Abstract
In North America the invasive winter moth (Operopthera brumata) has caused defoliation in forest and fruit crop systems in British Columbia, Nova Scotia, Oregon, and in the northeastern United States (the “Northeast”). In the Northeast, it was previously shown that hybridization is occurring with a native congener, Bruce spanworm (O. bruceata)—a species that has a broad distribution across much of North America. Whether hybridization among winter moth and Bruce spanworm populations has occurred in all of regions where winter moth established is unknown. One factor that might influence hybridization between these two species is the presence of reproductive manipulating endosymbionts, such a Wolbachia. To determine the geographic extent of hybridization among populations of these two species, we classified 1400 field-collected moths from Europe and North America as either being winter moth, Bruce spanworm, or hybrids using 10–12 polymorphic microsatellite loci. We then screened each individual for the presence of Wolbachia by PCR amplification of the wsp gene fragment. For all hybrids, we determined their maternal species-lineage by PCR amplification and sequencing of the mitochondrial locus cytochrome oxidase I. We find that winter moth x Bruce spanworm hybrid individuals appear to be present in all regions of North America that winter moth has invaded, and that hybrids are of both winter moth and Bruce spanworm maternal-origins. In addition, we find Wolbachia infected individuals from all species in North America, and that winter moth individuals in North America have a much lower infection rate (11.5%) than individuals in Europe (55.1%).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hybridization is an important driver of the evolutionary trajectories of species from across the tree of life (Allendorf et al. 2001; Harrison and Larson 2014) and has long been known to alter ecological interactions (Feder et al. 2003). When genetically distinct species or populations hybridize, a wide range of often contradictory outcomes are possible including, but not limited to, hybrid vigour (Szücs et al. 2012), hybrid breakdown (Ellison et al. 2008), speciation (Schwarz et al. 2007), and the potential to reverse the course of speciation (Garrick et al. 2014; Seehausen et al. 2008). Hybridization is particularly prominent among species of insects (Schwenk et al. 2008), where it may play an important role during the establishment of non-native species (Mesgaran et al. 2016; Prentis et al. 2008). For example, hybridization between native and non-native species can result in the formation of “mega-pests” (e.g., Anderson et al. 2018), and it can facilitate adaptation to novel and disturbed environments (e.g., Leite et al. 2017). From a conservation perspective, while hybridization can lead to increases in genetic diversity (Verhoeven et al. 2011), it can also result in the loss of genetic identity and eventual extinction of native species through the process of ‘hybridization to extinction’ (Ayres et al. 2004; Hinton 1975; Rhymer and Simberloff 1996; Wolf et al. 2001), a phenomenon well documented for plants and vertebrates, but not well studied for insects or other taxa (Todesco et al. 2016).
Over the past 100 years, invasive populations of the European winter moth, Operophtera brumata L. (Lepidoptera: Geometridae), have repeatedly become established in North America and have caused extensive defoliation to a wide range of deciduous trees and shrubs in Nova Scotia (Embree 1967; MacPhee 1967), Oregon (Kimberling et al. 1986), British Columbia (Gillespie et al. 1978), and in the northeastern United States (the “Northeast”) (Elkinton et al. 2010, 2014; Elkinton et al. 2015; Simmons et al. 2014). The biological control of winter moth in Nova Scotia represents one of the text-book examples of successful forest-insect biological control (Embree 1966; Kimberling et al. 1986; DeBach and Rosen 1991; Roland and Embree 1995), and efforts to release the biological control agent Cyzenis albicans (Fallén) (Diptera: Tachinidae) in the Northeast are currently underway (Elkinton et al. 2015; Broadley et al. 2018).
In the Northeast, winter moth has been hybridizing with a native congener the Bruce spanworm, O. bruceata Hulst (Elkinton et al. 2010, 2014; Havill et al. 2017), and while it is likely that hybridization has occurred in all of the invasive regions where winter moth has established, this has yet to be verified. One interesting aspect of the hybridization between winter moth and Bruce spanworm in the Northeast is that the hybridization rates have been shown to be asymmetrical towards winter moth (Havill et al. 2017)—a pattern often observed in insects when the reproductive manipulator Wolbachia Hertig (Rickettsiales: Rickettsiaceae) is present (Bordenstein et al. 2001; Jaenike et al. 2006; Shoemaker et al. 1999; Weeks et al. 2002; Weinert et al. 2015). Species of Wolbachia are known to infect a wide range of hosts (Werren et al. 2008) and tissue types (Dobson et al. 1999) and can cause cytoplasmic incompatibility (CI) (Moran et al. 2008; Werren 1997). For invasive species, Wolbachia infections can often facilitate their probability of establishment by providing fitness advantages to infected individuals over un-infected native congeners (Schuler et al. 2013, 2016), protection from parasites and pathogens (Zindel et al. 2011), and parthenogenesis induction (Guzman et al. 2012). Recently, anthropogenic manipulations of Wolbachia infections have opened novel opportunities for biological (Floate et al. 2006; Zabalou et al. 2004) and vector (Raghavendra et al. 2011; Turley et al. 2009; van den Hurk et al. 2012) control of pest species.
Whether Wolbachia infections are common among populations of winter moth in its native or invasive ranges, populations of Bruce spanworm, or hybrid individuals is unknown, however; recent genomic sequencing of a winter moth individual from the Netherlands indicated the presence of Wolbachia (Derks et al. 2015). Therefore, the objective of this work was to begin evaluating Wolbachia infections among individuals of these two pest-species and their hybrids. To achieve this goal, we screened over 1400 field-collected winter moths from Europe and field-collected moths from all of the regions in North America that winter moth has invaded (i.e., Nova Scotia, British Columbia, Oregon, and the Northeast) to classify them to species (i.e., winter moth, Bruce spanworm, or hybrids) using microsatellite loci. We then PCR-screened samples using the Wolbachia surface protein gene (wsp) to determine whether individuals are infected with Wolbachia and provide insights on regional and species-specific differences in Wolbachia infection rates. We discuss the implications of our findings in regards to the growing literature that suggests Wolbachia infections can be both spatially and temporally dynamic, and provide preliminary baseline data that can be used by future work aimed at studying changes in Wolbachia infection rates in response to the release of C. albicans in the Northeast for the biological control of winter moth populations.
Materials and methods
Sampling strategy and microsatellite genotyping
Using sex pheromone-baited traps (Elkinton et al. 2010, 2011), we collected adult male moths in Europe and North America. In addition, we collected 25 larvae from two sites in Maine in 2015 (five and 20, respectively) as well as 20 male and 27 female pupae from a site in Rhode Island in 2016. For adult males, prior to DNA extraction the uncus and wings were removed as morphological vouchers and were deposited in the Yale Peabody Museum of Natural History. The head, thorax, and remaining abdomen of the adult males, as well as the entire larva or pupa were then homogenized using 3/16” stainless steel beads (GlenMills Inc., Clifton, New Jersey) with a FastPrep-24 Sample Homogenizer (MP Biomedicals). DNA was extracted, and moths were genotyped either using 12 M13-labeled or 11 directly-labelled microsatellite loci using published protocols (Andersen et al. 2017; Havill et al. 2017). Due to differences in the fragment lengths between the M13- versus directly-labelled fragments, the datasets generated using these different approaches were analysed independently. In both cases, genotyping was conducted at the DNA Analysis Facility on Science Hill at Yale University using a 3730xl DNA Analyzer (Thermo Fisher Scientific, USA), and fragment lengths were scored in comparison to the GeneScan 500 LIZ size standard (Thermo Fisher Scientific, USA) using the microsatellite plugin in the software program Geneious v. 11.1.2 (https://www.geneious.com). Microsatellite genotype scores are provided as supplemental files, and complete collection information is provided in Table S1.
Species identification
Both datasets were filtered to only include individuals from which ≥ 10 loci were amplified. Using the software program NewHybrids v.1.1.b3 (Anderson 2008; Anderson and Thompson 2002) we calculated, for each dataset, the probability of assignment (Z) that an individual could be classified as being either “pure” winter moth or “pure” Bruce spanworm, or as one of four hybrid categories (F1, F2, winter moth backcross, or Bruce spanworm backcross). Four independent runs, each of one million generations, discarding the first 100,000 burn-in generations, were analysed using random starting values, and uniform priors for the estimates of Θ (i.e., allele frequencies) and π (i.e., mixing proportions) for each dataset. Results were then averaged across runs, and individuals receiving a score of Z ≥ 0.9 to any one category were classified as having strong support to that category, individuals receiving a score of 0.5 ≤ Z < 0.9 to any one category were classified as having moderate support to that category, and individuals receiving a score of Z < 0.5 to all categories were classified as having low support to the category with the highest Z score.
For all hybrid individuals, we then sequenced a portion of the mitochondrial locus cytochrome oxidase 1 (i.e., COI, the “DNA barcode” region) to determine their maternal lineage (i.e., winter moth or Bruce spanworm), using the primer pair LepF1 (5′-ATTCAACCAATCATAAAGATATTGG-3′) and LepR1 (5′-TAAACTTCTGGATGTCCAAAAAATCA-3′) and thermocycler protocols outlined in (Hebert et al. 2004). Each PCR reaction was run with the following conditions: 1 μl of eluted genomic extract, 5 μl of 5X Promega GoTaq® Buffer (Promega Corp., Maddison, WI), 0.5 μl of 10 mM dNTP (Promega), 0.5 μl each of 10 μM dilutions of the forward and reverse primers, 0.2 μl of taq (Promega), and the final volume was adjusted to 25 μl using HPCL grade H20. Reactions were run in 96-well plate format with a positive and a negative control included in each plate, and PCR products were visualized on a 1.5% agarose gel. Five μl of each PCR product was then cleaned using the Exo-SAP PCR product cleanup approach (0.5 μl Exo [Thermo Fisher Scientific, Walham, MA], 0.5 μl SAP [New England Biolabs, Ipswich, MA], 1 μl SAP buffer [New England Biolabs]), and DNA sequencing of both forward and reverse fragments was performed at the DNA Analysis Facility on Science Hill at Yale University. Sequence results were edited and visualized using Geneious. Individual DNA sequences are available on GenBank (accession numbers KY612991–KY613016, MH732738–MH732739). Each sequence was then compared to published sequences in the GenBank database using the ‘blastn’ search algorithm (Zhang et al. 2000), and whether the top match for each sequence was from winter moth (O. brumata), eastern Bruce spanworm (O. bruceata), or western Bruce spanworm (O. bruceata occidentalis) is presented in Table S2.
Surveys for Wolbachia
The presence of Wolbachia was determined by PCR amplification of a fragment of the Wolbachia surface protein gene (wsp) using the primer pair wsp81F (5′-TGGTCCAATAAGTGATGAAGAAAC-3′) and wsp691R (5′-AAAAATTAAACGCTACTCCA-3′) with the thermocycler protocols outlined in Zhou et al. (1998). Each PCR reaction was run with the following conditions: 1 μl of eluted genomic extract, 5 μl of 5X GoTaq® Buffer (Promega), 0.5 μl of 10 mM dNTP (Promega), 0.5 μl each of 10 μM dilutions of the forward and reverse primers, 0.2 μl of taq (New England BioLabs, Ippswich, MA), and the final volume was adjusted to 25 μl using HPCL grade H20. A preliminary survey of samples from Europe identified Wolbachia in several individuals. We then used one of these samples (06-196-01) as a positive control for all subsequent screenings. All reactions were run in 96-well plate format with a negative control and with sample 06-196-01 included as a positive control. PCR products were visualized on a 1.5% agarose gel. Samples with bands of the expected length (~ 600 bp) were scored as “Positive” for Wolbachia infection, and samples that either had an absence of bands or the presence of bands of different lengths than the positive control were scored as “Negative” for Wolbachia infection.
To verify that amplified fragments corresponded with the target fragment of the wsp gene, a subset of 48 positive PCR products representing moths collected in different regions and a mix of pure species and hybrids was haphazardly selected for sequencing. Five μl of each PCR product was cleaned using the Exo-SAP PCR product cleanup approach (0.5 μl Exo, 0.5 μl SAP, 1 μl buffer), and DNA sequencing of both forward and reverse fragments was performed at the University of California Berkeley DNA Sequencing Facility. Sequence results were edited and visualized using Geneious, and were compared to previously published sequences in the GenBank database using the blastn search algorithm. Individual DNA sequences are available on GenBank (accession numbers KY587618–KY587656).
Species and regional differences in Wolbachia infection rates
To provide preliminary indications as to whether Wolbachia infection rates differed between species (i.e., winter moth, Bruce spanworm, and hybrids), and for winter moth among geographic regions (i.e., Nova Scotia, British Columbia, Oregon, the Northeast, and Europe), the total number of Infected and Uninfected individuals for each species/region were compared using a χ2 test in the statistical package R v. 3.3.3 (R Core Team 2017) with post hoc analyses between each pair of species/regions conducted with an adjusted α based on Bonferroni’s correction for multiple comparisons. For European samples we also examined whether the probability that an individual was infected with Wolbachia was correlated with the genetic assignment (Q) of that individual as reported in Andersen et al. (2017) using a generalized linear model with a binomial distribution and a logit-link function in R.
Limitations of study design
For all analyses we caution that these results should be considered preliminary as we based our infection information from individuals collected over multiple years and primarily from field-collected males, which can have different infection rates than females due to a variety of factors including inefficient transmission, male killing, and feminisation (Duplouy and Hornett 2018). In addition, our removal of the male reproductive organs to act as voucher specimens prior to DNA extraction may have further influenced our results, as well as the uneven number of samples collected from different species and from different localities.
Results
Sampling strategy and microsatellite genotyping
After filtering, the final dataset included 1429 moths collected from 13 European countries, five Canadian provinces, and eight American states. These included 22 localities in Europe and 62 localities in North America (Supplemental Appendix Table S1).
Species identification
All 474 of the European samples were classified as winter moth with strong support after summarizing across the independent NewHybrids analyses. In North America, 530 individuals were classified as winter moth (all with strong support), 397 individuals were classified as Bruce spanworm (391 with strong support, five with moderate support, one with low support), and 28 individuals were classified as hybrids (21 F1 [15 with strong support, six with moderate support], six F2 [two with strong support, four with moderate support], and one winter moth backcross [moderate support]). These results are summarized graphically in Fig. 1. Hybrids were detected in all of the invaded regions in North America, including 22 individuals from the Northeast, one individual each from British Columbia and Nova Scotia, and three individuals from Oregon. In addition, we identified a hybrid individual in Ontario, Canada, a location from which winter moth has not yet been reported. Probabilities of assignment for each individual are presented in Supplemental Appendix Table S1. Based on COI sequencing of the 28 hybrids, eight individuals had eastern Bruce spanworm mtDNA, four had western Bruce spanworm mtDNA, and 16 had winter moth mtDNA (Supplemental Appendix Table S2).
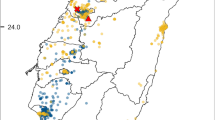
Geographic sampling localities and proportions of individuals infected with Wolbachia in western North America (a), the Great Lakes region (b), Europe (c), eastern North America (d), and along transects in Massachusetts (e) and Connecticut (f). Individual charts are drawn proportional to the number of surveyed individuals at each location. Dark and light blue, red, and orange shadings represent the proportion of winter moth, Bruce spanworm, and hybrid individuals infected and uninfected with Wolbachia, respectively. The figure was generated in ArcMap v.10.3.1 (Esri Co., Redlands) using the Europe and North America Albers Equal Area Conic projections. In instances when two charts overlap, a leading line is used to signify location of the displaced chart
Surveys for Wolbachia infections
Of the 1429 screened individuals, 333 were classified as infected with Wolbachia (23.3%). Sequencing of a haphazardly selected subset of 48 Wolbachia positive wsp PCR products, generated clean sequence reads from 39 individuals all with high scoring matches to published wsp sequences in GenBank (Supplemental Appendix Table S3). In North America, we classified six of 397 Bruce spanworm, five of 28 hybrids, and 61 of 530 winter moth as Wolbachia-infected (1.5%, 17.9%, and 11.5%, respectively). In Rhode Island, where both males and females were collected, one female pupa was infected with Wolbachia (3.7%), while no male pupae were infected. For hybrids, two of 16 individuals with winter moth mtDNA, two of eight individuals with eastern Bruce spanworm mtDNA, and one of four individuals with western Bruce spanworm mtDNA were infected with Wolbachia, respectively.
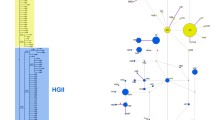
In Europe, we classified 261 of 474 (55.1%) winter moth individuals as Wolbachia-infected. In North America differences between species were highly significant (χ2 = 21.445, d.f. = 2, P < 0.0001). Post-hoc analyses were conducted using a corrected α of 0.0166, and pair-wise comparisons indicated that there were highly significant differences in Wolbachia infection levels between hybrids with Bruce spanworm mtDNA and pure individuals of that species (χ2 = 20.29, df = 1, P < 0.0001), but that there were no differences between hybrids with winter moth mtDNA and pure winter moth individuals (χ2 = 2.0237, d.f. = 1, P = 0.1549) or between the numbers of infected and uninfected pure Bruce spanworm and pure winter moth in North America (χ2 = 1.5388, d.f. = 1, P = 0.2148). For winter moth, differences between the numbers of Wolbachia-infected individuals in Europe versus North America were highly significant (χ2 = 111.68, d.f. = 1, P < 0.0001). Post-hoc analyses, indicated that there were significant differences between Europe and most of the North American invasive winter moth populations (using a corrected α of 0.0125). These included, Europe versus Nova Scotia (χ2 = 88.272, d.f. = 1, P < 0.0001), Europe versus the Northeast (χ2 = 164.44, d.f. = 1, P < 0.0001), and Europe versus British Columbia (χ2 = 7.3902, d.f. = 1, P = 0.0066). There was no significant difference between Europe and Oregon (χ2 = 2.6308, d.f. = 1, P = 0.1048). Among European samples we also found a highly significant correlation between the assignment (Q) of individuals to one of two genetic clusters previously published in Andersen et al. (2017) and the probability of infection with Wolbachia (z = 6.651, P < 0.0001) with the probability of infection increasing with the increased probability of assignment of an individual to the “Eastern European” genetic cluster (Fig. 2).
The presence or absence of Wolbachia infection was compared to the assignment (Q) of individuals to the Eastern European genetic cluster using a generalized linear model (GLM) with a binomial distribution conducted in R. Assignments (Q) are from Andersen et al. (2017), with histograms representing the frequency of individuals either infected or uninfected plotted along the x-axis using the ‘PopBio’ package
Discussion
Hybridization can play an important role in the establishment of invasive species (e.g., Prentis et al. 2008), particularly when coupled with infections by reproductive manipulators such as Wolbachia (Schuler et al. 2013). Building off previous reports of hybridization between winter moth and Bruce spanworm in the Northeast (Elkinton et al. 2010, 2014; Havill et al. 2017), here we provide the first genetic evidence of hybrid individuals in Oregon, British Columbia, and Nova Scotia. In addition, we identified a hybrid individual in Ontario, Canada, a region from which winter moth has not previously been reported from. This individual provides the first evidence that winter moth, or winter moth genomic material at least, is spreading into the interior regions of North America. More sampling in this region will be required to understand this phenomenon, and to identify whether this individual represents the presence of a recent migration event or an older and unreported population of winter moth in that region.
Often hybridization between native and non-native congeneric species occurs only during the immediate generations following the introduction of the non-native species (e.g., Roy et al. 2016), however; here we find evidence for continued hybridization as noted by the presence of multiple classes of hybrids including F1 hybrids in three of the four regions invaded by winter moth (one each in Nova Scotia and Oregon, and 20 in the Northeast), F2 hybrids in all invaded regions (one each in British Columbia, Nova Scotia, and Ontario; two individuals in the Northeast and three individuals in Oregon), as well as one backcrossed individual in the Northeast. The presence of these distinct hybrid classes builds on the recent finding of Havill et al. (2017) that multi-generational hybridization between winter moth and Bruce spanworm populations is occurring. As has been seen in other systems, this continued hybridization may have facilitated the establishment of each invasive North American population by increasing the performance of admixed individuals (e.g., Li et al. 2018), by aiding in establishment and expansion in these regions (e.g., Hirsch et al. 2017; Krojerová-Prokešová et al. 2017), or by promoting local adaptation in newly invaded regions (e.g., Vilatersana et al. 2016). As such, what role hybridization has played in the sustainability of existing and ongoing biological control efforts needs to be determined to better understand the factors that have influenced this textbook biological control program.
Hybridization among winter moth and Bruce spanworm populations may further have been influenced by the presence of the reproductive manipulator Wolbachia. Recent Wolbachia studies suggest that infection rates can be both temporally and spatially dynamic (Ahmed et al. 2015; Bing et al. 2014; Cattel et al. 2016; Kriesner et al. 2016; Michel-Salzat et al. 2001; Roy et al. 2015; Turelli et al. 2018; Zabal-Aguirre et al. 2010), and similarly, here we find that Wolbachia infections varied both among species (i.e., winter moth, Bruce spanworm, and their hybrids), and for winter moth among geographic regions. For invasive populations of winter moth in North America, we find preliminary evidence of a substantial reduction in Wolbachia infection rates (11.5% infection in North America vs. 55.1% infection in Europe). Reductions in Wolbachia infection rates have previously been observed as the results of bottlenecks associated with long-distance dispersal (Nguyen et al. 2016; Reuter et al. 2005; Yang et al. 2010), or differences in the survivability of Wolbachia due to differences in temperatures between geographic regions (Feder et al. 1999). Wolbachia infections may also impose fitness costs on their hosts (e.g., Fleury et al. 2000; Ross et al. 2016), in which case it would be beneficial to examine what the fitness costs of Wolbachia infections are for winter moth in order to better understand how the loss of Wolbachia might influence the invasive potential of this species in North America. However, due to the facts that we sampled almost exclusively adult males (from which we removed the reproductive organs prior to DNA extraction), and that our samples were collected over multiple years without a standardized sampling protocol in regards to the numbers of individuals collected at a specific location, we emphasize that these results should be viewed as preliminary, and we encourage future research into the role of Wolbachia in this exciting system.
In addition to supporting the findings of Derks et al. (2015) that some winter moths from Europe are infected with Wolbachia, we also find that several Bruce spanworm individuals and several hybrid individuals of both Bruce spanworm and winter moth maternal-origin are similarly infected. Unfortunately, without multi locus sequence typing (MLST; Baldo and Werren 2007) or genomic approaches (Bleidorn and Gerth 2017), it is unclear whether the Wolbachia found in pure Bruce spanworm and/or in hybrids of Bruce spanworm maternal-origin represents a native infection by Wolbachia, or suggests that Wolbachia may be moving from the invasive winter moth populations into native Bruce spanworm populations perhaps via horizontal transmission as has been seen in other systems (e.g., Schuler et al. 2013). This finding could have important implications both for understanding regional differences in hybridization rates among these two species or in altering the pest-status of the native Bruce spanworm given that Wolbachia infections can impede the effectiveness of parasites and parasitoids in suppressing the density of a pest species (e.g., Mochiah et al. 2002; Silva et al. 2000; White et al. 2015; Zindel et al. 2011). In addition, MLST or genomic sequencing could also explore whether Wolbachia causes cytoplasmic incompatibility in this system. However, our results suggest that this may be unlikely as our comparisons of mtDNA sequences from hybrid individuals indicated that infected hybrids are equally likely to have either winter moth or Bruce spanworm mtDNA (Supplemental Appendix Table S2).
For winter moth populations in Europe, widespread infections with Wolbachia might also help explain why previous mitochondrial-based phylogenetic studies have found limited biogeographic patterns (Gwiazdowski et al. 2013; Mannai et al. 2017) as Wolbachia infections have been shown to mask phylogenetic signals from mitochondrial markers (e.g., Hurst and Jiggins 2005; Schuler et al. 2016). In contrast, surveys using nuclear-loci have uncovered finer-scale biogeographic structure for this species that correspond with patterns of post-glacial recolonization (Andersen et al. 2017). While our results show that there is a clear correlation between Wolbachia infection levels and the assignment to the eastern or western European genetic lineages (Fig. 2), it would be beneficial to determine whether one of the two primary mtDNA haplotypes (presented in Gwiazdowski et al. 2013; Mannai et al. 2017) is more or less likely to be infect with Wolbachia.
Conclusions
Here we provide the first genetic evidence that hybridization between the invasive winter moth and native Bruce spanworm has occurred in all of the regions in North America that winter moth has established. We also present the first evidence that Wolbachia infection is widespread among European winter moth individuals, and that infection rates are both regionally dynamic within Europe and between European and invasive populations of winter moth. Future studies are need to better understand the role of Wolbachia in this system, particularly in regards to influencing hybridization rates and in altering the efficacy of introduced biological control agents.
Data accessibility
Genotype scores: Uploaded as two tab-delimited files in NewHybrid format. See the supplemental files titled “WMothBSpanwormNewHybridsM13.txt” and “WMothBSpanwormNewHybridsDL.txt”. Cytochrome oxidase I (COI) sequences: available on GenBank with the following accession numbers: KY612991-KY613016, MH732738–MH732739. Wolbachia surface protein (wsp) sequences: available on GenBank with the following accession numbers: KY587618-KY587656.
References
Ahmed MZ, Araujo-Jnr EV, Welch JJ, Kawahara AY (2015) Wolbachia in butterflies and moths: geographic structure in infection frequency. Front Zool 12:16. https://doi.org/10.1186/s12983-015-0107-z
Allendorf FW, Leary RF, Spruell P et al (2001) The problems with hybrids: setting conservation guidelines. Trends Ecol Evol 16:613–622. https://doi.org/10.1016/S0169-5347(01)02290-X
Andersen JC, Havill NP, Caccone A et al (2017) Postglacial recolonization shaped the genetic diversity of the winter moth (Operophtera brumata) in Europe. Ecol Evol 7:3312–3323. https://doi.org/10.1002/ece3.2860/
Anderson EC (2008) Bayesian inference of species hybrids using multilocus dominant genetic markers. Philos Trans R Soc B Biol Sci 363:2841–2850. https://doi.org/10.1098/rstb.2008.0043
Anderson EC, Thompson EA (2002) A model-based method for identifying species hybrids using multilocus genetic data. Genet 160:1217–1229
Anderson CJ, Oakeshott JG, Wee Tek T et al (2018) Hybridization and gene flow in the mega-pest lineage of moth, Helicoverpa. Proc Nat Acad Sci USA 115:5034–5039. https://doi.org/10.1073/pnas.1718831115
Ayres DR, Zaremba K, Strong DR (2004) Extinction of a common native species by hybridization with an invasive congener. Weed Technol 18:1288–1291
Baldo L, Werren JH (2007) Revisiting Wolbachia supergroup typing based on wsp: spurious lineages and discordance with MLST. Curr Microbiol 55:81–87. https://doi.org/10.1007/s00284-007-0055-8
Bing XL, Xia WQ, Gui JD et al (2014) Diversity and evolution of the Wolbachia endosymbionts of Bemisia (Hemiptera: Aleyrodidae) whiteflies. Ecol Evol 4:2714–2737. https://doi.org/10.1002/ece3.1126
Bleidorn C, Gerth M (2017) A critical re-evaluation of multilocus sequence typing (MLST) efforts in Wolbachia. bioRxiv 133710; https://doi.org/10.1101/133710
Bordenstein SR, O’Hara FP, Werren JH (2001) Wolbachia-induced incompatibility precedes other hybrid incompatibilities in Nasonia. Nature 409:707–710. https://doi.org/10.1038/35055543
Broadley HJ, Kelley EA, Elkinton JS et al (2018) Identification and impact of hyperparasitoids and predators affecting Cyzenis albicans (Tachinidae), a recently introduced biological control agent of winter moth (Operophtera brumata L.) in the northeastern U.S.A. Biol Control 121:99–108. https://doi.org/10.1016/j.biocontrol.2018.01.011
Cattel J, Kaur R, Gibert P et al (2016) Wolbachia in European populations of the invasive pest Drosophila suzukii: regional variation in infection frequencies. PLoS ONE 11:e0147766. https://doi.org/10.1371/journal.pone.0147766
DeBach P, Rosen D (1991) Biological control by natural enemies. Cambridge University Press, Cambridge
Derks MFL, Smit S, Salis L et al (2015) The genome of winter moth (Operophtera brumata) provides a genomic perspective on sexual dimorphism and phenology. Genome Biol Evol 7:2321–2332. https://doi.org/10.1093/gbe/evv145
Dobson SL, Bourtzis K, Braig HR et al (1999) Wolbachia infections are distributed throughout insect somatic and germ line tissues. Insect Biochem Mol Biol 29:153–160
Duplouy A, Hornett EA (2018) Uncovering the hidden players in Lepidoptera biology: the heritable microbial symbionts. PeerJ 6:e4629. https://doi.org/10.7717/peerj.4629
Elkinton JS, Boettner GH, Sremac M et al (2010) Survey for winter moth (Lepidoptera: Geometridae) in northeastern North America with pheromone-baited traps and hybridization with the native Bruce spanworm (Lepidoptera: Geometridae). Ann Entomol Soc Am 103:135–145. https://doi.org/10.1603/AN09118
Elkinton JS, Lance D, Boettner G et al (2011) Evaluation of pheromone-baited traps for winter moth and Bruce spanworm (Lepidoptera: Geometridae). J Econ Entomol 104:494–500
Elkinton JS, Liebhold A, Boettner GH et al (2014) Invasion spread of Operophtera brumata in northeastern United States and hybridization with O-bruceata. Biol Invasions 16:2263–2272. https://doi.org/10.1007/s10530-014-0662-9
Elkinton J, Boettener G, Liebhold A et al (2015) Biology, spread, and biological control of winter moth in the eastern United States. USDA Forest Service Publication, New York, p 22
Ellison CK, Niehuis O, Gadau J (2008) Hybrid breakdown and mitochondrial dysfunction in hybrids of Nasonia parasitoid wasps. J Evol Biol 21:1844–1851. https://doi.org/10.1111/j.1420-9101.2008.01608.x
Embree DG (1966) Role of introduced parasites in control of winter moth in Nova Scotia. Can Entomol 98:1159–1168
Embree DG (1967) Effects of winter moth on growth and mortality of red oak in Nova Scotia. For Sci 13:295–299
Feder ME, Karr TL, Yang W et al (1999) Interaction of Drosophila and its endosymbiont Wolbachia: natural heat shock and the overcoming of sexual incompatibility. Am Zool 39:363–373. https://doi.org/10.1093/icb/39.2.363
Feder JL, Berlocher SH, Roethele JB et al (2003) Allopatric genetic origins for sympatric host-plant shifts and race formation in Rhagoletis. Proc Natl Acad Sci USA 100:10314–10319. https://doi.org/10.1073/pnas.1730757100
Fleury F, Vavre F, Ris N et al (2000) Physiological cost induced by the maternally-transmitted endosymbiont Wolbachia in the Drosophila parasitoid Leptopilina heterotoma. Parasitology 121:493–500
Floate KD, Kyei-Poku GK, Coghlin PC (2006) Overview and relevance of Wolbachia bacteria in biocontrol research. Biocontrol Sci Technol 16:767–788. https://doi.org/10.1080/09583150600699606
Garrick RC, Benavides E, Russello MA et al (2014) Lineage fusion in Galapagos giant tortoises. Mol Ecol 23:5276–5290. https://doi.org/10.1111/mec.12919
Gillespie DR, Finlayson T, Tonks NV et al (1978) Occurrence of winter moth, Operophtera-brumata (Lepidoptera, Geometridae), on southern Vancouver-Island, British-Columbia. Can Entomol 110:223–224
Guzman NV, Lanteri AA, Confalonieri VA (2012) Colonization ability of two invasive weevils with different reproductive modes. Evol Ecol 26:1371–1390. https://doi.org/10.1007/s10682-012-9564-4
Gwiazdowski RA, Elkinton JS, DeWaard JR, Sremac M (2013) Phylogeographic diversity of the winter moths Operophtera brumata and O. bruceata (Lepidoptera: Geometridae) in Europe and North America. Ann Entomol Soc Am 106:143–151. https://doi.org/10.1603/AN12033
Harrison RG, Larson EL (2014) Hybridization, introgression, and the nature of species boundaries. J Hered 105:795–809. https://doi.org/10.1093/jhered/esu033
Havill NP, Elkinton JS, Andersen JC et al (2017) Asymmetric hybridization between non-native winter moth, Operophtera brumata (Lepidoptera: Geometridae), and native Bruce spanworm, O. bruceata, in the northeastern United States, assessed with novel microsatellites and SNPs. Bull Entomol Res 107:241–250. https://doi.org/10.1017/S0007485316000857
Hebert PDN, Penton EH, Burns JM et al (2004) Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proc Natl Acad Sci USA 101:14812–14817. https://doi.org/10.1073/pnas.0406166101
Hinton WF (1975) Natural hybridization and extinction of a population of Physalis-virginiana (Solanacea). Am J Bot 62:198–202
Hirsch H, Brunet J, Zalapa J et al (2017) Intra- and interspecific hybridization in invasive Siberian elm. Biol Invasions 19:1889–1904. https://doi.org/10.1007/s10530-017-1404-6
Hurst GD, Jiggins FM (2005) Problems with mitochondrial DNA as a marker in population, phylogeographic and phylogenetic studies: the effects of inherited symbionts. Proc R Soc B Biol Sci 272:1525–1534. https://doi.org/10.1098/rspb.2005.3056
Jaenike J, Dyer KA, Cornish C et al (2006) Asymmetrical reinforcement and Wolbachia infection in Drosophila. PLoS Biol 4:1852–1862. https://doi.org/10.1371/journal.pbio.0040325
Kimberling DN, Miller JC, Penrose RL (1986) Distribution and parasitism of winter moth, Operophtera-brumata (Lepidoptera, Geometridae), in western Oregon. Environ Entomol 15:1042–1046. https://doi.org/10.1093/ee/15.5.1042
Kriesner P, Conner WR, Weeks AR et al (2016) Persistence of a Wolbachia infection frequency cline in Drosophila melanogaster and the possible role of reproductive dormancy. Evolution 70:979–997. https://doi.org/10.1111/evo.12923
Krojerová-Prokešová J, Barančeková M, Kawata Y et al (2017) Genetic differentiation between introduced Central European sika and source populations in Japan: effects of isolation and demographic events. Biol Invasions 19:2125–2141. https://doi.org/10.1007/s10530-017-1424-2
Leite NA, Correa AS, Michel AP et al (2017) Pan-American similarities in genetic structures of Helicoverpa armigera and Helicoverpa zea (Lepidoptera: Noctuidae) with implications for hybridization. Environ Entomol 46:1024–1034. https://doi.org/10.1093/ee/nvx088
Li Y, Stift M, van Kleunen M (2018) Admixture increases performance of an invasive plant beyond first-generation heterosis. J Ecol 106:1595–1606
MacPhee AW (1967) Winter moth Operophtera brumata (Lepidoptera - Geometridae) a new pest attacking apple orchards in Nova Scotia and its coldhardiness. Can Entomol 99:829–834. https://doi.org/10.4039/Ent99829-8
Mannai Y, Ezzine O, Hausmann A et al (2017) Budburst phenology and host use by Operophtera brumata (Linnaeus, 1758) (Lepidoptera: Geometridae) in three Mediterranean oak species. Ann For Sci 74:3. https://doi.org/10.1007/s13595-016-0600-3
Mesgaran MB, Lewis MA, Ades PK et al (2016) Hybridization can facilitate species invasions, even without enhancing local adaptation. Proc Natl Acad Sci USA 113:10210–10214. https://doi.org/10.1073/pnas.1605626113
Michel-Salzat A, Cordaux R, Bouchon D (2001) Wolbachia diversity in the Porcellionides pruinosus complex of species (Crustacea: Oniscidea): evidence for host-dependent patterns of infection. Heredity 87:428–434
Mochiah MB, Ngi-Song AJ, Overholt WA et al (2002) Wolbachia infection in Cotesia sesamiae (Hymenoptera: Braconidae) causes cytoplasmic incompatibility: implications for biological control. Biol Control 25:74–80. https://doi.org/10.1016/S1049-9644(02)00045-2
Moran NA, McCutcheon JP, Nakabachi A (2008) Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet 42:165–190. https://doi.org/10.1146/annurev.genet.41.110306.130119
Nguyen DT, Spooner-Hart RN, Riegler M (2016) Loss of Wolbachia but not Cardinium in the invasive range of the Australian thrips species, Pezothrips kellyanus. Biol Invasions 18:197–214. https://doi.org/10.1007/s10530-015-1002-4
Prentis PJ, Wilson JRU, Dormontt EE et al (2008) Adaptive evolution in invasive species. Trends Plant Sci 13:288–294. https://doi.org/10.1016/j.tplants.2008.03.004
R Core Team (2017) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.r-project.org/. Accessed 6 Mar 2017
Raghavendra K, Barik TK, Reddy BPN et al (2011) Malaria vector control: from past to future. Parasitol Res 108:757–779. https://doi.org/10.1007/s00436-010-2232-0
Reuter M, Pedersen JS, Keller L (2005) Loss of Wolbachia infection during colonisation in the invasive Argentine ant Linepithema humile. Heredity 94:364–369. https://doi.org/10.1038/sj.hdy.6800601
Rhymer JM, Simberloff D (1996) Extinction by hybridization and introgression. Annu Rev Ecol Syst 27:83–109. https://doi.org/10.1146/annurev.ecolsys.27.1.83
Roland J, Embree DG (1995) Biological-control of the winter moth. Annu Rev Entomol 40:475–492. https://doi.org/10.1146/annurev.en.40.010195.002355
Ross PA, Endersby NM, Hoffman AA (2016) Costs of three Wolbachia infections on the survival of Aedes aegypti larvae under starvation conditions. PLoS Negl Trop Dis 10:e0004320. https://doi.org/10.1371/journal.pntd.0004320
Roy V, Girondot M, Harry M (2015) The distribution of Wolbachia in Cubitermes (Termitidae, Termitinae) castes and colonies: a modelling approach. PLoS ONE 10:e0116070. https://doi.org/10.1371/journal.pone.0116070
Roy D, Lucek K, Walter RP, Seehausen O (2016) Hybrid ‘superswarm’ leads to rapid divergence and establishment of populations during a biological invasion. Mol Ecol 24:5394–5411. https://doi.org/10.1111/mec.13405
Schuler H, Bertheau C, Egan SP et al (2013) Evidence for a recent horizontal transmission and spatial spread of Wolbachia from endemic Rhagoletis cerasi (Diptera: Tephritidae) to invasive Rhagoletis cingulata in Europe. Mol Ecol 22:4101–4111. https://doi.org/10.1111/mec.12362/
Schuler H, Koppler K, Daxbock-Horvath S et al (2016) The hitchhiker’s guide to Europe: the infection dynamics of an ongoing Wolbachia invasion and mitochondrial selective sweep in Rhagoletis cerasi. Mol Ecol 25:1595–1609. https://doi.org/10.1111/mec.13571
Schwarz D, Shoemaker KD, Botteri NL, McPheron BA (2007) A novel preference for an invasive plant as a mechanism for animal hybrid speciation. Evolution 61:245–256. https://doi.org/10.1111/j.1558-5646.2007.00027.x
Schwenk K, Brede N, Streit B (2008) Introduction. Extent, processes and evolutionary impact of interspecific hybridization in animals. Philos Trans R Soc Lond B Biol Sci 363:2805–2811. https://doi.org/10.1098/rstb.2008.0055
Seehausen O, Takimoto G, Roy D et al (2008) Speciation reversal and biodiversity dynamics with hybridization in changing environments. Mol Ecol 17:30–44. https://doi.org/10.1111/j.1365-294X.2007.03529.x
Shoemaker DD, Katju V, Jaenike J (1999) Wolbachia and the evolution of reproductive isolation between Drosophila recens and Drosophila subquinaria. Evolution 53:1157–1164. https://doi.org/10.1111/j.1558-5646.1999.tb04529.x
Silva I, Van Meer MMM, Roskam MM et al (2000) Biological control potential of Wolbachia-infected versus uninfected wasps: laboratory and greenhouse evaluation of Trichogramma cordubensis and T. deion strains. Biocontrol Sci Technol 10:223–238. https://doi.org/10.1080/09583150050044501
Simmons MJ, Lee TD, Ducey MJ et al (2014) Effects of invasive winter moth defoliation on tree radial growth in eastern Massachusetts, USA. Insects 5:301–318. https://doi.org/10.3390/insects5020301
Szücs M, Eigenbrode SD, Schwarzlaender M et al (2012) Hybrid vigor in the biological control agent, Longitarsus jacobaeae. Evol Appl 5:489–497. https://doi.org/10.1111/j.1752-4571.2012.00268.x
Todesco M, Pascual MA, Owens GL et al (2016) Hybridization and extinction. Evol Appl 9:892–908. https://doi.org/10.1111/eva.12367
Turelli M, Cooper BS, Richardson KM et al (2018) Rapid-global spread of wRI-like Wolbachia across multiple Drosophila. Curr Biol 28:963–971. https://doi.org/10.1016/j.cub.2018.02.015
Turley AP, Moreira LA, O’Neill SL et al (2009) Wolbachia infection reduces blood-feeding success in the dengue fever mosquito, Aedes aegypti. PLoS Negl Trop Dis 3:e516. https://doi.org/10.1371/journal.pntd.0000516
van den Hurk AF, Hall-Mendelin S, Pyke AT et al (2012) Impact of Wolbachia on infection with chikungunya and yellow fever viruses in the mosquito vector Aedes aegypti. PLoS Negl Trop Dis 6:e1892. https://doi.org/10.1371/journal.pntd.0001892
Verhoeven KJF, Macel M, Wolfe LM et al (2011) Population admixture, biological invasions and the balance between local adaptation and inbreeding depression. Proc R Soc B Biol Sci 278:2–8. https://doi.org/10.1098/rspb.2010.1272
Vilatersana R, Sanz M, Galian A, Castells E (2016) The invasion of Senecio pterophorus across continents: multiple, independent introductions, admixture and hybridization. Biol Invasions 18:2045–2065. https://doi.org/10.1007/s10530-016-1150-1
Weeks AR, Reynolds KT, Hoffmann AA et al (2002) Wolbachia dynamics and host effects: what has (and has not) been demonstrated? Trends Ecol Evol 17:257–262. https://doi.org/10.1016/S0169-5347(02)02480-1
Weinert LA, Araujo-Jnr EV, Ahmed MZ, Welch JJ (2015) The incidence of bacterial endosymbionts in terrestrial arthropods. Proc R Soc B Biol Sci 282:20150249. https://doi.org/10.1098/rspb.2015.0249
Werren JH (1997) Biology of Wolbachia. Annu Rev Entomol 42:587–609. https://doi.org/10.1146/annurev.ento.42.1.587
Werren JH, Baldo L, Clark ME (2008) Wolbachia: master manipulators of invertebrate biology. Nat Rev Microbiol 6:741–751. https://doi.org/10.1038/nrmicro1969
White JA, Richards NK, Laugraud A et al (2015) Endosymbiotic candidates for parasitoid defense in exotic and native New Zealand weevils. Microb Ecol 70:274–286. https://doi.org/10.1007/s00248-014-0561-8
Wolf DE, Takebayashi N, Rieseberg LH (2001) Predicting the risk of extinction through hybridization. Conserv Biol 15:1039–1053. https://doi.org/10.1046/j.1523-1739.2001.0150041039.x
Yang CC, Yu YC, Valles SM et al (2010) Loss of microbial (pathogen) infections associated with recent invasions of the red imported fire ant Solenopsis invicta. Biol Invasions 12:3307–3318. https://doi.org/10.1007/s10530-010-9724-9
Zabal-Aguirre M, Arroyo F, Bella JL (2010) Distribution of Wolbachia infection in Chorthippus parallelus populations within and beyond a Pyrenean hybrid zone. Heredity 104:174–184. https://doi.org/10.1038/hdy.2009.106
Zabalou S, Riegler M, Theodorakopoulou M et al (2004) Wolbachia-induced cytoplasmic incompatibility as a means for insect pest population control. Proc Natl Acad Sci USA 101:15042–15045. https://doi.org/10.1073/pnas.0403853101
Zhang Z, Schwartz S, Wagner L et al (2000) A greedy algorithm for aligning DNA sequences. J Comput Biol 7:203–214. https://doi.org/10.1089/10665270050081478
Zhou WG, Rousset F, O’Neill S (1998) Phylogeny and PCR-based classification of Wolbachia strains using wsp gene sequences. Proc R Soc B Biol Sci 265:509–515. https://doi.org/10.1098/rspb.1998.0324
Zindel R, Gottlieb Y, Aebi A (2011) Arthropod symbioses: a neglected parameter in pest- and disease-control programmes. J Appl Ecol 48:864–872. https://doi.org/10.1111/j.1365-2664.2011.01984.x
Acknowledgements
This work would not have been possible without the collaboration of many individuals who collected samples in both Europe and North America. A complete list of all the collectors is provided in Table S1. We would also like to thank D. Newman for laboratory assistance, and R. Crandall, M. Davis, B. Griffin, R. Gwiazdowski, M. Labbé, J. Lombardo, N.J. Mills, T. Murphy, and G.K. Roderick for their creative comments and suggestions throughout this study. Funding for this project was provided by USDA-FS 13-CA-11420004-236 awarded to JSE, 12-USDA-FS JV-11242303-096 awarded to AC. The authors would also like to thank Dr. James Fordyce and two anonymous reviewers who provided edits and suggestions to an earlier version of the manuscript.
Author information
Authors and Affiliations
Contributions
JSE directed the research. JSE and GHB coordinated the collection of samples. AC provided laboratory access and oversaw the molecular analyses. JCA, NPH, and HJB collected the molecular data. JCA analyzed the dataset and oversaw manuscript preparation. All authors contributed to the project design and in preparing the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Andersen, J.C., Havill, N.P., Broadley, H.J. et al. Widespread hybridization among native and invasive species of Operophtera moths (Lepidoptera: Geometridae) in Europe and North America. Biol Invasions 21, 3383–3394 (2019). https://doi.org/10.1007/s10530-019-02054-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10530-019-02054-1