Abstract
Objective
To ascertain the effect of chitin-binding domain (ChBD) and fibronectin type III domain (FN3) on the characterization of the intact chitinase from Bacillus thuringiensis.
Results
An intact chitinase gene (chi74) from B. thuringiensis HZP7 and its truncated genes (chi54, chi63 and chi66) were expressed in Escherichia coli BL21. The expression products were analyzed after purification. All chitinases were active from pH 4–7.5 and from 20 to 80 °C with identical optimal: pH 5.5 and 60 °C. The activity of colloid chitin degradation for Chi74 was the highest, followed by Chi66, Chi63 and Chi54. Ag+ reduced the activity of Chi74, Chi54, Chi63 and Chi66, but Mg2+ enhanced them. The effect of Ag+ and Mg2+ was more significant on the activity of Chi54 than on the activities of Chi63, Chi66 and Chi74.
Conclusion
ChBDChi74 and FN3Chi74 domains play a role in exerting enzymatic activity and can improve the stability of chitinase.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chitin, a homopolymer of unbranched chains of β-1,4-linked N-acetylglucosamines (GlcNAc), is one of the most abundant polysaccharides found in nature. It is the major structural component of fungal cell walls, and of the exoskeletons and peritrophic membranes of insects (Merzendorfer and Zimoch 2003). Chitinases (EC3.2.1.14) catalyze the conversion of insoluble chitin to a monosaccharide, disaccharide and oligosaccharide, and are produced by a large variety of organisms including bacteria, fungi, insects, plants and animals (Henrissat 1999).
Bacillus thuringiensis, is used worldwide as an agent for pest control in agriculture and forestry and for the control of various disease-related insect vectors. Its insecticidal activity is based on the effect of single or mixed Cry or Cyt protein. When a susceptible insect ingests these crystalline proteins, they are solubilized and proteolytically digested to yield the active toxic form. Toxins specifically bind to protein receptors in the epithelial insect midgut and produce pores, leading to the loss of normal membrane function (Schnepf et al. 1998). However, resistance to insecticidal crystal proteins by targeted pests has been reported (Liu and Tabashnik 1996).
With the development of resistance to Cry proteins, it is important to find approaches to improve the insecticidal activity of B. thuringiensis. B. thuringiensis produces a chitinase (Barboza-Corona et al. 1999) and although the precise mechanism by which chitinases exert their effect on insects is still unclear, several studies have demonstrated that co-expression of heterologous chi genes in B. thuringiensis can enhance its insecticidal activity (Wiwat et al. 1996). Chitinases destroy the peritrophic membrane of insects and assist in the invasion of protein toxin, which may account for their synergistic effect.
Interest in the chitinases of B. thuringiensis has increased due to their potential role as biological control agents for insects and plant pathogenic fungi (Barboza-Corona et al. 2008; Vega et al. 2006). Herein, we report the purification and partial biochemical characterization of intact and truncated chitinase from B. thuringiensis HZP7 expressed in E. coli. This information will aid our understanding of the ‘structure–function’ relation of chitinase.
Materials and methods
Bacterial strains, plasmid, and culture condition
Bacillus thuringiensis strain HZP7 was isolated from soil collected in Wuyi mountain (P.R. China). E. coli BL21 was used to clone and express the chitinase genes. Plasmid pMD-18T (TaKaRa, Japan) was used as a cloning vector and pGEX-KG as an expression vector. E. coli cells with recombinant plasmids were cultured in lysogeny broth (LB) agar containing 100 µg ampicillin/ml to screen transformants. The scale-up culture of transformants was carried out with LB broth supplemented with 100 µg ampicillin/ml.
Cloning of the chitinase genes
Total DNA from B. thuringiensis was extracted as described elsewhere (Lin and Guan 2004) and used as a template for PCR amplification of an intact chitinase gene (chi74) and three truncated chitinase genes (chi54, chi63 and chBD). The primers used in this study are shown in Supplementary Table 1 and were synthesized by Sangon Co. (Shanghai, P.R.China). PCR conditions consisted of an initial denaturation at 94 °C for 5 min followed by 30 cycles of 94 °C for 1 min, 55 °C for 1 min and 72 °C for 2 min, and a final extension at 72 °C for 10 min.
Using the Ultraclean 15 DNA Purification Kit (Mo Bio, USA), the PCR products were purified and ligated to form recombinant cloning vectors, generating four recombinant plasmids, pMD18-T-chi74, pMD18-T-chi54, pMD18-T-chi63 and pMD18-T-chi66. The recombinant plasmids were transformed into E. coli BL21 cells and the transformants were screened on LB agar supplemented with 100 µg ampicillin/ml.
Recombinant plasmids re-extracted from the transformants were sequenced by TaKaRa and the obtained sequences were analyzed by ExPASy.
Expression of the chitinase genes in E. coli
Using the Ultraclean 15 DNA Purification Kit, the amplified fragments of chi74, chi54, chi63 and chi66 were purified from the pMD18-T-chi74, pMD18-T-chi54, pMD18-T-chi63 and pMD18-T-chi66 digestions with EcoRI/HindIII, respectively, and were individually cloned into pGEX-KG vector to construct the recombinant plasmids, pKG-chi74, pKG-chi54, pKG-chi63 and pKG-chi66. The pGEX-KG vector carries a gene encoding a glutathione S-transferase (GST) located near the N-terminal of target protein product. The constructs were transformed into E. coli BL21 cells and the transformants were screened on LB agar containing 100 µg ampicillin/ml. To express the chitinases, the corresponding transformants were grown in LB broth containing 100 µg ampicillin/ml on a rotary shaker at 37 °C for 2 h and then induced with 1 mM IPTG at 27 °C for 18 h.
Purification of the chitinases
Cells were harvested by centrifugation at 12,000×g for 5 min. After washing with PBS buffer (pH 7.3) twice, the harvested cells were suspended in 5 ml PBS buffer with protease inhibitor (Roche, Swiss) and disrupted by sonication. The supernatant was collected by centrifugation at 12,000×g for 5 min. A GSTrap FF column with ÄKTA (GE,USA) was equilibrated with five column volumes of binding buffer (PBS, pH 7.3), and the supernatant was loaded onto the column at 0.2 ml/min. The column was washed with ten column volumes of binding buffer, after which the thrombin solution (GE, USA) was loaded onto the column, and incubated at 22 °C for 16 h to cut GST-tag. The GST-tag was still binding with column, and then were eluted by elution buffer (50 mM Tris/HCl, pH 8) with 10 mM reduced glutathione. The eluates were harvested and analyzed by 10 % (w/v) SDS-PAGE.
Chitinase activity assay and protein concentration determination
Using colloidal chitin as a substrate, chitinase activity was measured in a 2 ml containing purified chitinases 50 µg/ml, 0.5 % (w/v) colloidal chitin and 0.1 M phosphate buffer solution (PBS, pH 7). The reaction was performed at 37 °C for 1 h and stopped by adding an equal volume of 3,5-dinitrosalicylic acid (DNS) solution [20 % (w/v) potassium sodium tartrate, 1 % (w/v) NaOH, 0.2 % (w/v) phenol, 0.05 % (w/v) Na2SO3 and 1 % (w/v) DNS] and boiling for 10 min. After being cooled in ice, the reaction mixture was centrifuged at 12,000×g for 15 min to remove the remaining colloidal chitin. The supernatant was collected and its absorbance was measured at 530 nm. Readings were compared with a standard curve prepared with a series of dilutions of GlcNAc. One unit of chitinase activity was defined as the amount of enzyme required to produce 1 µmol GlcNAc per h under the above reaction conditions. Protein concentration was determined according to the Bradford method.
Effect of temperature, pH and metal ions on chitinase activity
For determination of the optimal temperature for chitinase activity, the enzyme was incubated in 0.1 M PBS (pH 7) for 1 h from 20 to 100 °C, using colloidal chitin as a substrate. The optimum pH was measured by incubating the enzyme in reaction buffers with pH values between pH 4 and 8 at the optimal temperature for 1 h. Effects of three different metal ions on enzyme activity were investigated by adding the corresponding chemicals at various concentrations to the reaction mixture, incubating the mixture for 1 h, then measuring the chitinase activity at optimal temperature and pH 7. All measurements were repeated three times.
Results
Characteristics of the intact and truncated chitinase genes
Due to our interest in the chitinolytic activity exhibited by B. thuringiensis HZP7, an intact chitinase gene, chi74, was cloned and sequenced. The sequence had been submitted to NCBI genebank under the accession number AY074882. It consists of 2025 bases and encodes the protein Chi74 having 674 amino acids residues and a predicted molecular weight of 74 kDa and pI 5.77. The analysis of the deduced amino acid sequence of Chi74 showed that the mature enzyme comprised a ChiA catalytic domain, a fibronectin type III (FN3) domain, and a chitin-binding (ChBD) domain.
To determine the roles of the domains that constitute Chi74, three truncated chitinase genes, chi54, chi63 and chi66, were constructed as summarized in Fig. 1, and were cloned and sequenced. The chi54 gene encodes a truncated chitinase (Chi54) with a predicted molecular weight of 54 kDa and pI 6.04, and lacks both FN3 and ChBD domains. The chi63 gene encodes a truncated chitinase (Chi63) with a predicted molecular weight of 63 kDa and pI 6.07, and lacks the ChBD domain. The chi66 gene encodes a truncated chitinase (Chi66) with a predicted molecular weight of 66 kDa and pI 5.74, and lacks the FN3 domain.
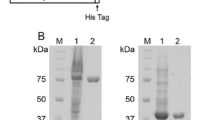
Expression and purification of Chi74, Chi54, Chi63 and Chi66 in E. coli
The expressed products from E. coli/pKG-chi74, E. coli/pKG-chi54, E. coli/pKG-chi63 and E. coli/pKG-chi66 with IPTG induction were the fusion proteins, containing GST from pGEX-KG. To obtain Chi74, Chi54, Chi63 and Chi66, proteins from sonication of cells cultured with IPTG were purified using a GSTrap FF column and cleaved by thrombin to delete GST. Results of SDS-PAGE (Fig. 2) showed that the molecular weights of Chi74, Chi54, Chi63 and Chi66 were about 74, 54, 63 and 66 kDa, respectively, corresponding with the predicted molecular weights by the sequences.
Characterization of chitinases
Chi74, Chi54, Chi63 and Chi66 showed activity from 20 to 80 °C (Fig. 3a). Above 60 °C, the activity of Chi54, Chi63 and Chi66 decreased faster than that of Chi74, indicating greater stability of the latter. Although the four chitinases all exhibited maximal activity at ~60 °C, only 39, 70 and 79 % of the maximal activity of Chi74 was detected for Chi54, Chi63 and Chi66, with 136, 53, 95 and 107 U/ml for Chi74, Chi54, Chi63 and Chi66, respectively.
The effects of temperature (a) and pH (b) on the activities of Chi74, Chi54, Chi63 and Chi66. Enzyme activity was determined at a range of temperature (20–80 °C) and pH (4–8) under the assay conditions described above, using colloid chitin as substrate. Data are presented as ΔNAG/µmol/h. Points indicate the average value of three independent experiments. Error bars indicate standard deviation
Chi74, Chi54, Chi63 and Chi66 showed activity between pH 4 and pH 8 (Fig. 3b). Activities of Chi74, Chi54, Chi63 and Chi66 were optimal at pH 5.5 and were 230, 72, 151, and 177 U/ml, respectively. There was no activity at pH 8 and above.
The activities of four chitinases were not affected by NaCl or NaNO3 (data are not shown). However, Ag+ at 0.5 mM decreased the activities of Chi74, Chi54, Chi63 and Chi66 by 13.7, 45.3, 36.8 and 30 %, respectively (Fig. 4a). Mg2+ increased the activities of the four chitinases, and at 6 mM the activities of Chi74, Chi54, Chi63 and Chi66 were increased by 141, 178, 156 and 152 %, respectively (Fig. 4b).
The effects of Ag+ (a) and Mg2+ (b) on the activities of Chi74, Chi54, Chi63 and Chi66. The assays were performed in presence of Ag+ and Mg2+ under assay conditions in the text using colloidal chitin as substrate. The values of vertical coordinates were calculated by compared without metal ions. All experiments were repeated three times along with control. Points indicate the average value of three independent experiments
Discussion
Chitinase improves the insecticidal and anti-fungal activity of B. thuringiensis (Liu et al. 2010; Smirnoff et al. 1973; Vega et al. 2006). Due to our interest in the chitinolytic activity of B. thuringiensis HZP7, the chitinase gene chi74 was expressed in E. coli and purified. The sequence of Chi74 is highly homologous to those from other strains (Barboza-Corona et al. 2003; Lin and Guan 2004). Conserved domains analysis showed that Chi74 comprises a ChiA catalytic domain, a FN3 domain and a ChBD domain. The structure of Chi74 is different than that of ChiA1 from Bacillus circulans WL-12, that contains two FN3 domains (Watanabe et al. 1994).
The activities of Chi54, Chi63, Chi66 and Chi74 were measured using colloidal chitin as substrate with that of Chi74 being the highest, (see Fig. 3). This showed that the ChBD and FN3 domains are not absolutely required for chitinolytic activity but do enhance the efficiency of degradation. As for the effect of ChBD domain on the chitinase activity, similar results were reported for ChiA1 from B. circulans WL-12 (Watanabe et al. 1994) but an opposite result was reported for chitinase Chi255 from B. thuringiensis subsp. kurstaki BUPM255 (Driss et al. 2007). Approx. 2 % of all animal proteins contain the FN3 repeat, including extracellular and intracellular proteins, membrane-spanning cytokine receptors, growth hormone receptors, tyrosine phosphatase receptors, and adhesion molecules (Benjamin et al. 2015), and that FN3-like domains are also found in bacterial glycosyl hydrolases including chitinase. However, there is still no report about the function of the FN3 domain on chitinases. The activity of Chi63 (with FN3 domain) was lower than that of Chi66 (with ChBD domain) but higher than that of Chi54 (without both FN3 and ChBD domains), indicating that the FN3 domain plays a role in chitin degradation, though a less important role than the ChBD domain.
The profiles of Chi74, Chi54, Chi63 and Chi66 activities at the tested temperatures, pH and metal ions showed similar variation. The monomodal distributions of enzyme activity were observed at the tested temperatures and pH range. Similar distribution behavior was also reported for ChiA74 from B. thuringiensis LBIT-82 (Barboza-Corona et al. 2003). Compared with Chi63, Chi66 and Chi74, the effect of Mg2+ and Ag+ on Chi54 activity was much more apparent, possibly because Chi54, lacking both ChBD and FN3 domains, allowed easier access of the metal ions to the catalytic domain.
In conclusion, The Chi74,Chi54,Chi63 and Chi66 were expressed in E. coli and purified. It can be concluded that ChBDChi74 and FN3 Chi74 domains play a role in exerting enzymatic activity and can improve the stability of intact chitinase. A comprehensive characterization of ChBDChi74 and FN3 Chi74 is in progress.
References
Barboza-Corona JE, Contreras JC, Velazquez-Robledo R et al (1999) Selection of chitinolytic strains of Bacillus thuringiensis. Biotechnol Lett 21:1125–1129
Barboza-Corona JE, Nieto-Mazzocco E, Velázquez-Robledo R et al (2003) Cloning, sequencing, and expression of the chitinase gene chiA74 from Bacillus thuringiensis. Appl Environ Microbiol 69:1023–1029
Barboza-Corona JE, Reyes-Rios DM, Salcedo-Hernandez R et al (2008) Molecular and biochemical characterization of an endochitinase (ChiA-HD73) from Bacillus thuringiensis subsp.kurstaki HD-73. Mol Biotechnol 39:29–37
Benjamin T, Porebski AA, Nickson DE et al (2015) Structural and dynamic properties that govern the stability of an engineered fibronectin type III domain. Protein Eng Des Sel 28:67–78
Driss F, Baanannou A, Rouis S et al (2007) Effect of the chitin binding domain deletion from Bacillus thuringiensis subsp. kurstaki chitinase Chi255 on its stability in Escherichia coli. Mol Biotechnol 36:232–237
Henrissat B (1999) Classification of chitinases modules. EXS 87:137–156
Lin Y, Guan X (2004) Molecular cloning and sequence analysis of the chitinase gene from Bacillus thuringiensis serovar alesti. Biotechnol Lett 26:635–639
Liu Y, Tabashnik BE (1996) Diamondback moth resistance to Bacillus thuringiensis toxin Cry1C in the field. Resist Pest Manag 8:44–46
Liu D, Cai J, Xie CC et al (2010) Purification and partial characterization of a 36-kDa chitinase from Bacillus thuringiensis subsp colmeri and its biocontrol potential. Enzyme Microb Technol 46:252–256
Merzendorfer H, Zimoch L (2003) Chitin metabolism in insects: structure, function and regulation of chitin syntheses and chitinases. J Exp Biol 206:4393–4412
Schnepf E, Crickmore N, Vanrie J et al (1998) Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol Mol Biol Rev 62:775–806
Smirnoff WA, Randall AP, Martineau R et al (1973) Field test of the effectiveness of chitinase additive to Bacillus thuringiensis against Choristoneura fumiferana. Can J For Res 3:228–236
Vega LM, Barboza-Corona JE, Aguilar-Uscanga MG et al (2006) Purification and characterization of an exochitinase from Bacillus thuringiensis subsp. aizawai and its action against phytopathogenic fungi. Can J Microbiol 52:651–657
Watanabe T, Ito Y, Yamada T et al (1994) The roles of the C-terminal domain and type III domains of chitinase A1 from Bacillus circulans WL-12 in chitin degradation. J Bacteriol 176:4465–4472
Wiwat C, Monton L, Patcharaporn S et al (1996) Expression of chitinase-encoding genes from Aeromonas hydrophila and Pseudomonas maltophilia in Bacillus thuringiensis subsp. israelensis. Gene 179:119–126
Acknowledgments
We thank Dr. Brian McGarvey for revising this manuscript. This work was supported by grants from Ministry of Science and Technology of the People’s Republic of China (No. 2011AA10A203), State Administration of Grain (No. 201313002-3) and Natural Science foundation of Fujian Province (No. 2013J01079).
Supporting information
Supplementary Table 1—Characteristics of primers.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sha, L., Shao, E., Guan, X. et al. Purification and partial characterization of intact and truncated chitinase from Bacillus thuringiensis HZP7 expressed in Escherichia coli . Biotechnol Lett 38, 279–284 (2016). https://doi.org/10.1007/s10529-015-1970-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-015-1970-6