Abstract
Flow-through aquifer columns were operated for 12 weeks to evaluate the benefits of aerobic biostimulation for the bioremediation of source-zone soil contaminated with chlorobenzenes (CBs). Quantitative Polymerase Chain Reaction (qPCR) was used to measure the concentration of total bacteria (16S rRNA gene) and oxygenase genes involved in the biodegradation of aromatic compounds (i.e., toluene dioxygenase, ring hydroxylating monooxygenase, naphthalene dioxygenase, phenol hydroxylase, and biphenyl dioxygenase). Monochlorobenzene, which is much more soluble than dichlorobenzenes, was primarily removed by flushing, and biostimulation showed little benefit. In contrast, dichlorobenzene removal was primarily due to biodegradation, and the removal efficiency was much higher in oxygen-amended columns compared to a control column. To our knowledge, this is the first report that oxygen addition can enhance CB source-zone soil bioremediation. Analysis by qPCR showed that whereas the biphenyl and toluene dioxygenase biomarkers were most abundant, increases in the concentration of the phenol hydroxylase gene reflected best the higher dichlorobenzene removal due to aerobic biostimulation. This suggests that quantitative molecular microbial ecology techniques could be useful to assess CB source-zone bioremediation performance.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Chlorobenzenes (CBs) represent a group of chemicals that are widely used as industrial solvents and degreasers, and are commonly encountered in the subsurface near industrial areas where they have been manufactured or used (Howard 1989). CBs include monochlorobenzene (MCB) and the dichlorobenzene (DCB) isomers (1,2-DCB; 1,3-DCB; and 1,4-DCB). These compounds have low USA federal drinking water standards (i.e., maximum contaminant levels [MCLs] ranging from 0.075 mg l−1 for 1,4-DCB to 0.6 mg l−1 for 1,2-DCB) and relatively high water solubility, ranging from ∼75 mg l−1 for 1,4-DCB to ∼500 mg l−1 for MCB.
Bioremediation and monitored natural attenuation (MNA) are among the most cost-effective approaches to manage soil and groundwater contamination by organic pollutants (Alvarez and Illman 2005; McDade et al. 2005). Past research shows that CBs can be degraded by a variety of both aerobic (Adrian et al. 2000; Dermietzel and Vieth 2002; Lorbeer et al. 2002; Vogt et al. 2002, 2004; Wenderoth et al. 2003) and anaerobic bacteria (Adrian et al. 2000; Kao and Prosser 1999; Kaschl et al. 2005; Wenderoth et al. 2003). Biodegradation generally proceeds faster aerobically (Wenderoth et al. 2003), particularly in the case of MCB and 1,4-DCB (Dermietzel and Vieth 2002), and oxygen availability is a common rate-limiting factor for microbial-mediated CB transformation. Several strategies have been developed that oxygenate the subsurface for biostimulating aerobic microorganisms to metabolize CBs, including the addition of hydrogen peroxide (Vogt et al. 2004) and air sparging (Balcke et al. 2004). However, most previous CB biodegradation research and bioremediation efforts have focused on groundwater treatment and plume management (Dermietzel and Vieth 2002; Lorbeer et al. 2002; Vogt et al. 2002, 2004; Wenderoth et al. 2003) rather than on treating the source zone. In the source zone, CBs occur as either adsorbed to the solid phase or as dense non-aqueous phase liquid (DNAPL) that slowly dissolve over many years and emanate groundwater plumes. Thus, plume-treatment technologies address the symptom rather than the cause, and there is a need to develop practical approaches to treat CB source-zones.
Early bioremediation research did not focus on directly treating source-zones due to concerns about the potential microbial toxicity of high contaminant concentrations. However, recent research has shown that microorganisms can increase the concentration gradient and dissolution flux of hydrophobic pollutants (Adamson et al. 2003; Cope and Hughes 2001; Zheng et al. 2001). This has promoted the acceptance of source-zone bioremediation (McGuire et al. 2006), which aims to increase the local flux of contaminants from DNAPLs or solid phase into the aqueous phase through biodegradation and the production of more soluble metabolites that can be more easily detoxified in situ or removed by alternative (plume management) technologies. The potential efficacy of source-zone bioremediation has been recently demonstrated under anaerobic conditions for chlorinated ethenes at the pilot (Adamson et al. 2003; Da Silva et al. 2006; Sleep et al. 2006) and field scales (Lendvay et al. 2003). However, there are no published reports on the use of bioremediation to treat CB source-zones, which provided motivation for this research.
A rigorous performance assessment of bioremediation requires documentation that contaminant removal is due to microbial rather than abiotic processes such as dissolution, dilution and volatilization. This is of particular importance in aerobic environments because of the difficulty in monitoring end-products (e.g., CO2) and the complications presented by quantifying contaminant loss in complex media where multiple phases may be present. Molecular microbial ecology techniques and specific biomarkers are increasingly being used to obtain supporting evidence of bioremediation (Abraham et al. 2005; Alfreider et al. 2002b; Balcke et al. 2004; Beller et al. 2002; Da Silva and Alvarez 2004; Da Silva et al. 2006; Futumata et al. 2001; Wenderoth et al. 2003), including genes coding for chlorocatechol dioxygenases (Alfreider et al. 2002b). Such efforts can be used to establish that specific genes associated with the degradation of target pollutants are present, and that their numbers are higher in the treatment zone compared to background samples. Numerous catabolic biomarkers have been used to quantify the presence of organisms that degrade aromatic compounds (Baldwin et al. 2003; Beller et al. 2002; Suzuki et al. 2000). These include the genes coding for toluene dioxygenase, ring hydroxylating monooxygenase, naphthalene dioxygenase, biphenyl dioxygenase, and phenol hydroxylase. These biomarker assays are relatively straightforward and powerful tools that are ideally suited for determining the efficacy of biostimulation within a source-zone. However, it is unknown whether such biomarkers could be applicable for a performance assessment of CB source-zone bioremediation. Given that the aerobic degradation of chlorinated benzenes is initiated by oxygenase-promoted hydroxylations prior to ring cleavage, these biomarkers represent promising candidates for further study.
In this paper, we report the results of a biostimulation experiment to evaluate the efficacy of aerobic CB source-zone bioremediation. Genotypic shifts associated with the proliferation of genes that code for various enzymes that initiate aerobic biodegradation of aromatic compounds (i.e., oxygenases) were quantified to establish candidate biomarkers for assessment of CB source-zone bioremediation.
Materials and methods
Biostimulation experiment
Flow-through columns were designed and operated to test the ability to enhance CB attenuation via aerobic biostimulation under conditions mimicking those present in a contaminated aquifer that is oxygen-limited. The studies were performed using aquifer material collected from a former chemical manufacturing facility located in the Midwestern United States. Environmental Visualization System software (EVS, Version 7.92) was used to identify the highest concentrations of MCB and DCB in the saturated zone source area to establish suitable locations for collection of soil samples. Samples for the biostimulation experiment were collected using rotasonic drilling techniques, and were packaged in sealed containers having minimal headspace for shipment at 4°C to Rice University. To provide baseline characterization data prior to biostimulation, additional sample aliquots were shipped at 4°C to Seven Trent Laboratories in Savannah, Georgia in laboratory-provided, method-specific containers for chemical and geotechnical analyses. This included soil samples used to characterize background microbial conditions, which were collected from a soil boring located approximately 700 m (cross-gradient) from the CB source-zone. This background soil sample was collected from the same stratigraphic horizon as the samples collected for the biostimulation experiment (12–15 m below ground surface). The boring was located adjacent to a separate mixed-waste disposal site, but chemical analysis of the soil indicated that the horizon used for background characterization did not contain detectable concentrations of volatile or semi-volatile organic compounds. The aquifer material used in these experiments (both biostimulated and background samples) uniformly consisted of fine to medium-grained sand, with an estimated bulk density of 1.7 g ml−1.
Flow-through aquifer columns were constructed using custom made (Specialty Glass, Houston, TX) 7.6 cm diameter and 15.2 cm long columns (total volume 695 ml) that were packed with site soil. Fitted caps at both ends were constructed to ensure gas-tight, non-reactive conditions. To mimic the in-flow of clean water from upgradient areas through the source zone at the site, water was introduced in an upflow mode via peristaltic pumps at a rate designed to match the groundwater velocity at the site (8.7 cm d−1). The influent solution was a bicarbonate-buffered mineral medium with geochemistry similar to the site groundwater, using deionized water supplemented with the following constituents (in mg l−1): MgSO4·7H2O (50), FeSO4·7H2O (3), (NH4)2SO4 (500), K2HPO4 (1,750), KH2PO4 (1,380), NaHCO3 (500), MnSO4·H2O (2), H3BO3 (0.1), CaSO4·5H2O (0.05), ZnSO4·7H2O (0.05), Na2MoO4·2H2O (0.05), H3BO3 (0.1), and CoSO4·7H2O (0.7). After a 2-week equilibration period, industrial grade pure oxygen (100% v/v) was sparged continuously into the influent reservoir providing oxygen-saturated water (∼48 mg l−1) for the duration of the experiment. The influent reservoir was maintained in a refrigerated environment to simulate the average site groundwater temperature of 13–16°C.
A total of 8 columns underwent aerobic biostimulation (i.e., fed with oxygen-saturated medium). In addition to these oxygen-amended columns, a control column was fed from a separate reservoir with N2-sparged medium (DO < 2 mg l−1) amended with sodium azide (10 mg l−1). The azide concentration used in the study was sublethal (Lichstein and Soule 1943) and resulted in bacteriostatic (rather than bactericidal) effects that decreased aerobic respiration and biodegradation activity. The azide-amended control column did not serve as a sterile control; it was used to elicit decreased biological activity that would occur under oxygen-limited conditions, which is the baseline condition at the site. Sodium azide was selected over other bacterial inhibitors because it does not change the structure and properties of the soil and because of its affordability and safe disposal.
The experiment was carried out over a 12-week period (following two weeks of equilibration). During the two-week equilibration period, deionized water flowed through all columns without the addition of oxygen and nutrients (for biostimulation columns) or nitrogen and azide (for the control column). This period served as a baseline to establish CB concentrations being flushed from the source zone. The effluent end of each column was fitted with an in-line sample collection reservoir, and weekly aqueous samples were collected for analysis of volatile organic compounds (VOC) by USEPA Method 8260 at Severn Trent Laboratories (Savannah, Georgia, USA). Dissolved oxygen (Oakton DO 110), oxidation–reduction potential (Cole-Parmer), temperature, pH, and specific conductivity (Hanna HI 991301) were measured directly by insertion of parameter-specific probes through a cap in the in-line reservoir. At two week intervals, a biostimulated (aerobic) column was sacrificed to obtain soil samples for posterior chemical and molecular analysis. The control column was sacrificed after 12 weeks. Column soil samples were collected in accordance with USEPA Method 3035 and shipped at 4°C to Seven Trent Laboratories (Savannah, Georgia, USA) for VOC analysis by USEPA Method 8260. A composite soil sample was also collected for molecular analysis performed at Rice University as described below.
DNA extraction
DNA was extracted from soil samples using MoBio power Soil DNA isolation kit (Carlsbad, CA, USA) according to manufacturer protocol. A bead-beating device (Model Mini Beadbeater-8; Biospec, Bartlesville, OK, USA) was utilized for cell lysis. The recovered DNA was collected in a 1.5-ml Eppendorf vial and stored in a freezer (Isotemp® Basic, Fischer Scientific, Rockville, MD, USA) at −75°C. The concentration and purity of the DNA was measured based on the wavelength absorbance ratio (absorbance of 260 nm for DNA and 280 nm for protein) A260/A280 using a spectrophotometer (Amersham Biosciences, Model Ultraspec 2100 Pro, Piscataway, NJ, USA). Each sampled was spiked with a 2-μl aliquot of bacteriophage λ DNA 500 bp (Sigma-Aldrich, St. Louis, MO, USA) prior to DNA extraction to serve as an internal standard for the determination of DNA efficiency recovery. When recovery was lower than 100%, gene copy numbers were normalized to the fraction recovered.
Real-time quantitative PCR (qPCR)
qPCR was used to quantify catabolic genes coding for toluene dioxygenase, naphthalene dioxygenase, ring hydroxylating monooxygenase, phenol hydroxylase, and biphenyl dioxygenase using the primers TOD, NAH, RMO, PHE, and BPH3, respectively, designed by Baldwin et al. (2003) (Table 1). These genes were selected as biomarkers because (i) to date, no qPCR primers have been designed to quantify CB degradation genes, and the chlorobenzene dioxygenase sequences in the NCBI database are highly similar and often indiscernible from the toluene and biphenyl dioxygenase genes. In fact, previous studies have demonstrated that genes involved in the degradation of CBs are evolutionarily linked to toluene/benzene and biphenyl dioxygenases (Beil et al. 1998; Van der Meer et al. 1998) and utilize similar metabolic routes (Gibson and Parales 2000); (ii) the targeted oxygenases are know to have broad substrate specificity and attack a wide variety of aromatic compounds (Wackett and Hershberger 2001) and (iii) these primers target a conserved region of the gene, permitting the detection of specific aromatic catabolic genotypes without excluding related but uncharacterized genes (Baldwin et al. 2003).
All primers and probes were obtained from Integrated DNA Technologies (Coralville, IA, USA). PCR mixtures contained 1× Taqman PCR Master Mix or SYBR GREEN (Applied Biosystems, Foster City, CA, USA); 500 nM forward and reverse primers, 250 nM of the probe (for reactions using Taqman) and sterile DNAase-free water to make up a final volume of 25 μl. PCR reactions were performed using a Sequence Detector (Model ABI 7500, Applied Biosystems, Foster City, CA, USA) with the following temperature conditions: 50°C for two min, followed by 95°C for 10 min and 40 cycles at 95°C for 15 s, and 60°C for one min. The genomic DNA sequences of various reference strains were utilized to prepare calibration curves for the targeted oxygenase genes. Pseudomonas putida F1 was used for toluene dioxygenase, Pseudomonas putida G7 for naphthalene dioxygenase, P. pseudoalcaligenes KF707 for biphenyl dioxygenase, R. picketti PK01 for ring hydroxylating monooxygenase, and Pseudomonas putida CF600 for phenol hydroxylase. Dilutions (101–108 gene copies μl−1) were prepared for all calibration curves, yielding r 2 values ≥ 0.99.
Gene copies in each of the dilutions were estimated based on the following equation:
This approach assumes that the approximate size of the bacterial genome used as the standard in the calibration curves was 6.18 × 106 base pairs (bp) (with approximately 9.12576 × 1014 bp μg−1 of DNA, equivalent to the size of the P. putida genome (http://www.genomesonline.org), and that there are seven gene copies for 16S rRNA gene per genome (1 copy for oxygenases) (http://www.rrndb.cme.msu.edu). The detection limits were on the order of 102 copy numbers g-soil−1 for oxygenase genes and 103 copy numbers g-soil−1 for 16S rRNA gene.
Results and discussion
Source-zone bioremediation
Aerobic biostimulation was tested as a method for enhancing the source-zone attenuation of a site contaminated with MCB and DCBs. A baseline characterization of unamended, homogenized soil indicated an average initial concentration of 420 mg kg−1 MCB, 2,000 mg kg−1 1,2-DCB, 470 mg kg−1 1,3-DCB, and 1,700 mg kg−1 1,4-DCB. These concentrations are consistent with historical high concentrations that have been measured at the site.
MCB concentrations in column effluents were stable or increased slightly over the 2-week equilibration period, and decreased exponentially during the treatment stage (Fig. 1). The decrease in effluent MCB concentration was slightly faster in the biostimulated columns, from an initial value of about 40 mg l−1 to below detection (<0.5 mg l−1) after 6 weeks of oxygen addition, compared to 9 weeks for the control column. The final MCB removal efficiency from the soil was greater than 99% for both biostimulated and control columns (Fig. 2), indicating that dissolution and advective flushing of this relatively soluble compound (500 mg l−1 water solubility) was an important removal mechanism.
In contrast to MCB, effluent concentrations of the DCB isomers remained relatively constant throughout the test, with the total DCB concentration ranging from 80 to 100 mg l−1 (Fig. 1). Yet, soil concentrations data provided evidence of DCB mass removal (Fig. 2). This suggests the presence of an organic phase (characteristic of source-zone soil), which was not depleted and continued to dissolve and sustain relatively high effluent DCB concentrations
The overall MCB mass reduction from the source-zone soil for the 12-week test period was greater than 99% in the biostimulated columns. A high degree of MCB removal was also observed in the control column (96%), consistent with effluent sampling data. DCB total mass removal in biostimulated columns was also high. At the end of the 12-week oxygenation period, total DCB mass removal was 89% for 1,2-DCB; 83% for 1,3-DCB; and 90% for 1,4-DCB. These removal efficiencies were much higher than those observed in the control column (34, 29 and 36% respectively) (Fig. 2), demonstrating the benefits of aerobic biostimulation for this less-soluble class of compounds. Oxygen consumption observed in the biostimulated columns (from an influent dissolved oxygen concentration of 48.6 mg l−1 to an effluent value of approximately 3 mg l−1) corroborates the finding aerobic biostimulation contributed to enhanced mass removal. Based on the observed mass removal over 12 weeks, the biostimulation enhancement factor (relative to the control) was 2.6-fold for 1,2-DCB, 2.9-fold for 1,3-DCB and 2.5-fold for 1,4-DCB.
Molecular analyses
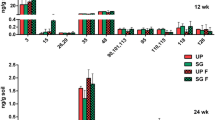
The total bacteria population (measured by qPCR as 16S-rRNA gene copies) was one order of magnitude higher in the source-zone sample than in the non-contaminated (background) soil, and increased by an additional order of magnitude following aerobic biostimulation (12 weeks) to about 107–108 copies g-soil−1 (Fig. 3). A comparison of background versus source-zone samples prior to biostimulation suggests that the presence of CBs promoted significant growth (p < 0.05) of bacteria harboring biphenyl dioxygenase, toluene dioxygenase and phenol hydroxylase genes. These genes also experienced an increase following aerobic biostimulation, but only the phenol hydroxylase gene was significantly enriched (p < 0.05) (Fig. 3). The biphenyl and toluene dioxygenase genes were already present at relatively high concentrations in the background soil (∼104 copies g-soil−1), making them less sensitive biomarkers with respect to monitoring changes following biostimulation. The possibility that their initial abundance was due to the proximity of the background sample location to a landfill for mixed waste (which might have promoted the growth of such genotypes) could not be ruled out. Nonetheless, naphthalene dioxygenase and ring hydroxylating monooxygenase genes were not detected in either the background or source-zone samples before or after biostimulation, indicating that these were not appropriate biomarkers for this soil and contaminant profile.
Biomarker gene concentrations measured by qPCR. Genes coding for biphenyl dioxygenase (BPH), phenol hydroxylase (PHE), and toluene dioxygenase (TOD) are displayed. Genes coding for naphthalene dioxygenase and ring hydroxylating monooxygenase were not detected. * denotes statistically significant increase (p < 0.05) relative to background control, and ** denotes statistically significant increase (p < 0.05) relative to both background and initial conditions (t = 0). Error bars depict the standard deviation from the mean of triplicate measurements. The inhibited control column (azide-amended, oxygen-free) was not considered a sterile control
DNA-based catabolic biomarkers such as those used in this work cannot provide unequivocal evidence of biodegradation activity because the presence of a gene does not guarantee its expression. Thus, instantaneous activity can be better inferred by mRNA (gene expression) rather than DNA (gene presence) analysis. Nevertheless, DNA gene copy numbers should be temporally quite responsive to biodegradation activity because bacterial growth supported by utilization of the target pollutant during bioremediation increases the number of pertinent catabolic genes relative to background levels. In addition, DNA analysis is generally more sensitive and easier to perform than mRNA analysis, primarily because mRNA is relatively unstable and its quantification is subject to variable reverse transcriptase efficiency and lower qPCR recovery.
Although an unequivocal etiology between biphenyl or toluene dioxygenase and CB degradation was not established, the notion that these enzymes played a role in CB biodegradation is supported by circumstantial evidence from previous studies. Specifically, regarding biphenyl dioxygenase, (i) bacteria harboring this enzyme have been reported to abound at sites contaminated with CBs (Abraham et al. 2005), and (ii) biphenyl dioxygenase is known to have a relaxed substrate specificity and attack chlorobiphenyls (http://www.brenda.uni-koeln.de), which are structural analogues of CBs. Therefore, biphenyl dioxygenase might either initiate the degradation of CBs or participate in the degradation of one or more of its byproducts. A similar argument can be postulated for toluene dioxygenase, because its gene is evolutionarily linked to biphenyl and chlorobenzene dioxygenase. In fact, the toluene dioxygenase gene sequence is difficult to discern from that of the chlorobenzene dioxygenase gene (Beil et al. 1998; Van der Meer et al. 1998).
The significant enrichment of the phenol hydroxylase gene following aerobic biostimulation suggests two possibilities: (a) phenols or chlorophenols, which are substrates for phenol hydroxylases, were produced following an initial attack on the CB molecule by another oxygenase, and some of these phenolics were subsequently degraded by organisms harboring phenol hydroxylase; or (b) phenol hydroxylase catalyzed both the first and second hydroxylation of the aromatic ring prior to oxidative cleavage. Regardless of whether the participation of organisms harboring phenol hydroxylase was direct or commensal, this biomarker was the most sensitive indicator of CB biodegradation in this work. This suggests the potential value of this biomarker (as well as those for biphenyl and toluene dioxygenases) for assessment of CB bioremediation potential.
Conclusion
This research demonstrates the potential efficacy of aerobic biostimulation to treat CB source-zones, and that quantification of specific catabolic DNA biomarkers can provide valuable insight as one of several converging lines to demonstrate bioremediation. Since the universal applicability of the oxygenase biomarkers tested in this work is unknown, further tests with samples from other contaminated sites are recommended to ascertain their reliability and evaluate their broad applicability as forensic tools to assess CB source-zone bioremediation performance.
Abbreviations
- CB:
-
Chlorobenzene
- DCB:
-
Dichlorobenzene
- DNAPL:
-
Dense non-aqueous phase liquid
- MCB:
-
Monochlorobenzene
- MCL:
-
Maximum contaminant level
- MNA:
-
Monitored natural attenuation
- qPCR:
-
quantitative Polymerase Chain Reaction
- rRNA:
-
ribosomal RNA
- VOC:
-
Volatile organic compounds
References
Abraham WR, Wenderoth DF, Glasser W (2005) Diversity of biphenyl degraders in a chlorobenzene polluted aquifer. Chemosphere 58:529–533
Adamson DT, McDade JM, Hughes JB (2003) Inoculation of DNAPL source-zone to initiate reductive dechlorination of PCE. Environ Sci Technol 37:2525–2533
Adrian L, Szewzyk U, Wecke J, Gorisch H (2000) Bacterial dehalorespiration with chlorinated benzenes. Nature 408:580–583
Alfreider A, Vogt C, Babel W (2002b) Expression of chlorocatechol 1,2-dioxygenase and chlorocatechol 2,3-dioxygenase genes in chlorobenzene-contaminated subsurface samples. Appl Environ Microbiol 69:1372–1376
Alvarez PJJ, Illman W (2005) Bioremediation and natural attenuation of groundwater contaminants: process fundamentals and mathematical models. John Wiley & Sons
Balcke GU, Turunen LP, Geyer R, Wenderoth DF, Schlosser D (2004) Chlorobenzene biodegradation under consecutive aerobic–anaerobic conditions. FEMS Microbiol Ecol 49:109–120
Baldwin BR, Nakatsu CH, Nies L (2003) Detection and enumeration of aromatic oxygenase genes by multiplex and real-time PCR. Appl Environ Microbiol 69:3350–3358
Beil S, Mason JR, Timmis KN, Pieper DH (1998) Identification of chlorobenzene dioxygenase sequence elements involved in dechlorination of 1,2,4,5-tetrachlorobenzene. J Bacteriol 180:5520–5528
Beller HR, Kane SR, Legler TC, Alvarez PJJ (2002) A real-time polymerase chain reaction method for monitoring anaerobic, hydrocarbon-degrading bacteria based on a catabolic gene. Environ Sci Technol 36:3977–3984
Cope N, Hughes JB (2001) Biologically-enhanced removal of PCE from NAPL source-zones. Environ Sci Technol 35:2014–2021
Da Silva MLB, Alvarez PJJ (2004) Enhanced anaerobic biodegradation of benzene-toluene-ethylbenzene-xylene-ethanol mixtures in bioaugmented aquifer columns. Appl Environ Microbiol 70:4720–4726
Da Silva MLB, Daprato RC, Gomez DE, Hughes JB, Ward CH, Alvarez PJJ (2006) Comparison of bioaugmentation and biostimulation for the enhancement of DNAPLS source-zone bioremediation. Water Environ Res 78:2456–2465
Dermietzel J, Vieth A (2002) Chloroaromatics in groundwater: chances of bioremediation. Environ Geol 41:683–689
Futumata H, Harayama S, Watanabe K (2001) Group-specific monitoring of phenol hydroxylase genes for a functional assessment of phenol-stimulated TCE bioremediation. Appl Environ Microbiol 67:4671–4677
Gibson DT, Parales RE (2000) Aromatic hydrocarbon dioxygenases in environmental biotechnology. Curr Opin Biotechnol 11:236–243
Howard PH (1989) Handbook of environmental fate and exposure data for organic chemicals. Lewis Publishers Chelsea, MI
Kao CM, Prosser J (1999) Intrinsic bioremediation of trichloroethylene and chlorobenzene: field and laboratory studies. J Hazard Mater B69:67–79
Kaschl A, Vogt C, Uhlig S, Nijenhuis I, Weiss H, Kastner M, Richnow HH (2005) Isotopic fractionation indicates anaerobic monochlorobenzene biodegradation. Environ Toxicol Chem 24:1315–1324
Lendvay JM, Löffler FE, Dollhopf M, Aiello MR, Daniels G, Fathepure BZ, Gebhard M, Heine R, Helton R, Shi J, Krajmalnik-Brown R, Major CL, Barcelona MJ, Petrovskis E, Hickey R, Tiedje JM, Adriaens P (2003) Bioreactive barriers: a comparison of bioaugmentation and biostimulation for chlorinated solvent remediation. Environ Sci Technol 37:1422–1431
Lichstein HC, Soule MH (1943) Studies of the effect of sodium azide on microbic growth and respiration: I. The effect of sodium azide on microbic growth. University of Michigan
Lorbeer H, Starke S, Gozan M, Tiehm A, Werner P (2002) Bioremediation of chlorobenzene-contaminated groundwater on granular activated carbon barriers. Water, Air, Soil Pollut 2:183–193
McDade JM, McGuire TM, Newell CJ (2005) Analysis of DNAPL source-depletion costs at 36 field sites. Remediat J 15:9–18
McGuire TM, McDade JM, Newell CJ (2006) Performance of DNAPL source depletion technologies at 59 chlorinated solvent-impacted sites. Ground Water Monit Remediat 26:73–84
Sleep BE, Seepersad DJ, Mo K, Heidorn CM, Hrapovic L, Morril PL, McMAster ML, Hood ED, Lebron C, Lollar BS, Major DW, Edwars EA (2006) Biological enhancement of tetrachloroethene dissolution and associated microbial community changes. Environ Sci Technol 40:3623–3633
Suzuki MT, Taylor LT, DeLong EF (2000) Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5′-nuclease assays. Appl Environ Microbiol 66:4605–4614
Van der Meer JR, Werlen C, Nishino SF, Spain JC (1998) Evolution of a pathway for chlorobenzene metabolism leads to natural attenuation in contaminated groundwater. Appl Environ Microbiol 64:4185–4193
Vogt C, Alfreider A, Lorbeer H, Ahlheim J, Feist B, Boehme O, Weiss H, Babel W, Wuensche L (2002) Two pilot plant reactors designed for the in situ bioremediation of chlorobenzene-contaminated ground water: hydrogeological and chemical characteristics and bacterial consortia. Water Air Soil Pollut Focus 2:161–170
Vogt C, Alfreider A, Lorbeer H, Hoffmann D, Wuensche L, Babel W (2004) Bioremediation of chlorobenzene-contaminated ground water in an in situ reactor mediated by hydrogen peroxide. J Contam Hydrol 68:121–141
Wackett LP, Hershberger CD (2001) Biocatalysis and biodegradation. ASM Press, Washington
Wenderoth DF, Rosenbrock P, Abraham WR, Pieper DH, Hofle MG (2003) Bacterial community dynamics during biostimulation and bioaugmentation experiments aiming at chlorobenzene degradation in groundwater. Micro Ecol 46:161–176
Zheng YR, Carr CS, Hughes JB (2001) Influence of hydraulic retention time on extent of PCE dechlorination and preliminary characterization of the enrichment culture. Bioremediat J 5:169–168
Acknowledgements
This study was performed with private support in conjunction with facilities and resources of the Civil and Environmental Engineering Department at Rice University. The authors thank Nathan Howell for providing laboratory and technical assistance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Dominguez, R.F., da Silva, M.L.B., McGuire, T.M. et al. Aerobic bioremediation of chlorobenzene source-zone soil in flow-through columns: performance assessment using quantitative PCR. Biodegradation 19, 545–553 (2008). https://doi.org/10.1007/s10532-007-9160-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10532-007-9160-4