EZH2 is a homolog of the Drosophila protein Enhancer of zeste, critical to development and cell differentiation [2–5]. The catalytic activity of EZH2 is contained in its SET domain, which methylates histone lysine residues, affecting chromatin structure and gene transcription. The catalytic activity of EZH2 appears to be regulated by its binding partners, since the multimeric complexes containing EZH2 change during differentiation, along with their substrate specificity [6]. EZH2 is critical for embryonic development, as demonstrated by the phenotype of EZH2-null mice, which is embryonic lethal [6]. Furthermore, EZH2 is preferentially expressed in embryonic tissues and is normally not expressed in terminally differentiated adult tissues. In contrast, EZH2 appears to be overexpressed in various malignancies, including steroid-dependent malignancies such as breast cancer, prostate cancer, multiple myeloma and lymphoma [7–18]. EZH2 overexpression also correlates strongly with tumor differentiation and pathologic grade for both breast and prostate cancer [7–9]. This observation may follow from the preferential expression of EZH2 in embryonic over terminally differentiated tissues. In addition to the finding of EZH2 overexpression in neoplastic cells, EZH2 overexpression is a powerful independent prognostic factor, correlating with metastatic disease and survival for both prostate and breast cancer [7, 8]. EZH2 also appears to be important in conferring a malignant phenotype to cells, and may be a bona fide oncogene. Knockdown of EZH2 dramatically reduces proliferation and induces growth arrest in prostate cancer and multiple myeloma cell lines [7, 10, 17]. Conversely, overexpression of EZH2 contributes to anchorage-independent growth, an invasive phenotype, and growth-factor independence [7, 10]. Finally, the overexpression of EZH2 is sufficient to induce oncogenic potential in a xenograft model [10].
Although EZH2 appears to play a role in neoplastic transformation, it remains unclear how EZH2 may contribute to the regulation of various developmental programs and their deregulation in cancer. Furthermore, although it has been shown that EZH2 expression correlates with differentiation of hormone-sensitive epithelial tissues such as breast and prostate, the mechanism underlying this correlation has not yet been determined. Since EZH2 requires a binding partner for its histone methyltransferase activity, we postulated that evaluating interacting proteins might shed light on how the activity of EZH2 is regulated. Using a biochemical screen to identify EZH2-interacting proteins, we have identified Repressor of Estrogen receptor Activity (REA) as a novel binding partner of EZH2 and confirmed this interaction in human cells. EZH2 was also found to repress estrogen-dependent signaling in a manner dependent on the presence of REA. Together, these data suggest that EZH2 and REA cooperate to regulate transcription of estrogen-dependent genes and may further our understanding of aberrant transcriptional regulation in hormonally regulated tumors such as breast and prostate cancer.
Materials and methods
Plasmids and RNA interference
The plasmids pEBB, pEBG, pEBB-FLAG, pEBB-HA, pEBB-CTAP, and pFG12 have been described previously [19–22]. EZH2 coding sequence was PCR amplified from an IMAGE EST clone 4521628 and subcloned into pEBB-CTAP, pEBB-FLAG and pEBG to construct pEBB-EZH2-TAP, pEBB-EZH2-FLAG, and pEBG-EZH2, respectively. The REA coding sequence was cloned from IMAGE EST clone 5728265 using PCR amplification and subcloned into pEBB-HA to generate pEBB-REA-HA. Sequences encoding a short-hairpin RNA targeting EZH2 or REA were subcloned into pFG12 for RNA-interference experiments using a lentiviral delivery system. All plasmids were sequenced to confirm correspondence with published data. Small interfering RNA oligonucleotides sequences were submitted to Qiagen for oligonucleotide synthesis. Sequences used for RNA interference are available upon request. pGL3-ERE2bf-TATA-luc and pSG5-ER were kindly provided by R. Koenig.
Cell culture, transfection, and co-precipitation
HEK293 and 293T cells were grown in Dulbecco’s Modified Eagle’s Medium (Cellgro, Herndon, VA, USA) and supplemented with 2 mM l-glutamine and 10% fetal bovine serum. MDA-MB-231 cells were grown in RPMI 1640 supplemented with 1% glutamax and 10% fetal bovine serum. When indicated, phenol-red free media and charcoal-stripped fetal bovine serum (Hyclone, Logan, UT, USA) were used to eliminate background estrogen signaling. A standard calcium phosphate method [23] was used to transfect both plasmid and siRNA into HEK293 cells and 293T cells. For transfection of siRNA, MDA-MB-231 cells were transfected with lipofectamine (Invitrogen, Carlsbad, CA, USA) using a protocol provided by the supplier. Alternatively, the Amaxa nucleofection system was employed, using program A-23 and nucleofector solution T, according to the recommendations of the manufacturer. Lentiviral production using 293T-cells was performed as previously described [22].
For co-precipitation experiments, HEK293 cells were transfected with pEBG-EZH2, pEBB-REA-HA or appropriate control plasmids. Cell lysates were prepared with Triton-X 100 lysis buffer. Glutathione sepharose beads (Amersham, Piscataway, NJ, USA) were added and the samples were incubated at 4°C for 3-6 h. After washing beads four times with lysis buffer, pelleted material was resuspended in LDS loading buffer (Invitrogen) and used for immunoblotting as described below.
Tandem affinity purification
HEK293 cells were transfected with 12 μg of pEBB-EZH2-TAP plasmid per 15-cm plate. After 2 days, cells were harvested and lysed in Triton lysis buffer (25 mM HEPES, 100 mM NaCl, 1 mM EDTA, 10% glycerol, 1% Triton X-100, protease inhibitors); cell lysate was supplemented with NaCl and Nonidet P-40. Lysate was applied to a chromatography column containing IgG-Sepharose beads (Amersham) and incubated for 2 h at 4°C. The column was drained and washed with IPP150 buffer (10 mM Tris–HCl, pH+ 8.0, 150 mM NaCl, 0.1% Nonidet P-40, protease inhibitors) and TEV cleavage buffer (10 mM Tris–HCl, pH+ 8.0, 150 mM NaCl, 0.1% Nonidet P-40, 0.5 mM EDTA, 1 mM dithiothreitol, protease inhibitors). After incubation for 2 h at 16°C in TEV cleavage buffer supplemented with TEV enzyme (Invitrogen), the eluate was collected and supplemented with CaCl2 and IPP150 calmodulin binding buffer (10 mM Tris–HCl, pH+ 8.0, 150 mM NaCl, 0.1% Nonidet P-40, 1 mM magnesium acetate, 1 mM imidazole, 2 mM CaCl2, 10 mM β-mercaptoethanol). The supplemented eluate was applied to a chromatography column containing calmodulin 4B beads (Amersham) and incubated at 4°C for 1 h. After this incubation, the column was drained and washed with IPP150 calmodulin binding buffer. A final eluate was collected after incubation at 4°C with IPP150 calmodulin elution buffer (10 mM Tris–HCl, pH+ 8.0, 150 mM NaCl, 0.1% Nonidet P-40, 1 mM magnesium acetate, 1 mM imidazole, 2 mM EGTA, 10 mM β-mercaptoethanol). Proteins were precipitated by adding cold 10% trichloroacetic acid in acetone and incubation overnight at −20°C. The samples were centrifuged at 4°C (10,000×g for 30 min) and precipitate was then rinsed in 100% acetone and allowed to air dry. These protein precipitates were submitted to the Proteomics Centre (http://www.proteincentre.com) at the University of Victoria for tryptic digestion, high-performance liquid chromatography separation, and tandem mass spectrometry (MS/MS) to determine peptide sequences. REA was identified as a putative EZH2-interacting protein based upon the peptide sequence QVAQQEAQR.
Western blot analyses
Whole cell extracts were prepared with Triton lysis buffer or RIPA lysis buffer (PBS, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) supplemented with 1 mM PMSF, one protease inhibitor tablet (Roche, Penzberg, Germany) per 10 ml of buffer, and 10 mM DTT. Proteins were separated by electrophoresis on 4–12% gradient Novex Bis–Tris gels (Invitrogen), transferred to nitrocellulose membranes (Invitrogen) and blocked with 5% milk solution in TBS containing between 0.05 and 0.1% Tween-20, dependent on the antibody. The membranes were incubated with the relevant primary and secondary antibodies followed by antibody detection. Antibody detection was performed with an enhanced chemiluminescence system according to the manufacturer’s directions (Amersham). Primary HRP-conjugated antibodies against FLAG and HA were obtained from Sigma, St.Louis, MO, USA. Other primary antibodies used included rabbit polyclonal antibodies against GST (Santa Cruz, CA, USA), REA and EZH2 (Upstate, NJ, USA), and a mouse monoclonal antibody against β-actin (Sigma). Secondary antibodies used were HRP-conjugated donkey anti-rabbit and HRP-conjugated sheep anti-mouse (Amersham).
Confocal and fluorescence microscopy
HEK293 cells (5 × 104 per well) were plated on poly lysine coated chambered coverglass slides. Cells were transfected by calcium phosphate precipitation with pEBB-EZH2-FLAG, pEBB-REA-HA or both. Twenty-four hours after transfection cells were washed with D-PBS (Dulbecco’s phosphate-buffered saline), fixed in 4% paraformaldehyde, and then permeablized in a solution of 0.1% Triton X-100/1% goat serum in D-PBS. Antibody staining was completed as follows: cells were blocked with 20% goat serum in D-PBS for 1 h at room temperature and then incubated with primary antibodies (anti-FLAG-FITC, anti-HA-Alexa594) for 1 h at room temperature in the dark. Following three washes with D-PBS, cells were stained with Hoechst 33342, chamber slides were allowed to air dry, and mounted with Pro-long Gold (Molecular probes, Eugene, OR, USA/Invitrogen) according to manufacturer’s instructions. Samples were then examined using a Zeiss Axiovert 100 M Confocal microscope equipped with a Zeiss LSM 510 spectrometer.
Luciferase assay
For luciferase reporter experiments, cells were seeded in 6-well plates and transfected with the relevant plasmids and siRNA, in addition to an estrogen receptor-expressing plasmid (pSG5-ER) and the reporter plasmid pGL3-ERE2bf-TATA-luc. The following day, cells were exposed to β-estradiol (Sigma) at a final concentration of 10−8 M, or an equivalent volume of ethanol, the vehicle used to solubilize β-estradiol. The Promega reporter lysis buffer and luciferase assay kit were used, with assays performed according to the manufacturer’s protocol. Cells were assayed for luciferase reporter activity 1 day after estrogen treatment.
Results
Although EZH2 contains the enzymatic domain responsible for histone methylation, its target specificity appears to be driven by its binding partners [6]. For this reason, and due to the apparent role of EZH2 in cell-identity and cancer progression, we were interested in identifying novel EZH2-binding partners. Using the tandem affinity purification (TAP) system [24], a variety of peptides were isolated and sequenced to identify candidate binding partners of EZH2. This screen suggested an interaction between EZH2 and the Repressor of Estrogen Activity (REA) protein. REA has been reported to act as a corepressor in estrogen-dependent transcription by binding to the estrogen receptor [25]. In addition, a decreased level of REA expression has been correlated with high-grade, poorly differentiated breast cancer [26]. Based on the novel nature of the interaction between EZH2 and REA and the potential role of REA in neoplasia, we chose to further investigate this interaction.
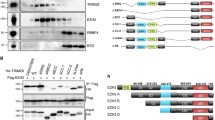
The interaction between REA and EZH2 was confirmed through a GST-fusion protein pull-down experiment. GST was fused to the EZH2 protein and co-transfected into cells with plasmid expressing REA fused to an HA tag. REA-HA was found to co-precipitate with EZH2-GST in extracts from cells transfected with both plasmids (Fig. 1a). REA-HA did not co-precipitate with GST alone, indicating the specific nature of this interaction. The interaction between EZH2 and REA was further analyzed in co-localization experiments. The subcellular localization patterns of both EZH2 and REA were examined using an immunofluorescence approach. HEK293 cells were transfected with plasmids encoding both EZH2-FLAG and REA-HA and stained with FITC-conjugated anti-FLAG and Alexa-594 conjugated anti-HA prior to analysis by confocal microscopy. As shown in Fig. 1b, REA displayed a predominantly cytoplasmic localization, with a small portion of the protein detected in the nucleus. In contrast, EZH2 was completely nuclear, with no detectable fluorescence in the cytoplasm. Interestingly, EZH2 localization within the nucleus was in the form of discrete foci, rather than diffuse within each nucleus.
Interaction between EZH2 and REA. (a) GST-pulldown assay using EZH2-GST. HEK293 cells were co-transfected with REA-HA and either EZH2-GST or GST. Cell lysates were prepared and purified using the affinity of GST for glutathione. Input lysates and pulldowns were immunoblotted and probed for either HA (top panels) or GST (bottom panels). (b) Co-localization of EZH2 and REA. HEK293 cells were co-transfected with separate plasmids constructed to express EZH2-FLAG and REA-HA fusion proteins. Immunofluorescence staining was performed using anti-FLAG (green, left upper panel) and anti-HA antibodies (red, right upper panel). Nuclei were stained with Hoescht (blue, left lower panel). The overlay image is shown in the right lower panel
Since REA has previously been shown to bind the estrogen receptor and repress transcription [25, 27], we asked whether EZH2 might also play a role in estrogen-dependent transcription. RNA interference was used to reduce levels of EZH2 and examine the role of EZH2 in this system. HEK293 cells were transfected with a siRNA oligonucleotide specific for the EZH2 transcript (siEZH2). The protein levels of endogenous EZH2 were specifically and effectively reduced with siEZH2, as shown in Fig. 2a. This siRNA was used to evaluate the effect of EZH2 depletion on the expression of a luciferase reporter construct whose promoter contained canonical estrogen-response elements. As expected, when transfected into both HEK293 cells and MDAMB231 cells, transcription was robustly activated by the addition of estradiol (E2) to the cell media (Fig. 2b, c, first three bars). In both of these systems, there was little basal activity in the absence of estradiol. When EZH2 levels were reduced in HEK293 cells, estrogen-dependent transcription was increased (Fig. 2b). This increase in estrogen-dependent transcription did not occur with a control siRNA oligonucleotide, indicating the specific nature of this result. Similar results were obtained from a parallel analysis in MDA-MB-231 cells, a breast cancer cell line. Estrogen-dependent transcription was increased when EZH2 levels were reduced using siEZH2 (Fig. 2c).
siRNA-mediated reduction in EZH2 expression increases ERE-dependent transcription. (a) HEK293 cells were transfected with either siEZH2 or siGFP using a calcium phosphate method. Lysates were prepared for Western blot analysis and probed using an antibody that recognized endogenous EZH2. Transfection of siEZH2 was able to efficiently decrease EZH2 expression in HEK293 cells. Similar results were achieved using siEZH2 to reduce the expression of EZH2 in MDA-MB-231 breast cancer cells. (b) Estrogen-dependent transcription was evaluated using a luciferase reporter system. Transcription of the luciferase gene from the plasmid pGL3-ERE2bf-TATA-luc is estrogen-dependent due to the two estrogen-response elements (ERE) in its promoter. pSG5-ER, which expresses the estrogen receptor (ER), was co-transfected with pGL3-ERE2bf-TATA-luc to enable estrogen-mediated transcription. Cells were cultured in phenol red-free media supplemented with charcoal-stripped serum to eliminate substances that would cross-react with the estrogen receptor. Luciferase transcription was confirmed to be estrogen dependent (+E2). Transcription increased with transfection of siEZH2 compared to siGFP control. (c) Estrogen-dependent transcription was also evaluated in MDA-MB-231 cells in a similar fashion. EZH2 levels were reduced with the introduction of a small interfering RNA oligonucleotide and effect on transcription of pGL2-ERE2bf-TAT-luc was examined.
Since decreasing the expression of EZH2 increased ERE-dependent transcription, we also asked if the converse was also true; that is, if overexpression of EZH2 would repress transcription in this system. EZH2- and REA-expressing plasmids were transfected into an MDA-MB-231 cell line. Consistent with previous reports, REA repressed transcription from the ERE-luciferase reporter plasmid. Moreover, overexpression of EZH2 was also shown to repress transcription from an estrogen-dependent promoter, a novel observation. The effects of REA and EZH2 on estrogen-dependent transcription were further examined to determine if their effects were additive. When REA and EZH2 were co-transfected, transcriptional levels were repressed, but only to the same level as with REA or EZH2 alone, indicating that the transcriptional effects of REA and EZH2 in this system were not additive (Fig. 3b).
Effects of EZH2 and REA expression on ERE-transcription in a lentiviral system. (a) MDA-MB-231 cells were infected with lentivirus constructs designed to express short hairpin RNA that interfere with either REA, EZH2 or GFP expression. Immunoblotting with anti-REA (left panel) or anti-EZH2 (right panel) demonstrates reduced expression of target proteins. (b) Cell lines were transfected and luciferase activity was determined under various conditions: control (shGFP and pEBB); EZH2 (shGFP and pEBB-EZH2-Flag); REA (shGFP and pEBB-REA-HA); EZH2 + REA (shGFP, pEBB-EZH2-Flag, and pEBB-REA-HA); shREA (shREA and pEBB); shEZH2 (shEZH2 and pEBB). (c) Cell lines were transfected as follows and luciferase activity was determined: control (shGFP and pEBB); EZH2 and sh control (shGFP and pEBB-EZH2-Flag); EZH2 and shREA (shREA and pEBB-EZH2-Flag); REA and sh control (shGFP and pEBB-REA-HA); REA and shEZH2 (shFZH2 and pEBB-REA-HA)
Finally, the repression of estrogen-dependent transcription by REA and EZH2 was investigated to assess whether REA and EZH2 acted independently or were mutually dependent. To answer this question, cell lines in which EZH2 or REA expression was stably reduced were established. A decrease in REA and EZH2 expression is demonstrated by Western blot of lysates from these cells (Fig. 3a). ERE-dependent transcription in these cell lines was examined. In a manner similar to our results with transient reduction of EZH2, transcription from the ERE-luciferase reporter construct increased when either REA or EZH2 levels were reduced in a stable fashion (Fig. 3b). The effect of EZH2 or REA overexpression was then evaluated in the absence of the other protein. Significantly, EZH2-mediated repression was abrogated in cell lines expressing the shREA knockdown construct (Fig. 3c). In addition, reducing the expression of EZH2 resulted in attenuation of REA-mediated repression. These results indicate that the repression of estrogen-dependent transcription by EZH2 is dependent on the presence of REA and that the transcriptional repression of REA also depends in part upon EZH2. Together, these data suggest a model in which EZH2 and REA are recruited together to promoters containing estrogen-response elements to modulate ER-dependent transcription.
Discussion
In this study, we report a novel interaction between the transcription factors EZH2 and REA. The protein REA was identified based on a peptide sequence obtained from a biochemical screen searching for EZH2-binding partners. This interaction was confirmed using full-length protein and a GST-pulldown technique (Fig. 1a). Examination of EZH2 and REA localization patterns also displayed significant overlap between their subcellular localization (Fig. 1b). EZH2 and REA were observed to co-localize within discrete nuclear foci. Interestingly, only a subset of REA was found to localize in these nuclear foci while the remainder was seen within the cytoplasm. On the other hand, EZH2 was only visualized within these discrete nuclear foci.
EZH2 was also shown to modulate the transcriptional activity of the estrogen receptor (Figs. 2, 3). The ability of EZH2 to methylate specific histone lysine residues has been associated with a compacted chromatin structure and transcriptional repression. It is possible that the transcriptional repression seen in response to overexpression of EZH2 may be a nonspecific effect. However, the repression of EZH2 of estrogen-repressor mediated transcription appears to be dependent on REA. When the expression of REA was silenced by an RNA interference approach, there was no repression of estrogen receptor transcription seen with EZH2 overexpression. In addition, silencing EZH2 repression partially reverses the transcriptional repression seen with REA. These data suggest that the interaction of REA and EZH2 is responsible for the recruitment of EZH2 to ERE-containing promoters and the dampening of ER transcription.
Kurtev et al. have previously shown that REA interacts with both classes I and II HDAC [28]. HDAC are also chromatin-modifying enzymes that have been implicated in development and carcinogenesis. Based on their observations, Kurtev et al. proposed that REA acts as a corepressor by recruiting HDAC to nuclear receptor target genes. They demonstrated that REA-mediated transcriptional repression of estrogen receptor activity is partially reversed by a specific histone deacetylase inhibitor, trichostatin A. Because this reversal was only partial, they suggested that the transcriptional effects of REA may be mediated in part by a HDAC-independent mechanism. This conjecture is further supported by their demonstration that REA contains a domain able to repress ER transcription but does not bind to HDAC. The interaction of REA with EZH2, a histone lysine methyltransferase, may, at least in part, explain the HDAC-independent repressive properties of REA.
Although EZH2 has been associated with embryonic development and the determination of cellular identity, it is unclear how EZH2-mediated repression coordinates the transcription of a specific set of genes involved in differentiation. REA also appears to play a critical role in embryonal development, as REA-deficiency in the mouse is embryonic lethal at embryonic day 9 [29]. REA acts a corepressor in estrogen-dependent transcription by binding to the estrogen receptor [25, 27]. Significantly, a decreased level of REA expression is correlated with high-grade, poorly differentiated breast cancer [26, 30]. This may be due to the repression of genes that are normally induced by estrogen and the estrogen receptor. The interaction of EZH2 and REA may, in a similar fashion, shed light on the association seen between EZH2 overexpression and the degree of differentiation seen in human breast cancer cases.
The association of nuclear receptors and chromatin remodeling proteins in cancer echoes findings of other model systems. For example, the remarkable clinical success of differentiation therapy in acute promyelocytic leukemia was dependent on the ability of retinoic acid to reverse the differentiation block induced by aberrant recruitment of HDAC to retinoic acid target genes [31]. This HDAC recruitment is mediated by N-Cor and SMRT, corepressors of the retinoic acid receptor. In a similar fashion, mounting evidence also points to the importance of histone-modifying enzymes in breast and prostate cancer, solid tumors, which are steroid hormone-dependent. The data presented here contribute to our understanding of the relationship between chromatin-modifying enzymes such as EZH2 in the differentiation of hormonally regulated tissues. Unraveling the key determinants of differentiation in these cell types and neoplasms may yield to the identification of potential targets for differentiation therapy in solid tumors.
References
Ting AH, McGarvey KM, Baylin SB (2006) The cancer epigenome—components and functional correlates. Genes Dev 20:3215–3231
Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y (2002) Role of histone H3 lysine 27 methylation in polycomb-group silencing. Science 298:1039–1043
Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V (2002) Drosophila enhancer of zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal polycomb sites. Cell 111:185–196
Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D (2002) Histone methyltransferase activity associated with a human multiprotein complex containing the enhancer of zeste protein. Genes Dev 16:2893–2905
Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O’Connor MB, Kingston RE, Simon JA (2002) Histone methyltransferase activity of a Drosophila polycomb group repressor complex. Cell 111:197–208
Kuzmichev A, Margueron R, Vaquero A, Preissner TS, Scher M, Kirmizis A, Ouyang X, Brockdorff N, Abate-Shen C, Farnham P, Reinberg D (2005) Composition and histone substrates of polycomb repressive group complexes change during cellular differentiation. Proc Natl Acad Sci USA 102:1859–1864
Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, Rubin MA, Chinnaiyan AM (2002) The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 419:624–629
Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, Ghosh D, Sewalt RG, Otte AP, Hayes DF, Sabel MS, Livant D, Weiss SJ, Rubin MA, Chinnaiyan AM (2003) EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci USA 100:11606–11611
Raaphorst FM, Meijer CJ, Fieret E, Blokzijl T, Mommers E, Buerger H, Packeisen J, Sewalt RA, Otte AP, van Diest PJ (2003) Poorly differentiated breast carcinoma is associated with increased expression of the human polycomb group EZH2 gene. Neoplasia 5:481–488
Croonquist PA, Van Ness B (2005) The polycomb group protein enhancer of zeste homolog 2 (EZH2) is an oncogene that influences myeloma cell growth and the mutant ras phenotype. Oncogene 24:6269–6280
Weikert S, Christoph F, Kollermann J, Muller M, Schrader M, Miller K, Krause H (2005) Expression levels of the EZH2 polycomb transcriptional repressor correlate with aggressiveness and invasive potential of bladder carcinomas. Int J Mol Med 16:349–353
Visser HP, Gunster MJ, Kluin-Nelemans HC, Manders EM, Raaphorst FM, Meijer CJ, Willemze R, Otte AP (2001) The polycomb group protein EZH2 is upregulated in proliferating, cultured human mantle cell lymphoma. Br J Haematol 112:950–958
van Kemenade FJ, Raaphorst FM, Blokzijl T, Fieret E, Hamer KM, Satijn DP, Otte AP, Meijer CJ (2001) Coexpression of BMI-1 and EZH2 polycomb-group proteins is associated with cycling cells and degree of malignancy in B-cell non-Hodgkin lymphoma. Blood 97:3896–3901
Sudo T, Utsunomiya T, Mimori K, Nagahara H, Ogawa K, Inoue H, Wakiyama S, Fujita H, Shirouzu K, Mori M (2005) Clinicopathological significance of EZH2 mRNA expression in patients with hepatocellular carcinoma. Br J Cancer 92:1754–1758
Raaphorst FM, van Kemenade FJ, Blokzijl T, Fieret E, Hamer KM, Satijn DP, Otte AP, Meijer CJ (2000) Coexpression of BMI-1 and EZH2 polycomb group genes in Reed–Sternberg cells of Hodgkin’s disease. Am J Pathol 157:709–715
Dukers DF, van Galen JC, Giroth C, Jansen P, Sewalt RG, Otte AP, Kluin-Nelemans HC, Meijer CJ, Raaphorst FM (2004) Unique polycomb gene expression pattern in Hodgkin’s lymphoma and Hodgkin’s lymphoma-derived cell lines. Am J Pathol 164:873–881
Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K (2003) EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J 22:5323–5335
Breuer RH, Snijders PJ, Smit EF, Sutedja TG, Sewalt RG, Otte AP, van Kemenade FJ, Postmus PE, Meijer CJ, Raaphorst FM (2004) Increased expression of the EZH2 polycomb group gene in BMI-1-positive neoplastic cells during bronchial carcinogenesis. Neoplasia 6:736–743
Duckett CS, Li F, Wang Y, Tomaselli KJ, Thompson CB, Armstrong RC (1998) Human IAP-like protein regulates programmed cell death downstream of Bcl-xL and cytochrome c. Mol Cell Biol 18:608–615
Lewis J, Burstein E, Birkey Reffey S, Bratton SB, Roberts AB, Duckett CS (2004) Uncoupling of the signaling and caspase-inhibitory properties of XIAP. J Biol Chem 279:9023–9029
Burstein E, Hoberg JE, Wilkinson AS, Rumble JM, Csomos RA, Komarck CM, Maine GN, Wilkinson JC, Mayo MW, Duckett CS (2005) COMMD proteins: a novel family of structural and functional homologs of MURR1. J Biol Chem 280:22222–22232
Maine GN, Mao X, Komarck CM, Burstein E (2007) COMMD1 promotes the ubiquitination of NF-kappaB subunits through a cullin-containing ubiquitin ligase. EMBO J 26:436–447
Duckett CS, Nava VE, Gedrich RW, Clem RJ, Van Dongen JL, Gilfillan MC, Shiels H, Hardwick JM, Thompson CB (1996) A conserved family of cellular genes related to the baculovirus iap gene and encoding apoptosis inhibitors. EMBO J 15:2685–2694
Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Seraphin B (2001) The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24:218–229
Delage-Mourroux R, Martini PG, Choi I, Kraichely DM, Hoeksema J, Katzenellenbogen BS (2000) Analysis of estrogen receptor interaction with a repressor of estrogen receptor activity (REA) and the regulation of estrogen receptor transcriptional activity by REA. J Biol Chem 275:35848–35856
Simon SL, Parkes A, Leygue E, Dotzlaw H, Snell L, Troup S, Adeyinka A, Watson PH, Murphy LC (2000) Expression of a repressor of estrogen receptor activity in human breast tumors: relationship to some known prognostic markers. Cancer Res 60:2796–2799
Montano MM, Ekena K, Delage-Mourroux R, Chang W, Martini P, Katzenellenbogen BS (1999) An estrogen receptor-selective coregulator that potentiates the effectiveness of antiestrogens and represses the activity of estrogens. Proc Natl Acad Sci USA 96:6947–6952
Kurtev V, Margueron R, Kroboth K, Ogris E, Cavailles V, Seiser C (2004) Transcriptional regulation by the repressor of estrogen receptor activity via recruitment of histone deacetylases. J Biol Chem 279:24834–24843
Park SE, Xu J, Frolova A, Liao L, O’Malley BW, Katzenellenbogen BS (2005) Genetic deletion of the repressor of estrogen receptor activity (REA) enhances the response to estrogen in target tissues in vivo. Mol Cell Biol 25:1989–1999
Murphy LC, Simon SL, Parkes A, Leygue E, Dotzlaw H, Snell L, Troup S, Adeyinka A, Watson PH (2000) Altered expression of estrogen receptor coregulators during human breast tumorigenesis. Cancer Res 60:6266–6271
Claus R, Lubbert M (2003) Epigenetic targets in hematopoietic malignancies. Oncogene 22:6489–6496
Acknowledgments
We thank Dr. Ron Koenig of the University of Michigan for his kind gift of the ER-expressing and ERE-luciferase reporter plasmids. We also acknowledge members of the Duckett lab for helpful discussions and critical reading of this manuscript. In addition, we specifically thank Karolyn Oetjen and David Kosoff for their contributions to this work. This work was supported by a grant from the Department of Defense (W81XWH-04-1-0314).
Author information
Authors and Affiliations
Corresponding author
Additional information
Clara Hwang and Veda N. Giri contributed equally to this work.
Rights and permissions
About this article
Cite this article
Hwang, C., Giri, V.N., Wilkinson, J.C. et al. EZH2 regulates the transcription of estrogen-responsive genes through association with REA, an estrogen receptor corepressor. Breast Cancer Res Treat 107, 235–242 (2008). https://doi.org/10.1007/s10549-007-9542-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-007-9542-7