Abstract
Objectives
This study aimed to screen, characterize, and annotate the genome along with the comparison of GABA synthesis genes presented in lactic acid bacteria (LAB).
Results
Thirty-five LAB isolates from fermented foods were screened for GABA production using thin-layer chromatography (TLC). Fifteen isolates produced GABA ranging from 0.07 to 22.94 g/L. Based on their GTG5 profiles, phenotypic, and genotypic characteristics, isolates LSI1-1, LSI1-5, LSI2-1, LSI2-2, LSI2-3, LSI2-5, and LSM3-1-4 were identified as Lactobacillus plantarum subsp. plantarum; isolate LSM1-4 was Lactobacillus argentoratensis; isolates CAB1-2, CAB1-5, CAB1-7, and LSI1-4 were Lactobacillus pentosus; and CAB1-1, LSM3-1-1 and LSM3-2-3 were Lactobacillus fermentum. Strains LSI2-1 and CAB1-7 from pickled vegetables were selected for genome analysis. The gadA gene (1410 bp, 470aa) was encountered in GABA production of both strains and no other glutamate decarboxylase (GAD) genes were found in the genomes when compared with other LAB strains. The presence of gadA is evidence for GABA production. Strains LSI2-1 and CAB1-7 produced 22.94 g/L and 11.59 g/L of GABA in GYP broth supplemented with 3% (w/v) MSG at 30 °C for 72 h, respectively.
Conclusions
Our report highlights the characterization of LAB and GABA production of L. plantarum LSI2-1 strain with its GABA synthesis gene.
Graphic abstract
GABA production of strains LSI2-1 and CAB1-7 in GYP broth with 3% (w/v) MSG and comparative GAD genes

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gamma-aminobutyric acid (GABA) is the major inhibitory neurotransmitter in the sympathetic nervous system (Mody et al. 1994) for controlling stress (Oh and Oh 2003), preventing Alzheimer’s disease (Nakashima et al. 2009), and reducing depression (Okada et al. 2000). It also plays a role in cardiovascular function (De Feudis 1983) and ameliorates diabetic conditions (Hagiwara et al. 2004). In addition, GABA has positive benefits for human health including treatment for diuretic and convulsive effects (Wong et al. 2003), sleeplessness (Okada et al. 2000) and chronic alcohol-related symptoms, while stimulating the immune system and hormone production (Oh and Oh 2003). GABA is synthesized from glutamate decarboxylase (GAD; EC 4.1.1.15), with pyridoxal 5′ phosphate (PLP) as a coenzyme that catalyzes the irreversible decarboxylation of l-glutamate into GABA (Ueno 2000). The GAD enzyme encoded by gadA or gadB is responsible for the synthesis of GABA in microbial cells, and the decarboxylated product is exported from the cell by an antiporter (Small and Waterman 1998).
Lactic acid bacteria (LAB) are used to produce GABA because they are considered safe and exhibit several health-promoting properties (Nejati et al. 2013). Strains of Lactobacillus brevis from goat cheese (Li et al. 2013), L. senmaizukei from traditional pickles (Hiraga et al. 2008), L. plantalum from Malaysian fermented foods (Zareian et al. 2015), L. rhamnosus from Chinese traditional food (Lin 2013), L. paracasei from Italian cheese and Japanese fermented fish (Komatsuzaki et al. 2005), and L. namurensis from Nham (Thai fermented pork sausage) (Ratanaburee et al. 2013) were all reported to produce GABA.
Different GABA production mechanisms can be explained by genome data of the cellular physiology of each bacterial species. Most reports on GABA production involved L. brevis strains, with little known about the genomes of L. pantarum and L. pentosus strains. Therefore, this study focused on screening, characterization, and gene description by comparing genome sequences of GABA-producing strains and the GAD systems of related Lactobacillus strains.
Materials and methods
Isolation of lactic acid bacteria (LAB)
Six Thai fermented food samples were collected from local markets including pickled Brassica oleracea from Bangkok, pickled Lasia spinosa (L.) Thwaites (Phak-nam dong) from Singburi Province, pickled Crateva adansonii subsp. trifoliata Roxb. (Phak-koom-dong) from Singburi Province, and fermented pork and pickled Allium ascalonicum (Hom-dong) from Samut Prakarn Province, Thailand (Table 1). Two hundred microliters or 1 g of sample were enriched in 5 mL of Man Rogosa Sharpe (MRS) broth (Difco) and incubated at 30 °C for 24 h. The culture broth was streaked on MRS agar plates with 0.3% (w/v) CaCO3 and cultivated at 30 °C for 24 h. Isolates that exhibited a clear zone were selected to purify on MRS with 0.3% (w/v) CaCO3 agar plates. The selected isolates were preserved in a freezer and by lyophilization until required for further study.
Screening and determination of GABA production
Active cultures were prepared by transfer from glycerol stock into MRS broth and incubated at 30 °C for 24 h. Glucose-yeast extract-peptone (GYP) broth supplemented with 3% (w/v) monosodium glutamate (MSG) (Lyu et al. 2018) was used as the production medium. A culture of each strain (10% v/v inoculum, cultivated at the same condition for 18 h) was inoculated in production medium and cultivated at 30 °C for 72 h. The culture broth was centrifuged at 10,000 rpm for 5 min and GABA production was analyzed qualitatively using thin-layer chromatography (TLC). The supernatant 2 µL was spotted onto TLC plates (aluminum TLC plates silica gel 60 F254 plates, Merck). The plates were developed in a mobile phase including acetic acid: n-butanol: distilled water (5:3:2) and subsequently sprayed with 0.4% (w/v) ninhydrin dissolved in ethanol. The plates were heated for several minutes until GABA spots appeared (Villegas et al. 2016).
GABA concentration was determined quantitatively by high-performance liquid chromatography (HPLC) following Silva et al. (2009) with slight modifications. Briefly, 400 µL of sample and 80 µL of 5 mg/mL ortho-phthaldialdehyde were dissolved in absolute methanol, 300 µL of 0.1 M borate buffer and 20 µL mercaptopropionic acid. The solution was mixed immediately, and left to stand at room temperature for 1 min before injection into the equipment as described by Kutlan and Molnar-Perl (2003). The HPLC instrument consisted of a Varian Prostar (Granite Quarry, NC, USA) with a 3 µm particle size (150 mm × 4.6 mm, ID), Luna 5U C18(2) column (Hibar-Futigsanle RT). The experiment was conducted at 25 °C and eluted with a mobile phase system of 85% (v/v) solution A [0.05 M sodium acetate, tetrahydrofuran, and methanol (50:1:49, v/v/v)] and 15% (v/v) solution B (methanol) at a flow rate of 1 mL/min. A fluorescence detector model 363 was set at an excitation wavelength of 337 nm and an emission wavelength of 454 nm.
Phenotypic characteristics
Morphological and cultural characteristics including Gram reaction, cell morphology, and colonial appearance of the isolates were determined after culturing the cells on MRS agar at 30 °C for 24 h. Physiological and biochemical characteristics consisted of growth at different parameters of pH (3, 6, and 9), temperatures (15, 30, and 45 °C), and NaCl concentrations (2, 4, 6, and 8% w/v). Catalase activity, nitrate reduction, gas production, arginine hydrolysis, and acid production from carbohydrates were performed as previously described (Tanasupawat et al. 1998).
Genotypic characteristics
The 16S rRNA gene sequences of isolates were PCR amplified as described by Phuengjayaem et al. (2017). Amplified 16S rRNA gene sequences were sequenced on a DNA sequencer (Macrogen, Korea) using universal primers as described earlier (Lane 1991). Sequence similarity values between the isolates and related reference strains were evaluated on the EzBiocloud server (Yoon et al. 2017). A phylogenetic tree based on the neighbor-joining (NJ) method (Saitou and Nei 1987) and GAD genes tree based on the Maximum-Likelihood were constructed using MEGA 7 (Kumar et al. 2016). Bootstrap analysis was performed to determine the confidence values of individual branches in the phylogenetic tree with 1000 replications (Felsenstein 1985). Identified sequences were deposited in DDBJ (DNA Data Bank of Japan, Mishima, Japan).
Crude DNA of isolates was extracted from cell pellets using the alkaline-polyethylene glycol-based method (Alkaline-PEG) (Chomczynski and Rymaszewski 2006). A DNA fingerprint was also performed based on the (GTG)5-PCR method. PCR amplification, gel electrophoresis, and analysis methods were conducted as previously described (Tolieng et al. 2018). L. plantarum subsp. plantarum NRIC 1067T and L. pentosus NRIC 1069T were used as reference strains.
Genome sequencing and assembly
Genomic DNA of representative strains was extracted by a phenol–chloroform method and purified according to the procedure of Tamaoka (1994). The genomes of each strain were sequenced with an Illumina Miseq platform (Illumina, Inc., San Diego, US-CA) using 2 × 250 bp paired-end reads assembled to contig using SPAdes 3.12 (Bankevich et al. 2012). Comparative genomic analyses were annotated using the RAST server (Aziz et al. 2008) in accordance with the NCBI prokaryotic genome annotation pipeline (PGAP) and performed for comparison using the SEED Viewer (Aziz et al. 2012). Draft genomes of the strains were publicly available from GenBank, and nucleotide data of closely related strains were obtained from the GenBank database.
Statistical analysis
All experiments were performed in triplicate, with data presented as mean ± standard deviation (SD). The statistical package for the social sciences (SPSS) for Windows program version 15 was used for data analysis, with one-way ANOVA utilized for comparison of each factor and Tukey’s multiple range test for pairwise comparison at p-value of 0.05.
Results and discussion
Isolation, screening, and determination of GABA production
Thirty-five isolates from fermented food were qualitatively screened for GABA production using the TLC method. Fifteen isolates including CAB1-1, CAB1-2, CAB1-5, CAB1-7, LSI1-1, LSI1-4, LSI1-5, LSI2-1, LSI2-2, LSI2-3, LSI2-5, LSM1-4, LSM3-1-1, LSM3-1-4, and LSM3-2-3 exhibited positive spots at Rf 0.57 compared with the spot of GABA standard (10 g/L). All isolates were quantitatively determined for GABA production using HPLC. The GABA standard curves showed a linear relationship (R2 = 0.991) between peak areas and GABA concentrations ranging from 2 to 16 mg/L (Supplementary Fig. S1). MSG and GABA chromatogram of HPLC are presented in Supplementary Fig. S2. Based on the statistical analysis results, different capabilities of GABA production by the 15 isolates were divided into five groups (Supplementary Tables S1 and S2). In the first group, isolates LSI2-1 and LSI2-3, exhibited GABA production of 22.94 and 17.45 g/L, respectively (Fig. 1). An increase in GABA production resulted in a decrease of MSG concentration. The second group, LSI2-3, CAB1-7, LSI1-1, LSI2-2, and LSI2-5 gave 17.45, 11.59, 10.77, 10.43, and 7.89 g/L of GABA, respectively. The remaining eight isolates produced GABA ranging from 0.07 to 7.22 g/L with high remaining MSG (Fig. 1). From previous reports, L. brevis strains IFO-12005 (Yokoyama et al. 2002) and HYE1 (Lim et al. 2017) produced GABA at 1.05 and 2.21 g/L respectively, while L. rhamnosus strain GG (Song and Yu 2018) and YS9 (Lin 2013), L. fermentum W1 (Rayavarapu et al. 2019), L. plantarum Taj-Apis 362 (Tajabadi et al. 2015) and Lactococcus lactis B (Lu et al. 2009) produced 1.13, 19.28, 5.34, 0.74, and 7.20 g/L of GABA, respectively. Here, strains LSI2-1, LSI2-3 displayed beneficial GABA production with no significant difference in quantity. Multiple comparison analysis using Tukey’s method at α 0.05 showed that isolates LSI2-1 and LSI2-3 (p-value 0.749) were classified in the same group (Supplementary Tables S1 and S2). Isolate LSI2-1 produced maximum GABA concentration at a higher efficiency rate.
Identification of isolates
All isolates consisted of catalase-negative rod-shaped bacteria and formed milky-white, circular, convex, and opaque colonies. The isolates produced acid from arabinose, fructose, glucose, maltose, ribose, sucrose, and trehalose with growth at 15–37 °C, pH 3–9 and 4–8% (w/v) NaCl. Their phenotypic characteristics are listed in Table 1.
The (GTG)5-PCR analysis was used to detect intraspecific variability of the strains. The number of detectable amplicons varied from 10 to 20, with DNA band size ranging from 300 to 10,000 bp. The UPGMA dendrogram relied on DNA fingerprinting using the (GTG)5 primer and was clustered into five groups (Fig. 2). Three isolates CAB1-2, CAB1-5, and CAB1-7 were clustered with L. pentosus NRIC 1069T, while isolates LSI2-1, LSI2-2, and LSI2-3 were clustered in the same group, and LSI1-1, LSI1-5 and LSI2-5 were clustered with L. plantarum NRIC 1067T. The profile of CAB1-1 was the same as LSM3-1-1 and LSM3-2-3, while isolates LSI1-4, LSM1-4, and LSM3-1-4 exhibited unique profiles.
On the basis of their phenotypic characteristics (Table 1), DNA fingerprinting pattern (Fig. 2), and the results of 16S rRNA gene sequences (Fig. 3) the isolates were divided into four groups as described below.
Group I consisted of seven isolates including LSI1-1, LSI1-5, LSI2-1, LSI2-2, LSI2-3, LSI2-5, and LSM3-1-4. Strains LSI1-1 (1385 bp), LSI1-5 (1428 bp), LSI2-1 (1419 bp), LSI2-2 (1410 bp), and LSM3-1-4 (1401 bp) were closely related to L. plantarum subsp. plantarum ATCC 14917T with 100% sequence similarity, while LSI2-3 (1441 bp) and LSI2-5 (1412 bp) were also closely related to L. plantarum subsp. plantarum ATCC 14917T (Bringel et al. 2005) with 99.93 and 99.72% sequence similarity, respectively (Fig. 3). Therefore, they were identified as Lactobacillus plantarum subsp. plantarum.
Group II contained only the strain LSM1-4 (1361 bp) that was closely related to L. argentoratensis DSM 16365T (Bringel et al. 2005) with 99.93% sequence similarity. L. argentoratensis DSM 16365T was previously allocated to Lactobacillus plantarum subsp. argentoratensis (Bringel et al. 2005). In this study, strain LSM1-4 exhibited a 16S rRNA gene sequence (Fig. 3) similar to strains in Group I but showed independent difference in DNA fingerprinting (GTG)5 (Fig. 2) from Group I strains. Therefore, it was identified as Lactobacillus argentoratensis.
The 16S rRNA gene sequence of isolates LSI1-1, LSI1-5, LSI2-1, LSI2-2, LSI2-3, LSI2-5, LSM3-1-4, and LSM1-4 were deposited in the DDBJ Database as accession numbers LC616682, LC616683, LC546823, LC546824, LC546825, LC546826, LC616687, and LC620497, respectively.
Group III consisted of four isolates including CAB1-2, CAB1-5, CAB1-7, and LSI1-4. Strains CAB1-2 (1253 bp), CAB1-5 (1337 bp), CAB1-7 (1415 bp), and LSI1-4 (1358 bp) were closely related to L. pentosus DSM 20314T (Zanoni et al. 1987) with 99.52, 100, 99.58, and 99.63% sequence similarity, respectively (Fig. 3). Therefore, they were identified as Lactobacillus pentosus. The 16S rRNA gene sequences of isolates CAB1-2, CAB1-5, CAB1-7, and LSI1-4 were deposited in the DNA DDBJ Database under accession numbers LC616686, LC546821, LC546822, and LC616684, respectively.
Group IV contained three isolates including CAB1-1, LSM3-1-1, and LSM3-2-3. Strains CAB1-1 (1429 bp), LSM3-1-1 (1373 bp), and LSM3-2-3 (1432 bp) were closely related to L. fermentum CECT 562T (Beijerinck 1901) with 99.86, 99.71, and 99.93% sequence similarity, respectively (Fig. 3). Therefore, they were identified as Lactobacillus fermentum. Sequences of CAB1-1, LSM3-1-1, and LSM3-2-3 were deposited in the DDBJ Database under accession numbers LC616685, LC577832, and LC616688, respectively.
From the results of GABA production, phenotypic characteristics, the (GTG)5-PCR profile, and16S rRNA gene sequences. LSI2-1 was selected as a representative strain of the L. plantarum group for genome analysis because it showed maximum efficiency of GABA production, while CAB1-7 was selected as a representative strain of the L. pentosus group to compare the genes based on genome sequencing analysis.
Genome annotation
Draft genome sequences of strains LSI2-1 and CAB1-7 were deposited in GenBank with accession numbers VSFJ00000000 and VSFK00000000, respectively (A genome circular maps are shown in supplementary Fig. S3). Genome features of strain LSI2-1 included 3,222,735 bp with GC content of 44.5 mol%, 32 contigs with protein-encoding genes (PEGs), 336 subsystems, 3087 coding sequences, and 64 RNAs, while the genome features of strain CAB1-7 were 3,201,382 bp with GC content of 44.6 mol%, 74 contigs with PEGs, 332 subsystems, 3055 coding sequences, and 69 RNAs. The gadA gene of LSI2-1 (470aa) was coding sequences (CDS) with no subsystem, 1410 bp (676,377–674,968 bp), and gadA (470 aa) of strain CAB1-7 was CDS with no subsystem. The size of gadA was 1410 bp located from 54,511 to 55,920 bp. The GAD gene of LSI2-1 was closely related to the GAD gene of L. plantarum subsp. plantarum ATCC 14917T with 100% similarity while the GAD gene of CAB1-7 was closely related to the GAD gene of L. plantarum subsp. plantarum ATCC 14917T with 99.79% similarity.
Comparative genomic analysis
Comparison of the GAD gene between strains LSI2-1, CAB1-7, and other Lactobacillus strains of 20 genomic sequencings using a maximum-likelihood tree indicated that both strains were closely related to L. plantarum in one branch (Fig. 4a). The gadA gene (No. 3 in Fig. 4b) of LSI2-1 and CAB1-7 was located between phosphoenolpyruvate carboxykinase gene (No.1 in Fig. 4b), hypothetical protein gene (No. 2 in Fig. 4b) and gamma-d-glutamate-meso-diaminopimelate muropeptidase gene (No.4 in Fig. 4b) the same as L. plantarum WCFS1 (Kleerebezem et al. 2003). However, only strain LSI2-1 had no hypothetical protein gene (No. 2 in Fig. 4b). Both gadA and gadB were found in the GAD gene of L. brevis 367T. The gadB gene was located next to the gadC gene (Fig. 4b) and the whole operon including gts and gadR, while gadA was located separately from the other gad genes (Lyu et al. 2018). The L. brevis strain was reported to show potential for GABA production based on gadA (Wu et al. 2017) while Lc. lactis subsp. lactis B produced maximum GABA of 7.2 g/L from 15 g/L MSG at 31.8 °C, pH 7.1 (Lu et al. 2009). By contrast, the gadA gene was absent in Lc. lactis subsp. lactis ll1403, but gadB and gadC were present. The GAD genes consisted of two isoforms: gadA or gadB, that exhibited similar functions to convert l-glutamate to GABA. Moreover, both isoforms of the GAD gene were biochemically indistinguishable (De Biase et al. 1996). Only gadA was found in strains LSI2-1and CAB1-7; however, they could produce GABA. Efficiency of GABA production depended on the levels of expression of the GAD gene and other enzymes involved in the glutamate pathway. Histone-like nucleoid-structuring (H-NS) depressed gad expression during the log phase, whereas RNA polymerase sigma (rpoS) relieved H-NS repression during the stationary phase; directly or indirectly accounting for transcription of GAD genes (De Biase et al. 1999). The rpoS gene, which induced gadA expression, was found in LSI2-1 and CAB1-7, whereas H-NS was absent. Temperature, pH, medium composition, and other factors also impacted GABA production. Therefore, expression of the gadA gene, enzyme activity and production conditions should be determined to enhance GABA production efficiency.
Comparison of GAD genes in strains CAB1-7 and LSI2-1 and related strains. a Maximum-likelihood tree based on amino acid sequences of GAD genes. Numbers on the branches indicate percentage bootstrap values of 1000 replicates. Accession number and length of GAD gene are shown in parentheses. b GAD genes of LSI2-1, CAB1-7 and related strains. 1, phosphoenolpyruvate carboxykinase gene; 2, hypothetical protein gene; 3, gadA gene; 4, gamma-d-glutamate-meso-diaminopimelate muropeptidase gene
Conclusions
Our phenotypic, genotypic and genome analysis results demonstrated that LAB isolated from fermented food as strains LSI2-1 and CAB1-7 showed capability of GABA biosynthesis at 22.94 g/L and 11.59 g/L, respectively. Strains LSI2-1 and CAB1-7 were identified as L. plantarum subsp. plantalum and L. pentosus based on their 16S rRNA gene sequences. The gadA gene was found in both strains as functionally evident GAD-encoding ability. The GABA optimization of strains LSI2-1 and CAB1-7 and their application in fermented food should be investigated in future studies.
References
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA et al (2008) The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:1–15. https://doi.org/10.1186/1471-2164-9-75
Aziz RK, Devoid S, Disz T, Edwards RA, Henry CS, Olsen GJ et al (2012) SEED Servers: high-performance access to the seed genomes, annotations, and metabolic models. PLoS ONE 7:e48053. https://doi.org/10.1371/journal.pone.0048053
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS et al (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. https://doi.org/10.1089/cmb.2012.0021
Beijerinck MW (1901) Sur les ferments lactiques de l’industrie. Arch Neerl Sci Exactes Nat (Sect 2) 6:212–243
Bringel F, Castioni A, Olukoya DK, Felis GE, Torriani S, Dellaglio F (2005) Lactobacillus plantarum subsp argentoratensis subsp. nov., isolated from vegetable matrices. Int J Syst Evol Micr 55:1629–1634. https://doi.org/10.1099/ijs.0.63333-0
Chomczynski P, Rymaszewski M (2006) Alkaline polyethylene glycol-based method for direct PCR from bacteria, eukaryotic tissue samples, and whole blood. Biotechniques 40:454–458. https://doi.org/10.2144/000112149
De Biase D, Tramonti A, John RA, Bossa F (1996) Isolation, overexpression, and biochemical characterization of the two isoforms of glutamic acid decarboxylase from Escherichia coli. Protein Expr Purif 8:430–438. https://doi.org/10.1006/prep.1996.0121
De Biase D, Tramonti A, Bossa F, Visca P (1999) The response to stationary-phase stress conditions in Escherichia coli: role and regulation of the glutamic acid decarboxylase system. Mol Microbiol 32:1198–1211. https://doi.org/10.1046/j.1365-2958.1999.01430.x
DeFeudis FV (1983) gamma-Aminobutyric acid and cardiovascular function. Experientia 39:845–849. https://doi.org/10.1007/BF01990401
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. https://doi.org/10.1111/j.1558-5646.1985.tb00420.x
Hagiwara H, Seki T, Ariga T (2004) The effect of pre-germinated brown rice intake on blood glucose and PAI-1 levels in streptozotocin-induced diabetic rats. Biosci Biotechnol Biochem 68:444–447. https://doi.org/10.1271/bbb.68.444
Hiraga K, Ueno Y, Sukontasing S, Tanasupawat S, Oda K (2008) Lactobacillus senmaizukei sp. nov., isolated from Japanese pickle. Int J Syst Evol Microbiol 58:1625–1629. https://doi.org/10.1099/ijs.0.65677-0
Kleerebezem M, Boekhorst J, van Kranenburg R, Molenaar D, Kuipers OP, Leer R et al (2003) Complete genome sequence of Lactobacillus plantarum WCFS1. Proc Natl Acad Sci USA 100:1990–1995. https://doi.org/10.1073/pnas.0337704100
Komatsuzaki N, Shima J, Kawamoto S, Momose H, Kimura T (2005) Production of gamma-aminobutyric acid (GABA) by Lactobacillus paracasei isolated from traditional fermented foods. Food Microbiol 22:497–504. https://doi.org/10.1016/j.fm.2005.01.002
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
Kutlan D, Molnar-Perl I (2003) New aspects of the simultaneous analysis of amino acids and amines as their o-phthaldialdehyde derivatives by high-performance liquid chromatography analysis of wine, beer and vinegar. J Chromatogr A 987:311–322. https://doi.org/10.1016/S0021-9673(02)01538-8
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. Wiley, New York, pp 115–175
Li H, Li W, Liu X, Cao Y (2013) gadA gene locus in Lactobacillus brevis NCL912 and its expression during fed-batch fermentation. FEMS Microbiol Lett 349:108–116. https://doi.org/10.1111/1574-6968.12301
Lim HS, Cha IT, Roh SW, Shin HH, Seo MJ (2017) Enhanced production of gamma-aminobutyric acid by optimizing culture conditions of Lactobacillus brevis HYE1 isolated from kimchi, a Korean fermented food. J Microbiol Biotechnol 27:450–459. https://doi.org/10.4014/jmb.1610.10008
Lin Q (2013) Submerged fermentation of Lactobacillus rhamnosus YS9 for gamma-aminobutyric acid (GABA) production. Braz J Microbiol 44:183–187. https://doi.org/10.1590/S1517-83822013000100028
Lu XX, Xie CY, Gu ZX (2009) Optimisation of fermentative parameters for gaba enrichment by Lactococcus lactis. Czech J Food Sci 27:433–442. https://doi.org/10.17221/45/2009-CJFS
Lyu C, Zhao W, Peng C, Hu S, Fang H, Hua Y et al (2018) Exploring the contributions of two glutamate decarboxylase isozymes in Lactobacillus brevis to acid resistance and γ-aminobutyric acid production. Microb Cell Fact 17:1–14. https://doi.org/10.1186/s12934-018-1029-1
Mody I, De Koninck Y, Otis TS, Soltesz I (1994) Bridging the cleft at gaba synapses in the brain. Trends Neurosci 17:517–525. https://doi.org/10.1016/0166-2236(94)90155-4
Nakashima Y, Ohsawa I, Konishi F, Hasegawa T, Kumamoto S, Suzuki Y et al (2009) Preventive effects of Chlorella on cognitive decline in age-dependent dementia model mice. Neurosci Lett 464:193–198. https://doi.org/10.1016/j.neulet.2009.08.044
Nejati F, Rizzello CG, Di Cagno R, Sheikh-Zeinoddin M, Diviccaro A, Minervini F et al (2013) Manufacture of a functional fermented milk enriched of Angiotensin-I converting enzyme (ACE)-inhibitory peptides and gamma-amino butyric acid (GABA). Lwt Food Sci Technol 51:183–189. https://doi.org/10.1016/j.lwt.2012.09.017
Oh CH, Oh SH (2003) Brown rice extracts with enhanced levels of gamma-aminobutyric acid inhibit cancer cell proliferation. FASEB J 17:A1157–A1157
Okada T, Sugishita T, Murakami T, Murai H, Saikusa T, Horino T et al (2000) Effect of the defatted rice germ enriched with GABA for sleeplessness, depression, autonomic disorder by oral administration. J Jpn Soc Food Sci 47:596–603. https://doi.org/10.3136/nskkk.47.596
Phuengjayaem S, Phinkian N, Tanasupawat S, Teeradakorn S (2017) Diversity and succinic acid production of lactic acid bacteria isolated from animals, soils and tree barks. Res J Microbiol 12:177–186. https://doi.org/10.3923/jm.2017.177.186
Ratanaburee A, Kantachote D, Charernjiratrakul W, Sukhoom A (2013) Enhancement of gamma-aminobutyric acid (GABA) in Nham (Thai fermented pork sausage) using starter cultures of Lactobacillus namurensis NH2 and Pediococcus pentosaceus HN8. Int J Food Microbiol 167:170–176. https://doi.org/10.1016/j.ijfoodmicro.2013.09.014
Rayavarapu B, Tallapragada P, Usha MS (2019) Statistical optimization of gamma-aminobutyric acid production by response surface methodology and artificial neural network models using Lactobacillus fermentum isolated from palm wine. Biocatal Agric Biotechnol 22:101362. https://doi.org/10.1016/j.bcab.2019.101362
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Silva DMD, Ferraz VP, Ribeiro AM (2009) Improved high-performance liquid chromatographic method for GABA and glutamate determination in regions of the rodent brain. J Neurosci Methods 177:289–293. https://doi.org/10.1016/j.jneumeth.2008.10.011
Small PL, Waterman SR (1998) Acid stress, anaerobiosis and gadCB: lessons from Lactococcus lactis and Escherichia coli. Trends Microbiol 6:214–216. https://doi.org/10.1016/s0966-842x(98)01285-2
Song HY, Yu RC (2018) Optimization of culture conditions for gamma-aminobutyric acid production in fermented adzuki bean milk. J Food Drug Anal 26:74–81. https://doi.org/10.1016/j.jfda.2016.11.024
Tajabadi N, Ebrahimpour A, Baradaran A, Rahim RA, Mahyudin NA, Manap MYA et al (2015) Optimization of gamma-aminobutyric acid production by Lactobacillus plantarum Taj-Apis362 from honeybees. Molecules 20:6654–6669. https://doi.org/10.3390/molecules20046654
Tamaoka J (1994) Determination of DNA base composition. In: Goodfellow M, O’Donnell AG (eds) Chemical methods in prokaryotic systematics. Wiley, Chichester, pp 463–470
Tanasupawat S, Okada S, Komagata K (1998) Lactic acid bacteria found in fermented fish in Thailand. J Gen Appl Microbiol 44:193–200. https://doi.org/10.2323/jgam.44.193
Tolieng V, Booncharoen A, Nuhwa R, Thongchul N, Tanasupawat S (2018) Molecular identification, l-lactic acid production, and antibacterial activity of Bacillus strains isolated from soils. J Appl Pharm 8:098–105. https://doi.org/10.7324/JAPS.2018.81013
Ueno H (2000) Enzymatic and structural aspects on glutamate decarboxylase. J Mol Catal B 10:67–79. https://doi.org/10.1016/S1381-1177(00)00114-4
Villegas JM, Brown L, de Giori GS, Hebert EM (2016) Optimization of batch culture conditions for GABA production by Lactobacillus brevis CRL 1942, isolated from quinoa sourdough. Lwt Food Sci Technol 67:22–26. https://doi.org/10.1016/j.lwt.2015.11.027
Wong CG, Bottiglieri T, Snead OC 3rd (2003) GABA, gamma-hydroxybutyric acid, and neurological disease. Ann Neurol 54(S6):S3–S12. https://doi.org/10.1002/ana.10696
Wu Q, Tun HM, Law Y-S, Khafipour E, Shah NP (2017) Common distribution of gad operon in Lactobacillus brevis and its gadA contributes to efficient GABA synthesis toward cytosolic near-neutral pH. Front Microbiol 8:206. https://doi.org/10.3389/fmicb.2017.00206
Yokoyama S, Hiramatsu J, Hayakawa K (2002) Production of gamma-aminobutyric acid from alcohol distillery lees by Lactobacillus brevis IFO-12005. J Biosci Bioeng 93:95–97. https://doi.org/10.1016/S1389-1723(02)80061-5
Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H et al (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol 67:1613–1617. https://doi.org/10.1099/ijsem.0.001755
Zanoni P, Farrow JAE, Phillips BA, Collins MD (1987) Lactobacillus pentosus (Fred, Peterson, and Anderson) sp. nov., nom. rev. Int J Syst Bacteriol 37:339–341. https://doi.org/10.1099/00207713-37-4-339
Zareian M, Oskoueian E, Forghani B, Ebrahimi M (2015) Production of a wheat-based fermented rice enriched with gamma-amino butyric acid using Lactobacillus plantarum MNZ and its antihypertensive effects in spontaneously hypertensive rats. J Funct Foods 16:194–203. https://doi.org/10.1016/j.jff.2015.04.015
Acknowledgements
We thank Dr. Nattakorn Kuncharoen for genome sequence analysis, and Mr. Engkarat Kingkaew and Ms. Saranporn Poothong for 16S rRNA gene sequence analysis.
Supplementary Information
Supplementary Fig. S1—Calibration curve of GABA concentration in the range of 0–16 μg/mL.
Supplementary Fig. S2—Chromatograms of MSG and GABA analysis using HPLC.
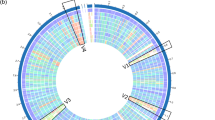
Supplementary Fig. S3—A genome circular maps of strain LSI2-1 (a) and CAB1-7 (b) created by the ‘circular viewer’ functionality implemented in the PATRIC web server.
Supplementary Table S1—ANOVA table showed the efficiency on GABA production of isolates.
Supplementary Table S2—The multiple comparisons of GABA production of isolates using Tukey’s multiple range tests.
Funding
This research was supported by a Grant for International Research Integration: Research Pyramid, Ratchadaphiseksomphot Endowment Fund (GCURP_58_01_33_01), and the Ratchadapiseksomphot Endowment Fund, Chulalongkorn University for a Postdoctoral Fellowship to Dr. Sukanya Phuengjayaem.
Author information
Authors and Affiliations
Contributions
SP performed the isolation, screening, identification, genome sequence analysis and writing the manuscript, AB contributed to the GTG5 profiles, and ST performed the identification and writing the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Phuengjayaem, S., Booncharoen, A. & Tanasupawat, S. Characterization and comparative genomic analysis of gamma-aminobutyric acid (GABA)-producing lactic acid bacteria from Thai fermented foods. Biotechnol Lett 43, 1637–1648 (2021). https://doi.org/10.1007/s10529-021-03140-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-021-03140-y