Abstract
Some insects are infected with maternally inherited bacterial endosymbionts that protect them against pathogens or parasitoids. The weevil Sitona obsoletus (=Sitona lepidus) is invasive in New Zealand, and suspected to contain such defensive symbionts, because it is particularly resistant to a Moroccan strain of the parasitoid Microctonus aethiopoides (which successfully attacks many other weevil species), and shows geographic variation in susceptibility to an Irish strain of the same parasitoid. Using 454 pyrosequencing, we investigated the bacterial community associated with S. obsoletus, two other exotic weevils (Sitona discoideus and Listronotus bonariensis) and two endemic New Zealand weevils (Irenimus aequalis and Steriphus variabilis). We found that S. obsoletus was infected by one strain of Wolbachia and two strains of Rickettsia, none of which were found in any other weevil species examined. Using diagnostic PCR, we found that S. obsoletus in the Northland region, where parasitism is highly variable, were primarily infected with Wolbachia and Rickettsia strain 2, indicating that these two symbionts should be investigated for potential defensive properties. In comparison, S. discoideus lacked any apparent maternally inherited bacterial endosymbionts. In the other weevil species, we found a different strain of Wolbachia and two different strains of Spiroplasma. Two weevil species (St. variabilis and L. bonariensis) were infected with distinct strains of Nardonella, the ancestral endosymbiont of weevils, whereas three weevil species (S. obsoletus, S. discoideus, and I. aequalis) lacked evidence for Nardonella infection. However, I. aequalis was consistently infected with a novel Enterobacteriaceae strain, suggesting that a symbiont replacement may have taken place, similar to that described for other weevil clades.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bacterial endosymbionts perform a wide array of services for their eukaryotic hosts, including provision of defense against natural enemies [1]. Among insect hosts, bacteria have been shown to protect insects against viruses [2, 3], infective nematodes [4], fungal pathogens [5–7], predators [8], and parasitoid wasps [9, 10]. One of the earliest documented examples of bacterial-provided defense was in the invasive alfalfa weevil, Hypera postica (Gyllenhal) (Coleoptera: Curculionidae). Three strains of this weevil became established in North America, only two of which were susceptible to the parasitoid, Microctonus aethiopoides Loan (Hymenoptera: Braconidae), which had been released in an attempt to provide biological control for the weevil [11]. Hsiao [12] found that M. aethiopoides performance in the resistant Western strain of H. postica improved significantly when the weevils had been pretreated with antibiotics. This strain of the weevil is naturally infected with the maternally inherited bacterium Wolbachia [13], which is best known for reproductively manipulating host insects to promote bacterial infection in host populations [14, 15]. The concomitant removal of Wolbachia infection and increase in parasitoid susceptibility led to the hypothesis that Wolbachia was the causative agent of parasitoid defense in H. postica [12].
However, unequivocal demonstration of Wolbachia-provided defense against M. aethiopoides has not been verified, as other strains of bacteria may have co-infected H. postica along with Wolbachia and been simultaneously cured by the antibiotic treatment. Curculionid weevils seem particularly prone to infection by facultative bacterial symbionts, and simultaneous infection by multiple facultative symbionts is common [16–18]. It therefore remains possible that a co-infecting symbiont, rather than Wolbachia, might be responsible for parasitism protection in Western strains H. postica.
Understanding which symbiont(s) provide weevils with defensive characteristics is important, because many weevil species are invasive or have the potential to be invasive worldwide. For example, weevils such as Otiorhynchus sulcatus (F.) (vine weevil), Rhynchophorus ferrugineus Olivier (red palm weevil), Sitona lineatus L. (pea leaf weevil), and H. postica (alfalfa weevil) can cause tremendous economic damage to crops if uncontrolled and consequently have been the focus of significant investment in biological control effort (e.g., [11, 19–21]).
To maximize the probability of success against invasive insect species, classical biological control programs consider many aspects of the target insect and putative biological control agent(s), including correct taxonomic identification, geographic origin, host plants, geographic and climatic range, phenology, and intraspecific genetic variation [22, 23], as well as non-target impacts once the biocontrol agent has been released (e.g., [24]). The potential impact of entomophagous biocontrol agents against target and non-target hosts could also be influenced by the symbiont communities within these host populations. Symbiont infection therefore could potentially be used to predict the efficacy of candidate control agents, if the defensive traits of various bacteria were reliably characterized.
In New Zealand, for example, bacterial symbionts could be responsible for the variable success of biological control programs to control exotic weevil pests in pasture ecosystems. The lucerne pest, Sitona discoideus Gyllenhal (Coleoptera: Curculionidae) [25], has been significantly controlled by a Moroccan strain of M. aethiopoides that was introduced in a classical biological control program [26, 27]. Unfortunately, this parasitoid strain was subsequently found to have a wide non-target host range and to be able to successfully parasitize and develop in many weevil species, including native and introduced beneficial weed biocontrol species [28]. Interestingly, however, this strain of M. aethiopoides readily attacked but was unable to develop in the clover root weevil, Sitona obsoletus Gyllenhal (Coleoptera: Curculionidae), a damaging exotic pest of white clover [29, 30]. This result was also observed by Sundaralingam et al. [31] on S. obsoletus collected in Pennsylvania. While genetic differences between M. aethiopoides strains associated with different Sitona species [32, 33] may explain the poor survival of Moroccan M. aethiopoides, this parasitoid strain’s ability to successfully develop in a wide range of novel hosts suggested that S. obsoletus might possess a defensive microbe that S. discoideus lacks. Additionally, subsequent biological control efforts for S. obsoletus with a more successful strain of M. aethiopoides sourced from Ireland [34] have shown geographic variation in success rates in New Zealand. In Northland (latitude S35), establishment of the parasitoid was variable across sites [35, 36] with high parasitoid egg and larval mortality in some sites but not all (P. Gerard, unpublished data). This was in contrast to observations in Canterbury (latitude S43) where there has been 100 % establishment and negligible larval mortality in the host (unpublished data). These patterns suggest that S. obsoletus shows intraspecific variation in defensive efficacy, which may or may not be associated with bacterial endosymbionts.
As a first step toward understanding the nature of S. obsoletus resistance to parasitism, the objective of this paper was to investigate bacterial diversity associated with this pest species versus other co-occurring exotic and native weevils in New Zealand, and to identify potential endosymbiotic associates of S. obsoletus and other weevil species that might confer resistance to parasitism.
Methods
Specimen Collection and Preparation
We investigated two populations of S. obsoletus: the Canterbury population (S43.70, E172.33), which typically is subject to high levels of parasitism from the introduced Irish parasitoid M. aethiopoides, and the Northland population (S35.00, E173.49), which typically exhibits lower mean levels of parasitism [36]. We collected adult specimens from each population and larval specimens from the Canterbury population, as indicated in Table S1. We also collected specimens from four other Curculionidae weevil species; two that have been accidently introduced to New Zealand (S. discoideus and Argentine stem weevil Listronotus bonariensis Kuschel) and two native New Zealand weevil species (Irenimus aequalis Broun and Steriphus variabilis Broun). Adults of S. obsoletus, L. bonariensis, I. aequalis, and St. variabilis were collected from ryegrass (Lolium perenne)/white clover (Trifolium repens) pasture while S. discoideus was collected from a lucerne (Medicago sativa) crop. Weevils were collected using a modified blower vac to suck the weevils into a collecting net fitted to the inlet that was then hand sorted to extract the weevils [37]. Larvae were collected from soil around the roots of white clover plants growing in pasture. Adult weevils were examined under a stereomicroscope (×10 magnification) and male and female weevils identified by the shape of the distal ventrite (e.g., [38–40]).
All specimens were preserved in 95 % ethanol. Between 8 and 18 specimens (as indicated in Table S1) were selected from each population, and individually surface sterilized in 5 % sodium hypochlorite solution (1 min), followed by three rinses in 95 % ethanol (1 min each), and a final rinse in sterile PCR grade water (1 min). For the two larger weevil species, S. obsoletus and S. discoideus, elytra were removed prior to homogenization. DNA was extracted from individual specimens using DNeasy blood and tissue kits (Qiagen, Valencia, CA) following manufacturer’s specifications. Proteinase K digestion was performed for 3 h at 56 °C. All specimens were re-eluted in two washes of 100 μl elution buffer, except for L. bonariensis specimens, which due to their smaller size were resuspended in two washes of 50 μl elution buffer.
454 Pyrosequencing
To characterize overall bacterial community diversity per species, eight pooled samples of DNA, representing three introduced and two native New Zealand weevil species (Table S1), were analyzed for bacterial diversity. For each sample, DNA from eight to ten specimens of each species was quantified using a Nanodrop 2000 spectrophotometer (Thermo Scientific, Wilmington, Delaware), a subsample of DNA per specimen was standardized to a concentration of 20 ng DNA/μl, and then pooled into a single sample. The samples were sent to Research and Testing Laboratory (Lubbock, TX) where they were multiplexed in bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP) using a 454 FLX instrument (Roche, Nutley, New Jersey) and analyzed as described in Brady and White [41]. Samples were run in two batches on different dates (Table S1). All samples were evaluated with 28 F (5′-GAGTTTGATCNTGGCTCAG-3′) and 519R (5′-GWNTTACNGCGGCKGCTG-3′) primers, which amplified approximately 500 bp of the 5′ end of bacterial 16S rRNA [42–44]. To determine whether results were sensitive to primer set selection, two of the S. obsoletus samples and one St. variabilis sample (identified in Table S1) were additionally evaluated with 939F (5′-TTGACGGGGGCCCGCAC-3′) and 1492R (5′-TACCTTGTTACGACTT-3′) primers, which amplified approximately 550 bp of the 3′ end of bacterial 16S rRNA [45].
Diagnostic Screening and Strain Typing
Based on the pyrosequencing results, DNA from individual specimens was diagnostically screened for four bacterial genera (Wolbachia, Rickettsia, Spiroplasma, and Nardonella), all of which have been associated with weevils as endosymbionts. Diagnostic PCR reactions were conducted using published bacterial primer sets, each specific to one of the endosymbiotic genera and performed as described by the original paper (Table S2). For Nardonella, we designed an alternate reverse primer that was better matched to bacterial strains discovered in this study (Table S2). PCR products were run on a 1 % agarose gel stained with either Gel-Red (Biotium, Hayward, CA) or RedSafe™ (iNtRON Biotechnology, Korea). All PCR sets included DNA from specimens of known infection status as positive controls, as well as a template-free reaction as a negative control. For populations in which any of these bacterial genera were detected, the PCR product from at least one specimen per population was sequenced at the University of Kentucky Advanced Genetic Technologies Center (UK-AGTC) using an ABI 3730 DNA Analyzer (Applied Biosystems) to verify bacterial identity.
Additional Sanger (BigDye®v 3.1, Applied Biosystems) sequencing was conducted at the UK-AGTC or at Macrogen (Korea) to (1) determine strain type distribution of Rickettsia symbionts across populations of S. obsoletus, (2) determine the multilocus sequence type (MLST) for Wolbachia strains detected in S. obsoletus and St. variabilis and (3) investigate a dominant but unusual symbiont detected in the native weevil I. aequalis.
-
1.
Because 16S pyrosequencing results suggested the presence of two different Rickettsia strains in S. obsoletus, we designed Rickettsia-specific primers to amplify two overlapping segments of 16S to yield almost the entire 1500 bp 16S sequence (Rick16S480R used with universal 16SA1F, Rick16S460F, and Rick16S1530R; Table S2). The majority of the resulting Rick16S460F and Rick16S1530R PCR products were sequenced to determine the Rickettsia strain type per individual; based on chromatogram inspection, we saw no evidence of multiple Rickettsia infections within a single individual.
-
2.
For the two Wolbachia positive weevil species (S. obsoletus and St. variabilis), we amplified and sequenced five multilocus sequence type (MLST) housekeeping genes from a single individual per species as described by Baldo et al. [46] (see also Table S2).
-
3.
On finding a dominant and novel Enterobacteriaceae sequence in I. aequalis, we designed specific primers to amplify the whole 16S gene of this potential endosymbiont by first aligning 16S gene sequences from a range of closely related endosymbiont bacteria from the same family and targeting the variable regions among genera. Forward primers specific for the novel sequence were combined with reverse primers that target Enterobacteriaceae in general (IaqEnteroF2, IaqEnteroF6, Entero16S760R, and Entero16S1500R; Table S2). Reactions contained Taq DNA polymerase (iNtRON Biotechnology i-StarTaq TM; at approx. 1 unit/ 10 μl PCR), 2 mM MgCl2, 1 mM dNTPs, 0.5 μM primers and template DNA at approx.4 ng per μl of PCR reaction. Cycling conditions started at 95 °C for 2 min, followed by 35 cycles of 95 °C for 30 s, 55 °C for 30 s and 72 °C for 60 s; the final step was 5 min at 72 °C. All I. aequalis specimens were diagnostically screened for the presence of this bacterial strain, and the PCR products from a single individual were sequenced.
Phylogenetic Analysis
16S sequences from this study were compared to previously published bacterial symbionts of weevils [18, 47], or their closest 16S rRNA sequences found in the GenBank ® nucleotide database [48] following use of the Basic Local Alignment Search Tool (BLAST; http://blast.ncbi.nlm.nih.gov/Blast). The individual and concatenated Wolbachia MLST genes were trimmed and compared to data provided on the MLST website (http://pubmlst.org/wolbachia/) developed by Keith Jolley and sited at the University of Oxford [49]. The development and maintenance of this site has been funded by the Wellcome Trust.
Multiple alignments of the nucleotide sequences were generated using the program Muscle [50]. The program Seaview [51] was used to visualize and edit the alignments when needed. Phylogenetic analyses were performed using maximum likelihood (ML) inference with Phyml v3.0.1 [52]. The model selected was GTR + I + G for Rickettsia (1476 sites), Spiroplasma (2426 sites), Wolbachia (2079 sites) and for the main Enterobacteriaceae tree (2679 sites). The robustness of the nodes was assessed with 500–1000 bootstrap replicates. This process was repeated using the model HKY85 + I but produced the same topologies, hence only the results for the GTR models are presented. Finally, because Wolbachia strains can undergo intergenic recombination [53], we also conducted individual analyses on each of the five Wolbachia housekeeping genes, using the same protocols. All nexus files are available in the supplemental material.
Results
Archived Sequences
Sequences resulting from this study have been deposited in the NCBI sequence read archive under project accession number SRP041582 (pyrosequences) and in GenBank under accession numbers KJ494864-8, and KJ522437-49 (Sanger sequences).
Sitona obsoletus
Pyrosequencing of the bacterial community across populations of the clover root weevil, S. obsoletus, indicated that the maternally inherited bacterial endosymbionts Wolbachia and Rickettsia were present in all pooled samples (Table 1). Reads from these two bacterial genera dominated all of the adult samples from Canterbury, composing 85–97 % of reads. S. obsoletus larvae from Canterbury additionally had strong representation from bacteria in the genus Rhizobium (25 %), presumably from consuming nodulated legume roots that were colonized by these bacteria. In contrast, the majority (49–64 %) of reads from the Northland sample aligned with bacteria in the genus Pantoea (Family Enterobacteriaceae), bacteria that are frequently associated with both plant and insect samples. Wolbachia and Rickettsia were also both present in the Northland sample, but Rickettsia, in particular, contributed a relatively minor proportion of reads (3–4 %) when compared to the two adult Canterbury samples, which had 25 and 52 % Rickettsia. The two adult Canterbury samples, which were collected on different dates and analyzed in different pyrosequencing runs, yielded a similar range of bacterial species, but the percentage composition was variable. For the two S. obsoletus samples that were analyzed using two different primer sets (28F–519R and 939F–1492R), the community composition appeared to be largely congruent.
Diagnostic PCR showed that most (97 %) of individual specimens of S. obsoletus from Canterbury and all from Northland were infected with Wolbachia (Table 2). This Wolbachia strain has not previously been described: three of the five MLST genes had novel alleles that were new accessions to the Wolbachia PubMLST database (coxA, gatB, hcpA; Figs S1-S5). A reconstructed phylogeny using the concatenated MLST sequences from five genes placed this Wolbachia strain in supergroup A, not too distant from Wolbachia strains in Ceutorhynchus weevils (Fig. 1). Neither the pyrosequencing nor Sanger sequencing of a subset of individuals gave any indication that multiple strains of Wolbachia might be present in S. obsoletus. Rickettsia was present in the majority of specimens from both populations; 90 % of Canterbury specimens and 66 % of Northland specimens were infected by Rickettsia. Furthermore, both pyrosequencing and Sanger sequencing indicated that two different strains of Rickettsia were present in both populations. The 16S sequence from these two strains diverged by 11/1294 bases (0.9 %), and both were strongly supported within a Rickettsial clade that has only been recovered from beetles, including many weevil species (Fig. 2). Both strains of Rickettsia were present in both populations, but their frequency of occurrence differed. Whereas the majority of Rickettsia-infected weevils in Canterbury had strain 1 (25/32 = 78 %), only 2/6 (33 %) Rickettsia-infected weevils from Northland had strain 1 (P = 0.047, Fisher’s exact test). The pyrosequencing results similarly showed a higher ratio of strain 1 relative to strain 2 in Canterbury (40–60× more strain 1 than strain 2 reads) than Northland (1.75–2.6× more strain 1 than strain 2 reads). Inspection of the Sanger sequencing chromatograms did not show evidence of multiple Rickettsia strains within an individual, but the possibility of multiple Rickettsia infection cannot be entirely excluded.
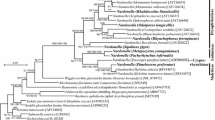
Phylogenetic placement of Wolbachia strains found in exotic and native weevils of New Zealand, based on five concatenated Wolbachia multilocus sequence typing (MLST) sequences. Supergroups and sequence types (ST) are as identified by the Wolbachia MLST website (http://pubmlst.org/wolbachia/). Endosymbionts are represented by ES before the general and Latin names of the host insect. Sequences obtained from this study are in bold type. Bootstrap values for the maximum likelihood analysis (×1000 replicates) are shown above or below nodes only where greater than 50
Phylogenetic placement of the two Rickettsia strains found in the exotic New Zealand weevil, Sitona obsoletus based on 16S ribosomal RNA. Genbank accession numbers are shown in brackets. Endosymbionts are designated by ES, followed by the general and Latin names of the host insect. Free-living bacteria appear on their own. Sequences obtained from this study are in bold type. Bootstrap values for the maximum likelihood analysis (×1000 replicates) are shown above or below nodes only where greater than 50
Other Invasive Weevils
In contrast with S. obsoletus, the invasive congener S. discoideus lacked any apparent maternally inherited bacterial endosymbionts. This weevil was characterized by a diverse community of environmental bacteria (Table 3). The highest prevalence bacterial strain fell within the Acidovorax/Diaphorobacter clade, of the family Comamondaceae, but almost 50 other bacterial genera were represented, including 30 genera at <1 % prevalence of reads within the pyrosequenced sample. The pyrosequenced bacterial community from the other invasive weevil, L. bonariensis, was much less diverse, with 89 % of reads belonging to a single bacterial taxon in the Enterobacteriaceae, corresponding to Candidatus Nardonella, the predominant bacterial associate of some weevil species (Fig. 3) [54]. We also detected 170 (4.5 %) reads of this same sequence associated with only one of the six S. obsoletus samples, which we suspect was the result of contamination during the pyrosequencing process or sample handling. Aside from Nardonella, 16 bacterial genera were detected in L. bonariensis, but none were from known endosymbiotic clades. Diagnostic screens corroborated the presence of Nardonella in all L. bonariensis specimens (Table 2), and the absence of Rickettsia, Spiroplasma, and Wolbachia in L. bonariensis and S. discoideus specimens. Nardonella was not detected in any S. obsoletus or S. discoideus specimens (Table 2).
Phylogenetic placement of potential nutritional endosymbionts found in exotic and native New Zealand weevils based on 16S ribosomal RNA. Genbank accession numbers are shown in brackets. Latin names are given where the bacteria has been identified; endosymbiotic strains are designated by ES, followed by the general and Latin names of the host insect. Free-living bacteria appear on their own. Sequences obtained from this study are in bold type. Bootstrap values for the maximum likelihood analysis (×500 replicates) are shown above or below nodes only where greater than or equal to 50
Native Weevils
We detected different facultative and apparently obligate endosymbiotic bacteria from both native weevils. St.variabilis showed a similar range of bacterial diversity regardless of the primer set used, but in this case the percentage composition was markedly different between primer sets. For 28F–519R, the total read count was small (426 reads), and most came from environmental bacteria (Table 3). However, both Wolbachia and Nardonella were detected, albeit at relatively low prevalence. These same bacteria were also detected using the 939F–1492R primer set, but here Nardonella was the dominant bacterial strain, comprising 76 % of the 3146 reads. Diagnostic screening showed this bacterium to be present in all but one St. variabilis specimen (Table 2). Phylogenetic analysis of a 1294 bp segment of 16S rRNA strongly supported both this and the dominant L. bonariensis Enterobacteriaceae strains as members of the Nardonella clade.
The Wolbachia in St. variabilis was found in every individual (Table 2) and a reconstructed phylogeny using the concatenated MLST sequences from five genes placed it in supergroup B, revealing that this strain was quite distinct from the Wolbachia found in S. obsoletus (Fig. 1). Four of the five MLST genes had novel alleles that were new accessions to the Wolbachia PubMLST database (coxA, ftsZ, gatB, hcpA; Figs S1–S5). Phylogenies based on the individual MLST genes all supported placement of the St. variabilis Wolbachia in supergroup B and the S. obsoletus Wolbachia in supergroup A (Figs. S1–S5).
In I. aequalis, the most highly represented bacterial clade was Spiroplasma, constituting 42 % of the 23,000 reads (Table 3). These reads could clearly be broken into two distinct strains, differing from one another by 82/486 bp (17 %). Using Spiroplasma-specific diagnostic PCR of individual specimens, we found that two individuals out of eight, 1 male and 1 female, were infected with Spiroplasma. Sanger sequencing indicated that each of these individuals was infected with a different strain of Spiroplasma. The Spiroplasma in the female was very similar (99.5 %) to endosymbionts recovered from other weevil species in the genus Curculio (accession numbers AB545038, JQ692307, JN100091 [16, 18]). The endosymbiotic nature of this strain from I. aequalis was strongly corroborated by its clustering with sequences from a clade composed entirely of endosymbiotic Spiroplasma strains (Fig. 4). The Spiroplasma recovered from the male I. aequalis specimen, on the other hand, fell within a clade that contains both endosymbiotic associates of insects and an insect-vectored plant pathogen (Spiroplasma citri) (Fig. 4).
Phylogenetic placement of the two Spiroplasma strains found in the native New Zealand weevil Irenimus aequalis based on 16S ribosomal RNA. Genbank accession numbers are shown in brackets. Latin names are given where the bacteria has been classified; Endosymbiotic strains are designated by ES, followed by the general and Latin names of the host insect. Free-living bacteria appear on their own. Sequences obtained from this study are in bold type. Bootstrap values for the maximum likelihood analysis (×1000 replicates) are shown above or below nodes only where greater than 50
We also detected an unknown bacterium within I. aequalis, constituting 13 % of reads (Table 3). Diagnostic PCR using newly designed primers (Table S2) found that this bacterial strain was present in all I. aequalis specimens. There were no close matches for this bacterium in Genbank, with the closest matches for a 1486-bp segment of 16S rRNA being an unidentified bacteria at ~93 % similarity, and bacteria within the Xenorhabdus genus (Class Gammaproteobacteria, Order Enterobacteriales Family Enterobacteriaceae) at 89.6 % similarity. The reconstructed phylogeny, using known endosymbionts from Enterobacteriaceae, placed this novel bacterium quite distantly from the Nardonella clade of weevil endosymbionts [54] (Fig. 3). However, the presence of the novel bacteria in all specimens, as well as the absence of Nardonella (Table 2), suggest that these bacteria might play a similar nutritional role [17, 55].
Discussion
Maternally Inherited Facultative Symbionts with Defensive Potential
We found three maternally inherited endosymbiotic bacterial taxa in S. obsoletus that plausibly might provide defense against parasitoids: one strain of Wolbachia, and two different strains of Rickettsia. Bacteria in the genus Wolbachia are obligate inhabitants of arthropod hosts and are often reproductive parasites [15]. However, multiple recent studies have found that Wolbachia can protect insect hosts against viruses [2, 3, 56], bolstering the possibility that strains of this versatile bacterium might be capable of protecting weevils such as S. obsoletus and H. postica from parasitism as originally postulated by Hsiao [12]. Bacteria in the genus Rickettsia are nearly as diverse in their documented effects on arthropod hosts as Wolbachia (reviewed in [57, 58]), including a strain that has recently been shown to protect an aphid from a fungal pathogen [7]. Moreover, both Wolbachia and Rickettsia can be found in host hemolymph [59, 60], where parasitoids such as M. aethiopoides insert their eggs (e.g., [61]). Bacteria that are restricted to only a small portion of the host’s anatomy (e.g., gut bacteria such as Pantoea, or bacteria that are restricted to bacteriomes) are likely of limited utility in defending the host against parasitoids that insert their eggs elsewhere in the host’s body.
It is not yet clear if any of these symbiont strains are involved with parasitoid defense of S. obsoletus. These strains represent prospects for further investigation. To attribute symbiont causality, mechanistic tests of parasitoid success in differentially infected hosts will be necessary. In the meantime, correlative data from the present study suggests that two of the three symbiont strains may be particularly worth investigating for defensive properties against parasitoids. S. obsoletus throughout New Zealand is highly resistant to a Moroccan strain of the parasitoid M. aethiopoides [30, 40], but shows geographic variation in susceptibility to the Irish strain of the same parasitoid [62]. The latter effect is not driven by parasitoid genetic variation, as parasitoid populations throughout New Zealand are derived from the same genetically homogeneous source colony [63, 64]. Wolbachia is the best candidate for a protective role against the Moroccan M. aethiopoides strain, given its nearly universal presence in tested individuals. In contrast, Rickettsia strain 2 is the best candidate for protection against the Irish parasitoid strain, as this symbiont was found at higher prevalence in Northland, where parasitism rates are lower. Other studies have documented that symbiont strains can vary in their efficacy against different parasitoid strains [65, 66], corroborating the possibility that multiple symbiont strains may be involved in defense in S. obsoletus. Furthermore, the invasive congener of this species, S. discoideus, lacks Rickettsia and Wolbachia and is highly susceptible to both Moroccan and Irish strains of the parasitoid (M. McNeill, unpublished data). Should both symbionts prove to be defensive, S. obsoletus would be a useful study system for examining complementary defensive roles of multiple symbionts in an economically important host.
Neither Wolbachia nor Rickettsia were widespread in other weevils examined in this study, despite their relatively high prevalence in weevils overall [16–18], suggesting that horizontal transmission among co-occurring weevils is not frequent within this system. No other weevil species in this study had Rickettsia, and only one other species had Wolbachia. The Wolbachia in St. variabilis was clearly distinct from that of S. obsoletus, however, belonging to a different clade (supergroup B). Across weevils, both supergroup A and B Wolbachia strains have previously been reported [16, 18], but functional roles have not been attributed to any sequenced strains from weevils. The early work on potentially defensive Wolbachia from H. postica [12] was conducted before easy sequencing and strain-typing methodologies were developed, thus sequence information from the Wolbachia from H. postica is not available as a benchmark for comparison.
We also found two strains of Spiroplasma in I. aequalis. As a symbiont, Spiroplasma can induce reproductive manipulations in some insect hosts [67], but has been associated with parasitoid and nematode defense in others [4, 10]. However, some Spiroplasma strains are horizontally transmitted insect pathogens, insect-vectored plant pathogens or free-living in the environment [68]. Phylogenetic placement supports the likelihood that Spiroplasma strain 1 is a maternally transmitted facultative symbiont, given that it falls with high support in a clade entirely composed of symbiotic associates of beetles. The clade for Spiroplasma strain 2 was more diverse, containing both free-living and symbiotic Spiroplasma strains.
While this survey has focused on bacteria that are obligate and heritable associates of insects, the particularly high prevalence of Pantoea within the Northland S. obsoletus sample is also worth noting. Pantoea strains are frequent gut associates of insects, but typically are not maternally inherited: they are cultivable bacteria that are also common in plant-associated samples, and are sometimes pathogenic [69–72]. We found strains of Pantoea associated with all of the weevil species we examined, and all strains were very similar to environmental isolates, suggesting that all were environmentally acquired. Nevertheless, some environmentally acquired gut bacteria have been shown to have major phenotypic effects on their insect hosts [73], and Pantoea has been shown to influence the fitness of the insects whose guts they inhabit [74]. While relatively unlikely to play a defensive role, Pantoea may be worth further investigation as a potentially important associate of S. obsoletus.
Potential Nutritional Symbionts
Many weevil species are infected with maternally inherited symbionts that aid nutrition and promote normal growth and development [17, 55, 75]. These bacteria are typically housed in specialized organs, bacteriomes, that are associated with the larval gut (reviewed in [76]). The ancestral weevil endosymbiont, Candidatus Nardonella, has co-evolved with its weevil hosts for an estimated 125 million years [53, 75]. However, various weevil clades lack Nardonella. Some of these weevils are instead infected with bacteria from alternate clades [47, 77, 78], and others apparently lack nutritional endosymbionts entirely [17, 54].
In the present study, we found evidence of Nardonella in some of the weevil species examined, but not others. Both L. bonariensis and St. variabilis were infected with bacterial strains that fell firmly within the Nardonella clade. These strains were distinct from one another (90 % similarity), as would be expected for vertically transmitted symbionts that have codiversified along with their hosts [79]. In contrast, Nardonella was not detected at all from S. discoideus or I. aequalis, and the few pyrosequencing reads detected from one S. obsoletus sample were exactly identical to those from L. bonariensis, suggesting that they resulted from sample contamination or sequencing barcode errors, rather than a true infection. Diagnostic tests using Nardonella-specific primers were consistently negative for all specimens from all three of these species.
In the case of the endemic weevil I. aequalis, it is possible that a nutritional symbiont replacement has taken place. An unclassified strain of Enterobacteriaceae was substantially represented among the sequences (~13 %), and was diagnostically found to be represented in all specimens tested. The 16S sequence of this bacterium was distinct from any Genbank accessions (<93 % similarity), corroborating the hypothesis that this is a specialized insect associate, rather than a general environmental bacterium. This bacterium is dissimilar to any previously described obligate weevil symbionts (Fig. 3), and thus may represent an independent symbiont replacement event at some point within the evolutionary history of I. aequalis, similar to the replacements that have taken place in Curculio and Sitophilus weevil genera [17, 75, 77]. However, validation of the symbiont replacement hypothesis for I. aequalis awaits physiological studies of bacterial localization, and/or functional tests of bacterial function within the host.
For the two Sitona species, it remains to be determined if any maternally inherited bacteria play a nutritional role. Lefevre et al. [54] found that nutritional endosymbionts had been lost from Sitophilus linearis, a weevil species that had shifted hosts from an ancestral cereal to a more nutritious legume seed. As Sitona weevils also feed on legumes [80], this clade of weevils may not need nutritional assistance, and may have lost their ancestral symbiont as a consequence.
Conclusions
The present work reinforces that weevils are host to a diverse array of bacterial associates, some of which may play a role in parasitoid defense, and others in host nutrition. To date, research on symbiont-derived defense among insect hosts has largely focused on aphids [1] and Drosophila [10]. Despite the fact that a weevil was the earliest suggested example of symbiont-provided defense against parasitoids [12], it remains unclear exactly which bacterial strains might provide such defense, which weevil species might benefit from it, and what mechanism provides the defense. Here, we identify three bacterial strains that infect S. obsoletus, as candidate defensive symbionts. Based on their sequences, none of these bacteria are particularly unusual, belonging to clades that are widespread endosymbionts of insect in general, and weevils in particular. Should any of these symbionts prove to be defensive, the implications would be far-reaching, given that weevils are one of the most diverse clades of animals on earth, with numerous species of economic import as either pests or agents of biological control [81].
References
Oliver KM, Moran NA (2009) Defensive symbionts in aphids and other insects. In: White JF, Torres MS (eds) Defensive mutualism in microbial symbiosis. Taylor & Francis, London, pp 129–147
Hedges LM, Brownlie JC, O’Neill SL, Johnson KN (2008) Wolbachia and virus protection in insects. Science 322(5902):702. doi:10.1126/science.1162418
Teixeira L, Ferreira A, Ashburner M (2008) The bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster. PLoS Biol 6(12):2753–2763. doi:10.1371/journal.pbio.1000002
Jaenike J, Unckless R, Cockburn SN, Boelio LM, Perlman SJ (2010) Adaptation via symbiosis: recent spread of a Drosophila defensive symbiont. Science 329(5988):212–215. doi:10.1126/science.1188235
Kaltenpoth M, Göttler W, Herzner G, Strohm E (2005) Symbiotic bacteria protect wasp larvae from fungal infestation. Curr Biol 15(5):475–479. doi:10.1016/j.cub.2004.12.084
Scarborough CL, Ferrari J, Godfray HCJ (2005) Aphid protected from pathogen by endosymbiont. Science 310(5755):1781–1781. doi:10.1126/science.1120180
Lukasik P, van Asch M, Guo HF, Ferrari J, Godfray HCJ (2013) Unrelated facultative endosymbionts protect aphids against a fungal pathogen. Ecol Lett 16(2):214–218. doi:10.1111/ele.12031
Kellner RLL (2002) Molecular identification of an endosymbiotic bacterium associated with pederin biosynthesis in Paederus sabaeus (Coleoptera : Staphylinidae). Insect Biochem Mol 32(4):389–395. doi:10.1016/S0965-1748(01)00115-1
Oliver KM, Russell JA, Moran NA, Hunter MS (2003) Facultative bacterial symbionts in aphids confer resistance to parasitic wasps. Proc Natl Acad Sci U S A 100(4):1803–1807. doi:10.1073/pnas.0335320100
Xie JL, Vilchez I, Mateos M (2010) Spiroplasma bacteria enhance survival of Drosophila hydei attacked by the parasitic wasp Leptopilina heterotoma. PLoS ONE 5(8):e12149. doi:10.1371/journal.pone.0012149
Radcliffe EB, Flanders KL (1998) Biological control of alfalfa weevil in North America. Integr Pest Manag Rev 3(4):225–242. doi:10.1023/A:1009611219360
Hsiao T (1996) Studies of interactions between alfalfa weevil strains, Wolbachia endosymbionts and parasitoids. In: Symondson WOC, Liddell JE (eds) The ecology of agricultural pests. Chapman & Hall, London, pp 51–72
Leu S-JC, Li JK-K, Hsiao TH (1989) Characterization of Wolbachia postica, the cause of reproductive incompatibility among alfalfa weevil strains. J Invert Pathol 54(2):248–259. doi:10.1016/0022-2011(89)90035-9
Stouthamer R, Breeuwer JA, Hurst GD (1999) Wolbachia pipientis: microbial manipulator of arthropod reproduction. Annu Rev Microbiol 53:71–102. doi:10.1146/annurev.micro.53.1.71
Werren JH, Baldo L, Clark ME (2008) Wolbachia: master manipulators of invertebrate biology. Nat Rev Microbiol 6(10):741–751. doi:10.1038/nrmicro1969
Toju H, Fukatsu T (2011) Diversity and infection prevalence of endosymbionts in natural populations of the chestnut weevil: relevance of local climate and host plants. Mol Ecol 20(4):853–868. doi:10.1111/j.1365-294X.2010.04980.x
Toju H, Tanabe AS, Notsu Y, Sota T, Fukatsu T (2013) Diversification of endosymbiosis: replacements, co-speciation and promiscuity of bacteriocyte symbionts in weevils. ISME J 7(7):1378–1390. doi:10.1038/ismej.2013.27
Merville A, Venner S, Henri H, Vallier A, Menu F, Vavre F, Heddi A, Bel-Venner M-C (2013) Endosymbiont diversity among sibling weevil species competing for the same resource. BMC Evol Biol 13(1):28. doi:10.1186/1471-2148-13-28
Vankosky MA, Carcamo HA, Dosdall LM (2011) Identification of potential natural enemies of the pea leaf weevil. Sitona lineatus L. in western Canada. J Appl Entomol 135:293–301. doi:10.1111/j.1439-0418.2010.01542.x
Moorhouse E, Charnley A, Gillespie A (1992) A review of the biology and control of the vine weevil, Otiorhynchus sulcatus (Coleoptera: Curculionidae). Ann Appl Biol 121(2):431–454. doi:10.1111/j.1744-7348.1992.tb03455.x
Murphy S, Briscoe B (1999) The red palm weevil as an alien invasive: biology and the prospects for biological control as a component of IPM. Biocontrol News Inf 20:35N–46N
Gurr GM, Wratten SD (2000) Biological control: measures of success. Kluwer, Dordrecht, p 429
Phillips CB, Vink CJ, Blanchet A, Hoelmer KA (2008) Hosts are more important than destinations: what genetic variation in Microctonus aethiopoides (Hymenoptera: Braconidae) means for foreign exploration for natural enemies. Mol Phylogenet Evol 49(2):467–476. doi:10.1016/j.ympev.2008.08.005
Babendreier D, Bigler F, Kuhlmann U (2005) Methods used to assess non-target effects of invertebrate biological control agents of arthropod pests. Biocontrol 50(6):821–870. doi:10.1007/s10526-005-3633-3
Goldson S, Dyson C, Proffitt J, Frampton E, Logan J (1985) The effect of Sitona discoideus Gyllenhal (Coleoptera: Curculionidae) on lucerne yields in New Zealand. B Entomol Res 75(3):429–442. doi:10.1017/S000748530001453X
Stufkens M, Farrell J, Goldson S (1987) Establishment of Microtonus aethiopoides, a parasitoid of the sitona weevil in New Zealand. Proc New Zealand Weed and Pest Control Conference. New Zealand Weed and Pest Control Society Inc., pp. 31–35
Kean JM, Barlow ND (2000) Long-term assessment of the biological control of Sitona discoideus by Microctonus aethiopoides and test of a model. Biocontrol Sci Technol 10(3):215–221. doi:10.1080/09583150050044493
Barratt B, Evans A, Ferguson C, Barker G, McNeill M, Phillips C (1997) Laboratory nontarget host range of the introduced parasitoids Microctonus aethiopoides and M. hyperodae (Hymenoptera: Braconidae) compared with field parasitism in New Zealand. Environ Entomol 26(3):694–702
Barratt B, Evans A, Ferguson C, O’Callaghan M (1997) Potential for biocontrol of Sitona lepidus Gyllenhal by Microctonus spp. Proc New Zealand Plant Protection Conference. New Zealand Plant Protection Society Inc, pp. 37–40
McNeill M, Barratt B, Evans A (2000) Behavioural acceptability of Sitona lepidus (Coleoptera: Curculionidae) to the parasitoid Microctonus aethiopoides (Hymenoptera: Braconidae) using the pathogenic bacterium Serratia marcescens Bizio. Biocontrol Sci Technol 10(3):205–213. doi:10.1080/09583150050044484
Sundaralingam S, Hower A, Kim K (2001) Host selection and reproductive success of French and Moroccan populations of the parasitoid, Microctonus aethiopoides (Hymenoptera: Braconidae). BioControl 46(1):25–41. doi:10.1023/A:1009914907209
Phillips C, Cane R, Mee J, Chapman H, Hoelmer K, Coutinot D (2002) Intraspecific variation in the ability of Microctonus aethiopoides (Hymenoptera: Braconidae) to parasitise Sitona lepidus (Coleoptera: Curculionidae). N Z J Agric 45(4):295–303. doi:10.1080/00288233.2002.9513519
Vink C, Phillips C, Mitchell A, Winder L, Cane R (2003) Genetic variation in Microctonus aethiopoides (Hymenoptera: Braconidae). Biol Control 28(2):251–264. doi:10.1016/S1049-9644(03)00103-8
Goldson SL, McNeill MR, Proffitt JR, Barratt BIP (2005) Host specificity testing and suitability of a European biotype of the braconid parasitoid Microctonus aethiopoides as a biological control agent against Sitona lepidus (Coleoptera : Curculionidae) in New Zealand. Biocontrol Sci Technol 15(8):791–813. doi:10.1080/09583150500136444
Gerard P, Eden T, Hardwick S, Mercer C, Slay M, Wilson D (2007) Initial establishment of the Irish strain of Microctonus aethiopoides in New Zealand. N Z Plant Protect 60:203
Gerard P, Wilson D, Eden T (2010) Clover root weevil biocontrol distribution in the North Island—release tactics and outcomes. Proc N Z Grassl Assoc 72:85–89
Goldson SL, McNeill MR, Gerard PJ, Proffitt JR, Phillips CB, Cane RP, Murray PJ (2004) British-based search for natural enemies of the clover root weevil, Sitona lepidus in Europe. N Z J Zool 31(3):233–240
Bright DE (1994) Revision of the genus Sitona (Coleoptera, Curculionidae) of North America. Ann Entomol Soc Am 87(3):277–306
Goldson SL, Emberson RM (1981) Reproductive morphology of the argentine stem weevil, Hyperodes bonariensis (Coleoptera, Curculionidae). N Z J Zool 8(1):67–77. doi:10.1080/03014223.2004.9518375
Barratt BIP, Kuschel G (1996) Broad-nosed weevils (Curculionidae: Brachycerinae: Entimini) of the Lammermoor and Rock and Pillar Ranges in Otago, with descriptions of four new species of Irenimus. N Z J Zool 23(4):359–374. doi:10.1080/03014223.1996.9518096
Brady CM, White JA (2013) Cowpea aphid (Aphis craccivora) associated with different host plants has different facultative endosymbionts. Ecol Entomol 38(4):433–437. doi:10.1111/een.12020
Dowd SE, Callaway TR, Wolcott RD, Sun Y, McKeehan T, Hagevoort RG, Edrington TS (2008) Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP). BMC Microbiol 8(1):125. doi:10.1186/1471-2180-8-125
Medina RF, Nachappa P, Tamborindeguy C (2011) Differences in bacterial diversity of host-associated populations of Phylloxera notabilis Pergande (Hemiptera: Phylloxeridae) in pecan and water hickory. J Evol Biol 24(4):761–771. doi:10.1111/j.1420-9101.2010.02215.x
Ishak HD, Plowes R, Sen R, Kellner K, Meyer E, Estrada DA, Dowd SE, Mueller UG (2011) Bacterial diversity in Solenopsis invicta and Solenopsis geminata ant colonies characterized by 16S amplicon 454 pyrosequencing. Microb Ecol 61(4):821–831. doi:10.1007/s00248-010-9793-4
Steelman SM, Chowdhary BP, Dowd S, Suchodolski J, Janečka JE (2012) Pyrosequencing of 16S rRNA genes in fecal samples reveals high diversity of hindgut microflora in horses and potential links to chronic laminitis. BMC Vet Res 8(1):231. doi:10.1186/1746-6148-8-231
Baldo L, Hotopp JCD, Jolley KA, Bordenstein SR, Biber SA, Choudhury RR, Hayashi CY, Maiden MCY, Tettelin H, Werren JH (2006) Multilocus sequence typing system for the endosymbiont Wolbachia pipientis. Appl Environ Microbiol 72(11):7098–7110. doi:10.1128/AEM. 00731-06
Toju H, Hosokawa T, Koga R, Nikoh N, Meng XY, Kimura N, Fukatsu T (2010) “Candidatus Curculioniphilus buchneri,” a novel clade of bacterial endocellular symbionts from weevils of the genus Curculio. Appl Environ Microbiol 76(1):275–282. doi:10.1128/AEM. 02154-09
Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW (2013) GenBank. Nucleic Acids Res 41:D36–D42. doi:10.1093/nar/gks1195
Jolley KA, Maiden MCJ (2010) BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinforma 11(1):595. doi:10.1186/1471-2105-11-595
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32(5):1792–1797. doi:10.1093/nar/gkh340
Gouy M, Guindon S, Gascuel O (2010) SeaView Version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 27(2):221–224. doi:10.1093/molbev/msp259
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59(3):307–321. doi:10.1093/sysbio/syq010
Baldo L, Bordenstein S, Wernegreen JJ, Werren JH (2006) Widespread recombination throughout Wolbachia genomes. Mol Biol Evol 23(2):437–449. doi:10.1093/molbev/msj049
Lefevre C, Charles H, Vallier A, Delobel B, Farrell B, Heddi A (2004) Endosymbiont phylogenesis in the Dryophthoridae weevils: evidence for bacterial replacement. Mol Biol Evol 21(6):965–973. doi:10.1093/molbev/msh063
Kuriwada T, Hosokawa T, Kumano N, Shiromoto K, Haraguchi D, Fukatsu T (2010) Biological role of Nardonella endosymbiont in its weevil host. PLoS ONE 5(10):e13101. doi:10.1371/journal.pone.0013101
Bian GW, Xu Y, Lu P, Xie Y, Xi ZY (2010) The endosymbiotic bacterium Wolbachia induces resistance to Dengue virus in Aedes aegypti. PLoS Pathog 6(4):e1000833. doi:10.1371/journal.ppat.1000833
Perlman SJ, Hunter MS, Zchori-Fein E (2006) The emerging diversity of Rickettsia. Proc R Soc B Biol Sci 273(1598):2097–2106. doi:10.1098/rspb.2006.3541
White JA (2011) Caught in the act: rapid, symbiont-driven evolution. Bioessays 33(11):823–829. doi:10.1002/bies.201100095
Dobson SL, Bourtzis K, Braig HR, Jones BF, Zhou WG, Rousset F, O’Neill SL (1999) Wolbachia infections are distributed throughout insect somatic and germ line tissues. Insect Biochem Mol 29(2):153–160. doi:10.1016/s0965-1748(98)00119-2
Gottlieb Y, Ghanim M, Chiel E, Gerling D, Portnoy V, Steinberg S, Tzuri G, Horowitz AR, Belausov E, Mozes-Daube N, Kontsedalov S, Gershon M, Gal S, KatZir N, Zchori-Fein E (2006) Identification and localization of a Rickettsia sp in Bemisia tabaci (Homoptera : Aleyrodidae). Appl Environ Microbiol 72(5):3646–3652. doi:10.1128/aem. 72.5.3646-3652.2006
Loan C, Holdaway F (1961) Microctonus aethiops (Nees) auctt. and Perilitus rutilus (Nees)(Hymenoptera: Braconidae), European parasites of Sitona weevils (Coleoptera: Curculionidae). Can Entomol 93(12):1057–1079. doi:10.4039/Ent931057-12
Gerard P, Eden T, Wilson D, Burch G (2008) Distribution of the clover root weevil biocontrol agent in the North Island of New Zealand. N Z Plant Protect 60:24–30
McNeill M, Proffitt J, Gerard P, Goldson S (2006) Collections of Microctonus aethiopoides Loan (Hymenoptera: Braconidae) from Ireland. N Z Plant Protect 59:290
Phillips C, Iline I, Vink C, Winder L, McNeill M (2006) Methods to distinguish between the Microctonus aethiopoides strains that parasitise Sitona lepidus and Sitona discoideus. N Z Plant Protect 59:297
Rouchet R, Vorburger C (2012) Strong specificity in the interaction between parasitoids and symbiont-protected hosts. J Evol Biol 25(11):2369–2375. doi:10.1111/j.1420-9101.2012.02608.x
Rouchet R, Vorburger C (2014) Experimental evolution of parasitoid infectivity on symbiont-protected hosts leads to the emergence of genotype specificity. Evolution 68(6):1607–1616. doi:10.1111/evo.12377
Hurst GDD, von der Schulenburg JHG, Majerus TMO, Bertrand D, Zakharov IA, Baungaard J, Volkl W, Stouthamer R, Majerus MEN (1999) Invasion of one insect species, Adalia bipunctata, by two different male-killing bacteria. Insect Mol Biol 8(1):133–139. doi:10.1046/j.1365-2583.1999.810133.x
Regassa LB, Gasparich GE (2006) Spiroplasmas: evolutionary relationships and biodiversity. Front Biosci 11:2983–3002. doi:10.2741/2027
de Vries EJ, Jacobs G, Breeuwer JAJ (2001) Growth and transmission of gut bacteria in the western flower thrips, Frankliniella occidentalis. J Invert Pathol 77(2):129–137. doi:10.1006/jipa.2001.5010
Morris CE, Monier JM (2003) The ecological significance of biofilm formation by plant-associated bacteria. Annu Rev Phytopathol 41:429–453. doi:10.1146/annurev.phyto.41.022103.134521
Barash I, Manulis-Sasson S (2009) Recent evolution of bacterial pathogens: the gall-forming Pantoea agglomerans case. Annu Rev Phytopathol 47:133–152. doi:10.1146/annurev-phyto-080508-081803
Moro CV, Tran FH, Raharimalala FN, Ravelonandro P, Mavingui P (2013) Diversity of culturable bacteria including Pantoea in wild mosquito Aedes albopictus. BMC Microbiol 13(1):70. doi:10.1186/1471-2180-13-70
Kikuchi Y, Hayatsu M, Hosokawa T, Nagayama A, Tago K, Fukatsu T (2012) Symbiont-mediated insecticide resistance. Proc Natl Acad Sci U S A 109(22):8618–8622. doi:10.1073/pnas.1200231109
de Vries EJ, Jacobs G, Sabelis MW, Menken SBJ, Breeuwer JAJ (2004) Diet-dependent effects of gut bacteria on their insect host: the symbiosis of Erwinia sp. and western flower thrips. Proc R Soc B Biol Sci 271(1553):2171–2178. doi:10.1098/rspb.2004.2817
Conord C, Despres L, Vallier A, Balmand S, Miquel C, Zundel S, Lemperiere G, Heddi A (2008) Long-term evolutionary stability of bacterial endosymbiosis in curculionoidea: additional evidence of symbiont replacement in the dryophthoridae family. Mol Biol Evol 25(5):859–868. doi:10.1093/molbev/msn027
Buchner P (1965) Endosymbiosis of animals with plant microorganisms. Interscience, New York
Nardon P, Grenier A-M (1991) Serial endosymbiosis theory and weevil evolution: the role of symbiosis. In: Margulis L, Fester R (eds) Symbiosis as a source of evolutionary innovation: speciation and morphogenesis. MIT Press, Cambridge, pp 153–169
Heddi A, Nardon P (2005) Sitophilus oryzae L.: a model for intracellular symbiosis in the Dryophthoridae weevils (Coleoptera). Symbiosis 39(1):1–11
Moran NA, McCutcheon JP, Nakabachi A (2008) Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet 42:165–190. doi:10.1146/annurev.genet.41.110306.130119
de Castro AJV, Alonso-Zarazaga MA, Outerelo R (2007) Systematics of Sitonini (Coleoptera : Curculionidae : Entiminae), with a hypothesis on the evolution of feeding habits. Syst Entomol 32(2):312–331. doi:10.1111/j.1365-3113.2006.00368.x
McKenna DD, Sequeira AS, Marvaldi AE, Farrell BD (2009) Temporal lags and overlap in the diversification of weevils and flowering plants. Proc Natl Acad Sci U S A 106(17):7083–7088. doi:10.1073/pnas.0810618106
Acknowledgments
We thank A. Dehnel and A. Maldonado for technical support, and three anonymous reviewers for helpful commentary on earlier versions of the manuscript. This research was funded by the University of Kentucky Department of Entomology, USDA National Institute of Food and Agriculture, Hatch project KY008052, the New Zealand Ministry for Business, Innovation and Employment through contract LINX0304, Ecosystems Bioprotection, and the AgResearch Curiosity Fund (A18906).
Author information
Authors and Affiliations
Corresponding author
Additional information
The nucleotide sequence data reported in this manuscript have been submitted to the DDBJ/EMBL/GenBank databases under accession numbers KJ494864-8, KJ522437-49, and the sequence read archive under project accession number SRP041582; all have been publicly released as of 31 Aug, 2014.
Rights and permissions
About this article
Cite this article
White, J.A., Richards, N.K., Laugraud, A. et al. Endosymbiotic Candidates for Parasitoid Defense in Exotic and Native New Zealand Weevils. Microb Ecol 70, 274–286 (2015). https://doi.org/10.1007/s00248-014-0561-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-014-0561-8