Abstract
Objectives
To construct an Escherichia coli strain capable of producing riboflavin with high titer and yield.
Results
A low copy number plasmid pLS01 containing a riboflavin operon under the control of a constitutive promoter was constructed and introduced into Escherichia coli MG1655. Subsequently, the pfkA, edd and ead genes were disrupted, and the resulting strain LS02T produced 667 mg riboflavin/l in MSY medium supplied with 10 g glucose/l in flask cultivation. In a fed-batch process, riboflavin production of the strain reached 10.4 g/l with a yield of 56.8 mg riboflavin/g glucose.
Conclusion
To our knowledge, this is the first report of engineered E. coli strains that can produce more than 10 g riboflavin/l in fed-batch cultivation, indicating that E. coli has potential for riboflavin production.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Riboflavin is the precursor of coenzymes flavin mononucleotide (FMN) and flavin adenine dinucleotide (FAD), both of which act as oxidation–reduction cofactors involved in a wide range of biological reactions. It has been widely applied in many fields such as pharmaceuticals and cosmetics, as well as human and animal nutrition. Riboflavin is commercially produced mainly by biological processes, which have many advantages such as lower costs, less waste and lower energy consumption compared to chemical manufacturing processes.
A number of riboflavin overproducers have been reported, including fungi such as Ashbya gossypii (Buey et al. 2015) and Candida famata (Dmytruk et al. 2014), as well as bacteria (Duan et al. 2010; Stahmann et al. 2000). These overproducers have been developed using classical mutagenesis and/or rational metabolic engineering, including down-regulation of flavokinase/FAD synthetase activity (Mack et al. 1998), overexpression of the riboflavin operon involved in riboflavin synthesis from GTP and ribulose 5-phosphate (Marx et al. 2008), deregulation of purine biosynthesis (Shi et al. 2009; Xu et al. 2015), and engineering central pathways such as the gluconeogenesis pathway and pentose phosphate pathway (Wang et al. 2014, 2011).
In our previous work, we have engineered E. coli for riboflavin production (Lin et al. 2014; Xu et al. 2015). One strain, RF05S-M40, produced 2.7 g riboflavin/l in flask cultivation with a yield of 0.13 g riboflavin/g glucose (Lin et al. 2014). However, this strain has a number of disadvantages which hinder its further application in riboflavin production. Firstly, it requires IPTG to induce the expression of the riboflavin operon, which is unfavorable for industrial processes. Secondly, loss of the riboflavin operon expression plasmid, p20C-EC10, caused the strain to be unstable in a fed-batch process. In addition, extra glycine is needed in the cultivation medium, which increases the producing cost to some extent. To resolve these problems, we have reconstructed a riboflavin-producing E. coli strain, in which pfkA, edd and eda genes were deleted to enhance the flux through the PP pathway. Additionally, the strain harbors an riboflavin operon expression plasmid with higher stability. In fed-batch cultivation, the engineered strain produced more than 10 g riboflavin/l with a yield of 56.8 mg riboflavin/g glucose.
Materials and methods
Strains, media and cultivation conditions
Strains used in this study are listed in Table 1. E. coli K-12 MG1655 was used to engineer riboflavin-producing strains. E. coli K12 DH5α was selected as the host to propagate vector DNA, and all E. coli cells were cultured at 30 or 37 °C in lysogeny broth (LB) (10 g tryptone/l, 5 g yeast extract/l, 10 g NaCl/l) with the addition of antibiotics. Three media were used for riboflavin production in shake-flasks: LBG medium (LB medium with 10 g glucose/l), M9Y medium (M9 medium with the addition of 5 g yeast extract/l and 10 g glucose/l), and MSY medium (10 g glucose/l, 3.8 g Na2HPO4/l, 1.5 g KH2PO4/l, 1.0 g(NH4)2SO4/l, 0.2 g MgSO4/l, 5 g yeast extract/l, 2 % (v/v) trace element solution (see Qiu et al. 2005). LBG medium and 500 g glucose/l were picked as initial medium and feed for fed-batch fermentations. MSY medium was selected as the final medium and the feed medium contained 700 g glucose/l with 150 % concentrated MSY medium. When needed, 5 mg chloramphenicol/l or 100 mg ampicillin/l was added to the media.
Plasmid construction and genome engineering
All plasmids used in this study are listed in Table 2. The riboflavin operon was amplified by PCR from p20C-EC10 using the primers rib-F (GCCGCGAGCTCAAGTCCTCAACTACCAAGGAGAAAAC) and rib-R (CAGCCAAGCTTTCAGGCTTCTGTGC) and cloned into pZY48 using the SacI and HindIII restriction sites, creating plasmid pLS01. Both restriction sites were provided on the primers (restriction sites are underlined). All primers used in this study are listed in Supplementary Table 1. The genome engineering technique used in this study was as described previously (Lin et al. 2014).
Testing plasmid stability
To verify the stability of pLS01 and p20C-EC10, the strains RF03S and RF03T were tested using the same procedure as published previously (Sohoni et al. 2015). Both strains were cultured in shake flasks with MSY medium. The numbers of colonies on plates with and without antibiotics were defined as Y and X respectively. The plasmid loss rate was defined as \( {\text{R}} = \left( {{\text{X}} - {\text{Y}}} \right)/{\text{X}}. \)
Riboflavin production in shake-flasks and bioreactors
Batch fermentation was identical to a previous study (Lin et al. 2014) except for the absence of IPTG for strains harboring the plasmid pLS01.
For fed-batch cultivation, seed culture cultivation was performed in two stages termed pre-seed and seed phase, respectively. The pre-seed phase was the same as the process of seed preparation for batch fermentation. In the seed stage, 1 % (v/v) of pre-seed was transferred into 50 ml MSY (or LBG) medium with 10 g glucose/l in a 500 ml shake flask and shaken at 220 rpm and 37 °C for 8–10 h, after which 10 % (v/v) seed culture was inoculated into a 5 l bioreactor containing 2 l MSY (or LBG) medium and 5–10 g/l glucose. Fed-batch fermentation was performed at 37 °C with aeration at 1 vvm. pH was maintained at 7 by automatically adding 10 % (v/v) NH4OH. Dissolved O2 was maintained at over 20 % of air saturation via a cascaded control of the feed rate.
Analytical methods
The measurements of biomass, glucose and riboflavin concentration were as described by Lin et al. (2014). The results represent the mean ± SD of three independent experiments.
Results and discussion
Construction of riboflavin operon expression plasmid pLS01
We previously constructed the riboflavin operon expression plasmid, p20C-EC10, with a high copy number, bearing the pBR322 replication origin and an artificial riboflavin operon under the control of the IPTG-induced Trc promoter (Lin et al. 2014). Introducing this plasmid into E. coli MG1665 resulted in the highest riboflavin production among the tested operon expression plasmids with different copy numbers. However, high copy number plasmids also inflict a much higher metabolic burden on the host strain than those with a lower copy number. In addition, the induced expression of the riboflavin operon is very inconvenient and costly in fermentation processes.
Thus, we inserted the artificial riboflavin operon EC10 into the low copy number plasmid pZY48, creating pLS01 (Supplementary Fig. 1). Operon transcription was driven by the constitutive Trc promoter, which was modified by deleting the lacO region of the original inducible promoter.
Plasmid stability and its effect on riboflavin production
To test the contribution of pLS01 to riboflavin production, MG1665 and RF03 (Lin et al. 2014) were selected as host strains. When using the wild-type MG1665 as host strain, p20C-EC10 performed markedly better than pLS01. Riboflavin production of RF01S was 220 mg/l, which was 293 % higher than that of RF01T. However, when using RF03 as host, the p20C-EC10 harboring strain RF03S produced 563.8 mg riboflavin/l, which was only 12 % higher than that of RF03T, which was harboring pLS01 (Fig. 1). We also compared the stability of the two plasmids during the cultivation process. As shown in Fig. 2, the plasmid loss rate of pLS01 was much lower than that of p20C-EC10 throughout the cultivation process. As for RF03T, only 3 % of all cells lost the plasmid at 9 h, and about 78 % cells still harbored the plasmid at the end of cultivation. In contrast, about 75 % of RF03S cells lost the plasmid p20C-EC10 at 9 h, and plasmid containing cells comprised no more than 7 % of the total population at 23 h. Correspondingly, RF03S produced 415 mg riboflavin/l at 12 h with a productivity of 34.6 mg l−1 h−1, but only produced 149 mg riboflavin/l with a much lower productivity of 13.5 mg l−1 h−1 from 12–23 h. In contrast, RF03T accumulated 291 mg riboflavin/l in the first 12 h and 225 mg/l riboflavin in the next 11 h, with a productivity of 24.3 mg l−1 h−1 and 20.5 mg l−1 h−1, respectively. These results indicated that the much lower plasmid loss rate of pLS01 only led to a slight decrease in riboflavin productivity, which was much better than the case of p20C-EC when using RF03 as host. Consequently, pLS01 was selected as the riboflavin operon expression plasmid for further study.
The effect of pfkA, edd and eda knockout on riboflavin production
In our previous study, the deletion of the pgi gene (encoding phosphoglucose isomerase) in RF01S led to a 72 % increase in riboflavin production (Lin et al. 2014). However, pgi knockout strains showed significantly reduced glucose uptake and maximum specific growth rates. In addition, pgi deletion, together with edd and eda knockout, appeared to result in insufficient supply of glycine for purine and riboflavin biosynthesis. For example, riboflavin production of RF05S-M40 was 1630 ± 34 mg/l without glycine, and reached 2543 ± 69 mg/l when 2 g glycine/l was added to the medium (Lin et al. 2014).
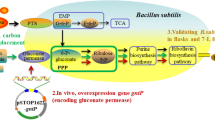
To verify this, we analyzed the effect of glycine addition on riboflavin production of strain RF03T. As shown in Table 3, with the increase of additional glycine, both biomass and riboflavin production were improved. We speculated that deletion of the three genes completely blocked the formation of glyceraldehyde 3-phosphate (GAP) from the EMP and ED pathways (Fig. 3), which reduced the flux from GAP to glycerate-3P too much and caused the insufficient glycine supply in vivo. To solve this, we reconstructed the host strain LS01 by deleting pfkA (encoding 6-phosphofructokinase I) instead of the pgi gene. In E. coli, the phosphorylation of fructose-6-phosphate is catalyzed by two isozymes, 6-phosphofructokinase I (pfkA) and 6-phosphofructokinase II (pfkB). More than 90 % of the phosphofructokinase activity relie on pfkA (Kotlarz et al. 1975). Thus, disruption of pfkA only partially blocked GAP formation from the EMP pathway, which might alleviate the insufficiency of glycine supply. Interestingly, the pfkA deletion resulted in a significant increase in riboflavin production. LS01T accumulated 442 ± 15 mg riboflavin/l, which was almost 720 % higher than that of the control strain RF01T. Considering that disruption of the ED pathway also contributed much to improvement of riboflavin production (Lin et al. 2014), eda and edd were further deleted in LS01, resulting in strain LS02. The riboflavin production BY LS02T was further enhanced to 605 ± 7.8 mg/l, which was 37 % higher than that of LS01T. In contrast to RF03T, riboflavin production decreased markedly with the addition of glycine (Table 3), especially when glycine was >2 g/l. It can thus be concluded that the effects of glycine addition can be quite different depending on the background of the strain involved, and the reason for this may be the subject of further research. In addition to this, the biomass yields of both strains increased slightly with the addition of glycine.
Schematic overview of the relevant pathways and engineering strategies for riboflavin production. Dashed lines indicate multiple enzymatic steps. A cross indicates the deletion of the corresponding gene. The enzymes encoded by the indicated genes are: zwf, glucose-6-phosphate-1-dehydrogenase; pgl, 6-phosphogluconolactonase; pgi, phosphoglucose isomerase; pfkA, 6-phosphofructokinase I; pfkB, 6-phosphofructokinase II; prs, ribose-5-phosphate diphosphokinase; purF, amidophosphoribosyl transferase; purD, phosphoribosylamine-glycine ligase; edd, phosphogluconate dehydratase; eda, multifunctional 2-keto-3-deoxygluconate 6-phosphate aldolase and 2-keto-4-hydroxyglutarate aldolase and oxaloacetate decarboxylase; glyA, serine hydroxymethyltransferase. Other non-standard abbreviations: FBP, fructose 1,6-bisphosphate; TCA, tricarboxylic acid cycle; PRPP, 5-phospho-α-d-ribose 1-diphosphate; PRA, 5-phospho-β-d-ribosylamine; GAR, N1-(5-phospho-β-d-ribosyl)glycinamide; GAP, d-glyceraldehyde 3-phosphate; KDGP, 2-dehydro-3-deoxy-d-gluconate 6-phosphate; glycerate-3P, 3-phospho-d-glycerate; Ser, l-serine; Gly, glycine
Culture media optimization and fed-batch fermentation
In order to select a better medium for riboflavin fermentation, three different media were tested for their effect on riboflavin production. As shown in Table 4, LS03T produced 667 ± 9.4 mg riboflavin/l in MSY medium, which was the highest among the three selected media (riboflavin production, biomass and glucose consumption are shown in Supplementary Fig. 2). Thus, MSY medium was further used for fed-batch fermentations in a 5L bioreactor.
To further investigate the potential of LS02T for riboflavin production, fed-batch fermentations were carried out in a 5 l bioreactor. As shown in Fig. 4, glucose was maintained at ~1 g/l during the greatest part of the fermentation process, and the maximal biomass reached 32 g/l at the end of fermentation. LS02T produced 10.4 g riboflavin/l with a yield of 56.8 mg riboflavin/g glucose after 71 h. We also performed fed-batch fermentations with glycine addition (2 and 10 g glycine/l were added into the initial medium and feed medium, respectively). Similar to the results of the flask fermentations, glycine addition led to a significant decrease in riboflavin production, with a titer of only 5.8 g riboflavin/l.
Conclusion
Plasmid pLS01 with a constitutively expressed riboflavin operon was constructed and exhibited increased stability compared to previously published research. A knockout of 6-phosphofructokinase I and blocking of the Entner–Doudoroff pathway efficiently increased riboflavin production in E. coli. The final stain LS02T harboring pLS01 accumulated 10.4 g riboflavin/l in a fed-batch fermentation, which demonstrates that E. coli has a great potential for industrial production of riboflavin.
References
Buey RM, Ledesma-Amaro R, Balsera M, de Pereda JM, Revuelta JL (2015) Increased riboflavin production by manipulation of inosine 5′-monophosphate dehydrogenase in Ashbya gossypii. Appl Microbiol Biotechnol 99:9577–9589
Dmytruk K, Lyzak O, Yatsyshyn V, Kluz M, Sibirny V, Puchalski C, Sibirny A (2014) Construction and fed-batch cultivation of Candida famata with enhanced riboflavin production. J Biotechnol 172:11–17
Duan YX, Chen T, Chen X, Zhao XM (2010) Overexpression of glucose-6-phosphate dehydrogenase enhances riboflavin production in Bacillus subtilis. Appl Microbiol Biotechnol 85:1907–1914
Kotlarz D, Garreau H, Buc H (1975) Regulation of the amount and of the activity of phosphofructokinases and pyruvate kinases in Escherichia coli. Biochim Biophys Acta 381:257–268
Kuhlman TE, Cox EC (2010) Site-specific chromosomal integration of large synthetic constructs. Nucleic Acid Res 38:e92. doi:10.1093/nar/gkp1193
Lin Z, Xu Z, Li Y, Wang Z, Chen T, Zhao X (2014) Metabolic engineering of Escherichia coli for the production of riboflavin. Microb Cell Factor 13:104. doi:10.1186/s12934-014-0104-5
Mack M, van Loon AP, Hohmann H-P (1998) Regulation of riboflavin biosynthesis in Bacillus subtilis affected by the activity of the flavokinase/flavin adenine dinucleotide synthetase encoded byribC. J Bacteriol 180:950–955
Marx H, Mattanovich D, Sauer M (2008) Overexpression of the riboflavin biosynthetic pathway in Pichia pastoris. Microb Cell Factor 7:23. doi:10.1186/1475-2859-7-23
Qiu YZ, Han J, Guo JJ, Chen GQ (2005) Production of poly(3-hydroxybutyrate-co-3-hydroxyhexanoate) from gluconate and glucose by recombinant Aeromonas hydrophila and Pseudomonas putida. Biotechnol Lett 27:1381–1386
Shi S, Shen Z, Chen X, Chen T, Zhao X (2009) Increased production of riboflavin by metabolic engineering of the purine pathway in Bacillus subtilis. Biochem Eng J 46:28–33
Sohoni SV, Nelapati D, Sathe S, Javadekar-Subhedar V, Gaikaiwari RP, Wangikar PP (2015) Optimization of high cell density fermentation process for recombinant nitrilase production in E. coli. Bioresour Technol 188:202–208
Stahmann KP, Revuelta JL, Seulberger H (2000) Three biotechnical processes using Ashbya gossypii, Candida famata, or Bacillus subtilis compete with chemical riboflavin production. Appl Microbiol Biotechnol 53:509–516
Wang Z, Chen T, Ma X, Shen Z, Zhao X (2011) Enhancement of riboflavin production with Bacillus subtilis by expression and site-directed mutagenesis of zwf and gnd gene from Corynebacterium glutamicum. Bioresour Technol 102:3934–3940
Wang G, Bai L, Wang Z, Shi T, Chen T, Zhao X (2014) Enhancement of riboflavin production by deregulating gluconeogenesis in Bacillus subtilis. World J Microbiol Biotechnol 30:1893–1900
Xu Z, Lin Z, Wang Z, Chen T (2015) Improvement of the riboflavin production by engineering the precursor biosynthesis pathways in Escherichia coli. Chin J Chem Eng 23:1834–1839
Acknowledgments
The authors thank Dr. Thomas E. Kuhlman and Dr. Zhenquan Lin for kindly providing some of the plasmids used in this study. This work was supported by the National 973 Project (2012CB725203), and National High-tech R&D Program of China (2012AA022103, 2012AA02A702).
Supporting information
Supplementary Table 1—Primers used in this study.
Supplementary Figure 1—pLS01 map.
Supplementary Figure 2—Time profiles of cell growth, glucose consumption and riboflavin production of LS02T cultivated in LBG, M9 and MS media.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Liu, S., Kang, P., Cui, Z. et al. Increased riboflavin production by knockout of 6-phosphofructokinase I and blocking the Entner–Doudoroff pathway in Escherichia coli . Biotechnol Lett 38, 1307–1314 (2016). https://doi.org/10.1007/s10529-016-2104-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-016-2104-5